Abstract
HIV-1–associated nephropathy (HIVAN) is a common complication of HIV-1 infection, and its skewed incidence in certain ethnic groups suggests that there is a genetic basis to HIVAN susceptibility. In their study reported in this issue of the JCI, Papeta and colleagues used a combination of gene expression profiling and linkage analysis to identify three genomic loci that regulate a network of genes expressed by podocytes — cells that are crucial to the filtration of fluid and waste by the kidney (see the related article beginning on page 1178). Surprisingly, two of these loci confer disease susceptibility in a transgenic mouse model of HIVAN. This report confirms the central role of podocytes in the pathogenesis of HIVAN and demonstrates the power of this combination of genomic analysis techniques in elucidating the pathogenesis of glomerular disease.
HIV-1–associated nephropathy (HIVAN) is a leading cause of morbidity and mortality in HIV-1–infected patients. There is a significant component of genetic risk in HIVAN, as it occurs almost exclusively in patients of African descent (1). Histologically, the renal lesion is characterized by collapsing glomerulopathy, epithelial cell proliferation, tubuloreticular inclusions, microcystic dilation of the tubules, and tubulointerstitial inflammation (reviewed in refs. 2, 3). Over the past decade, much has been learned about the pathogenesis of HIVAN at a cellular level (reviewed in ref. 2). Although the involvement of tubular epithelial cells, endothelial cells, and T cells has been reported, emerging data have supported a major role for the podocyte in the development of HIVAN (4–6).
Podocytes and the glomerular filtration barrier
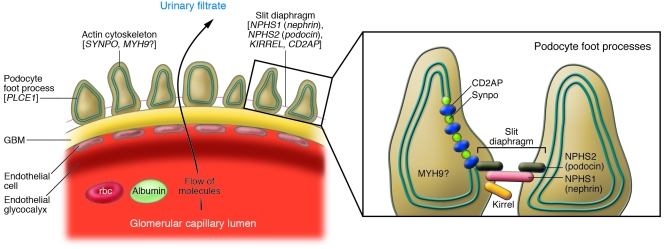
Podocytes (also known as glomerular visceral epithelial cells) are unusual cells that are only found in the renal glomerulus. Together with glomerular endothelial cells and the intervening glomerular basement membrane, they form the glomerular filtration barrier, a permselective sieve and the site of formation of the primary urinary filtrate (7) (Figure 1). The identification of mutations in genes that are expressed by podocytes has explained the cause of a number of Mendelian glomerular diseases (8–13) and has placed the podocyte in a central position in studies of glomerular disease. In patients with HIVAN, direct HIV-1 infection of podocytes has been documented, and viral proteins induce podocyte proliferation, loss of cell contact inhibition, and upregulation of growth factors in immortalized podocytes in culture (14–17). The concept of a dysregulated podocyte phenotype in HIVAN has been proposed (15), and this phenotype is characterized by loss of expression of many genes typically seen in differentiated podocytes, such as synaptopodin (SYNPO), nephrosis 1 homolog (NPHS1; which encodes the slit-diaphragm protein nephrin), and NPHS2 (which encodes the podocyte slit diaphragm protein podocin), and with the re-expression of embryonic genes such as paired box gene 2 (PAX2) that are normally only expressed in presumptive podocytes and podocyte progenitors during glomerular development.
Figure 1. The glomerular filtration barrier.
The glomerular filtration barrier consists of podocytes, endothelial cells, and an intervening glomerular basement membrane (GBM). The slit diaphragm is a specialized intercellular junction and signaling center that connects adjacent podocyte foot processes. Blood entering the glomerular capillaries is filtered across the endothelium and the GBM and through the filtration slits to produce the primary urinary filtrate. Genomic regions that contain HIVAN susceptibility loci regulate a network of podocyte-expressed genes that encode proteins found in the slit diaphragm, including nephrin, podocin, CD2-associated protein (CD2AP), and kirrel as well as actin cytoskeleton–associated proteins, including Synpo, and possibly MYH9. Phospholipase C, ε1 (PLCE1) is an enzyme found in podocytes. Inset adapted with permission from Nature Genetics (36).
Genetic susceptibility and HIV
In North America, HIVAN is largely restricted to patients of African descent, suggesting that genetic factors are important in the development of or susceptibility to this disease. Furthermore, the relative risk for developing end-stage renal disease is increased 51-fold in black patients compared with patients of European descent diagnosed with AIDS or an AIDS-defining illness (18). Recently, SNP polymorphisms in the myosin, heavy chain 9, non-muscle (MYH9) gene, which expresses a protein that likely forms part of the actin cytoskeleton in podocytes, were associated with non-diabetic glomerular disease, including HIVAN, in African Americans (19, 20). The significance of these SNPs with respect to podocyte or glomerular function is not yet clear, although coding mutations in MYH9 have been identified in rare syndromes associated with glomerulonephritis (21–23).
Genetic mapping for HIVAN susceptibility loci
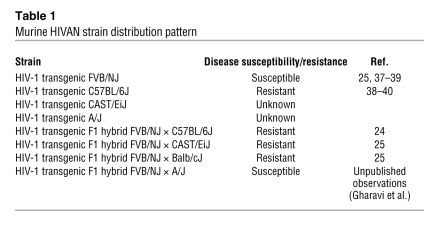
In order to identify modifying genes responsible for the development and progression of HIVAN, Papeta, Gharavi, and associates, in their study in this issue of the JCI, used an established transgenic mouse model of HIVAN with major, strain-specific differences in disease susceptibility depending on the genetic background of the mouse (24) (Table 1). In this murine model, a replication-deficient strain of HIV-1 is randomly integrated as a transgene into the genome of the mouse (4). On an FVB/NJ background, HIV-1 TgFVB/NJ mice develop glomerular disease with many features of HIVAN, including microcystic tubular dilation, epithelial cell proliferation, and glomerulosclerosis. When crossed to a number of other inbred mouse strains (25) the renal disease was modified, ranging from severe renal disease to no renal disease at all.
Table 1 .
Murine HIVAN strain distribution pattern
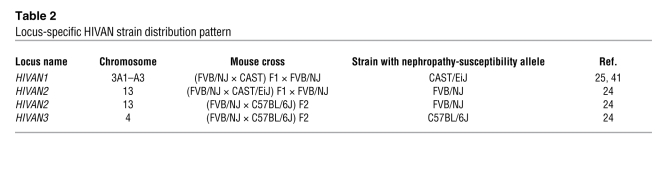
Using genetic linkage analysis of backcrosses between HIV-1 TgFVB/NJ mice and one of these inbred strains, CAST/EiJ mice, Gharavi et al. previously found a major susceptibility locus for nephropathy on chromosome 3 (called HIVAN1) (Table 2) (25). The risk of more severe renal disease mapped to a susceptible CAST allele. In their current JCI study (24), the authors extended their previous mapping studies and produced an F2 intercross between C57BL/6J and the HIV-1 TgFVB/NJ mouse strains, allowing the identification of two additional loci regulating the severity of renal disease, called HIVAN2 and HIVAN3, on chromosome 13 and chromosome 4, respectively (Table 2). Interestingly, at HIVAN2 the FVB allele conferred increased risk of renal injury, while at HIVAN3 the C57BL/6J allele conferred increased risk of renal injury. More often than not in expression quantitative trait locus (eQTL) mapping studies, both susceptibility and resistance loci for the trait under investigation may be identified to originate in the same strain of mouse, regardless of whether the mouse strain on a pure inbred background is resistant or susceptible to the trait overall; this is a reflection of the complexity of modifier genes and phenotypes.
Table 2 .
Locus-specific HIVAN strain distribution pattern
Successful eQTL mapping depends upon a robust quantifiable trait that can be used to distinguish or score a mouse as susceptible or resistant to a disease. In this case, the investigators used histologic scoring of glomerular disease, which included assessments of podocyte hypertrophy, tubular dilation, and glomerulosclerosis (24).
eQTL analysis
Over recent years, as the efficiency of generating gene expression profiles from specific tissues or cells has improved, the technique of eQTL analysis has emerged. With this approach, expression of a gene(s) or a gene set is treated as a quantitative trait (a trait that does not follow patterns of Mendelian inheritance and is attributable to 2 or more genes and their interaction with the environment) in much the same way as histologic scoring or phenotyping has been used in the past. The obvious benefit of such an approach is that alterations in gene expression are intermediate between genomic variation and a phenotype; genes already have a “genomic address,” whereas clinical phenotypes do not. Statistical models have been developed allowing the investigator to predict whether changes in gene expression are reactive to or causal for disease. In complex diseases, such as obesity, this approach has proven very successful, allowing the identification of new genetic variants that are causally associated with obesity (26, 27). Furthermore, eQTL analysis can identify networked responses, reflected as changing transcript levels, underlying complex phenotypes. While eQTL analysis has been used to identify genes promoting hypertension in the spontaneously hypertensive rat (SHR) model (28), this approach has not been used previously to identify susceptibility loci for glomerular diseases.
Both cis-eQTLs and trans-eQTLs can be identified. A cis-eQTL is defined as a region of the genome that is acting locally or is within the region of the gene whose expression is altered. As an example, this might include a change in the promoter region of the gene, which results in altered expression levels. On the other hand, a trans-eQTL regulates the gene(s) of interest from a distance. The current study by Papeta et al. (24) provides what is believed to be the first eQTL analysis of a primary glomerular disease.
Analysis of podocyte gene expression
Rather than performing genome-wide expression analysis, Papeta et al. examined a number of candidate genes that are expressed by podocytes and associated with glomerular disease (24). In kidneys from mice with HIVAN, they found a 3-fold difference in the level of transcript of Nphs2 that was highly strain dependent, making it an attractive gene for eQTL analysis. NPHS2 was originally identified as the gene responsible for a heritable form of autosomal recessive, steroid-resistant nephrotic syndrome (12). In later studies, it was also associated with sporadic cases of focal segmental glomerulosclerosis (29). NPHS2 interacts with proteins of the slit diaphragm and is required for proper trafficking of NPHS1 to the lipid raft (Figure 1), which is also dependent upon cholesterol biosynthesis (30–32). In the current study (24), when the Nphs2 transcript level was treated as a quantifiable trait for genome-wide mapping, a cis-eQTL was identified that accounts for approximately 40% of variability in NPHS2 expression. Direct sequencing showed tremendous variation in the promoter and regulatory region of the Nphs2 gene. Phylogenetic analysis of sequence variants showed that these alterations were not random but rather fell into 3 ancestral mouse haplotypes. This is not the first time that Nphs2 has been linked to a variable phenotype. Maternal strain has a major effect on the phenotype of Nphs2-knockout mice, and although modifying QTLs for proteinuria and glomerular injury have been identified for this maternal influence on phenotype (33), they are distinct from the variants identified here. While intriguing, the reason for the high degree of genetic variation in Nphs2 is unclear and awaits future study.
Generation of a consomic mouse strain (an inbred strain with one of its chromosomes replaced by the homologous chromosome of another inbred strain) by transferring the portion of chromosome 1 containing the “low-expressing” Nphs2 cis-eQTL from the A/J strain into the “high-expressing” C57BL/6J strain confirmed that this cis-eQTL regulates Nphs2 expression (24). Moreover, levels of a number of slit diaphragm proteins known to interact with NPHS2 as well as the products of other podocyte genes including Nphs1, Synpo, Kirrel, Myh9, and Plce1 were upregulated in these mice, suggesting that this eQTL functions in trans to regulate a network of podocyte genes, perhaps as a compensatory response to maintain glomerular integrity.
Along with this locally acting cis-eQTL, 2 trans-eQTLs were also identified for Nphs2 that surprisingly mapped to the HIVAN1 and HIVAN2 loci, suggesting that susceptibility to HIVAN may be determined by a network of genes expressed in podocytes (24). To test this possibility, the authors generated congenic strains (produced by transferring a portion of a chromosome containing a genomic variant from one genetic background to a specific inbred strain through repeated backcrossing) or consomic strains of mice carrying the HIVAN1 and HIVAN2 susceptibility regions and determined the expression of Nphs2 and other genes in the podocyte network. Similar to the results of cis-eQTL consomic experiments, the authors observed alterations in the expression of multiple genes, although the expression patterns were not identical in both sets of experiments. Expression levels in mice without glomerular disease were also examined to exclude secondary or reactive changes due to podocyte depletion in kidneys with scarred glomeruli. Indeed, it appears that at least two of the three identified HIVAN loci (i.e., HIVAN1 and HIVAN2) control a network of podocyte-expressed genes. Given the central role of the podocyte in the glomerular pathology of HIVAN, this genetic model provides an appealing explanation of the pathophysiology of this disorder (Figure 2). In the absence of HIV-1 infection, mice carrying the trans-eQTLs exhibited altered levels of expression of NPHS2 and other podocyte genes but did not develop glomerular disease. When infected with HIV-1, mice carrying these trans-eQTLs or susceptibility loci for HIVAN could not compensate for the added stress and presumed podocyte injury and developed overt glomerular disease.
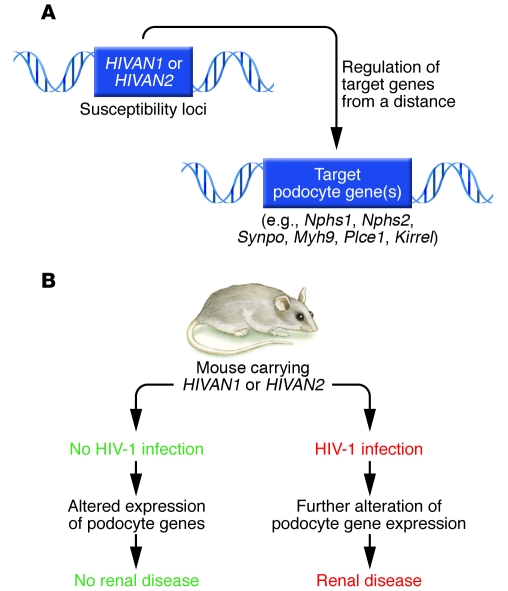
Figure 2. Genetic susceptibility to HIVAN.
(A) In this issue of the JCI, Papeta et al. report that HIVAN1 and HIVAN2 represent susceptibility loci for the development of renal disease in a transgenic mouse model of HIVAN (24). These same two genomic loci regulate a network of genes expressed by podocytes, including Nphs1, Nphs2, Synpo, Kirrel, and Myh9, and thus represent trans-eQTLs — regions of the genome that regulate genes of interest from a distance. (B) In the absence of HIV-1 infection, mice carrying the trans-eQTLs HIVAN1 or HIVAN2 have altered expression levels of podocyte genes but do not develop renal disease. However, when infected with HIV-1, there is further perturbation of podocyte gene expression in mice carrying HIVAN1 or HIVAN2 susceptibility loci and overt renal disease develops.
Types of genomic variation that result in eQTLs
What might the genomic fragments comprising these eQTLs contain that might explain the observed alterations in podocyte gene expression? In the case of trans-regulatory elements, perhaps they encode a transcription factor that has effects on multiple podocyte genes. However, no known transcription factors regulating podocyte gene expression, such as Lmx1b, Tcf21, Wt1, Foxc2, or MafB (reviewed in ref. 34), map to these regions. Furthermore, in exhaustive eQTL studies in yeast, transcription factors are not generally responsible for a trans-eQTL (35). Instead, the effects of a trans-eQTL are typically mediated by complex regulatory mechanisms including microRNAs or changes in chromatin structure that affect other factors such as protein folding.
The study by Papeta et al. is believed to be the first report using eQTL analysis to successfully identify susceptibility loci for a glomerular disease (24). With this approach, the investigators have discovered a novel genetic network controlling podocyte genes that affects susceptibility to kidney disease following HIV-1 infection. Further studies to identify the specific mutation or genetic variant affecting expression of these podocyte genes will provide insight into the mechanisms of susceptibility to HIVAN and other glomerular diseases. Characterization of syntenic regions (conserved regions of the genome carrying common gene and intergenic regions between two species) in the human genome and determination of their effects on the severity of renal disease will be useful for understanding genetic susceptibility to kidney disease in patients with HIVAN and likely other glomerulopathies.
Acknowledgments
The author’s work in this area is supported by the Canadian Institutes of Health Research (grants MOP-62931and MOP-77756) and a Kidney Foundation of Canada grant.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Nonstandard abbreviations used: eQTL, expression quantitative trait locus; HIVAN, HIV-1–associated nephropathy; Myh9, myosin, heavy chain 9, non-muscle; Nphs1, nephrosis 1 homolog; Synpo, synaptopodin.
Citation for this article: J. Clin. Invest. 119:1085–1089 (2009). doi:10.1172/JCI39254
See the related article beginning on page 1178.
References
- 1.Wyatt C.M., Klotman P.E. HIV-associated nephropathy in the era of antiretroviral therapy. Am. J. Med. 2007;120:488–492. doi: 10.1016/j.amjmed.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt C.M., Klotman P.E. HIV-1 and HIV-associated nephropathy 25 years later. Clin. J. Am. Soc. Nephrol. 2007;2(Suppl. 1):S20–S24. doi: 10.2215/CJN.03561006. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt C.M., Klotman P.E., D’Agati V.D. HIV-associated nephropathy: clinical presentation, pathology, and epidemiology in the era of antiretroviral therapy. Semin. Nephrol. 2008;28:513–522. doi: 10.1016/j.semnephrol.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruggeman L.A., et al. Nephropathy in human immunodeficiency virus-1 transgenic mice is due to renal transgene expression. J. Clin. Invest. 1997;100:84–92. doi: 10.1172/JCI119525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conaldi P.G., et al. Human immunodeficiency virus-1 tat induces hyperproliferation and dysregulation of renal glomerular epithelial cells. Am. J. Pathol. 2002;161:53–61. doi: 10.1016/S0002-9440(10)64156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Husain M., et al. HIV-1 nef induces proliferation and anchorage-independent growth in podocytes. . J. Am. Soc. Nephrol. 2002;13:1806–1815. doi: 10.1097/01.ASN.0000019642.55998.69. [DOI] [PubMed] [Google Scholar]
- 7.Pavenstadt H., Kriz W., Kretzler M. Cell biology of the glomerular podocyte. Physiol. Rev. 2003;83:253–307. doi: 10.1152/physrev.00020.2002. [DOI] [PubMed] [Google Scholar]
- 8.Winn M.P., et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308:1801–1804. doi: 10.1126/science.1106215. [DOI] [PubMed] [Google Scholar]
- 9.Reiser J., et al. TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat. Genet. 2005;37:739–744. doi: 10.1038/ng1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinkes B., et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat. Genet. 2006;38:1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan J.M., et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat. Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 12.Boute N., et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 13.Kestila M., et al. Positionally cloned gene for a novel glomerular protein — nephrin — is mutated in congenital nephrotic syndrome. Mol. Cell. 1998;1:575–582. doi: 10.1016/S1097-2765(00)80057-X. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz E.J., et al. Human immunodeficiency virus-1 induces loss of contact inhibition in podocytes. J. Am. Soc. Nephrol. 2001;12:1677–1684. doi: 10.1681/ASN.V1281677. [DOI] [PubMed] [Google Scholar]
- 15.Barisoni L., Kriz W., Mundel P., D’Agati V. The dysregulated podocyte phenotype: a novel concept in the pathogenesis of collapsing idiopathic focal segmental glomerulosclerosis and HIV-associated nephropathy. J. Am. Soc. Nephrol. 1999;10:51–61. doi: 10.1681/ASN.V10151. [DOI] [PubMed] [Google Scholar]
- 16.Korgaonkar S.N., et al. HIV-1 upregulates VEGF in podocytes. J. Am. Soc. Nephrol. 2008;19:877–883. doi: 10.1681/ASN.2007050629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winston J.A., et al. Nephropathy and establishment of a renal reservoir of HIV type 1 during primary infection. N. Engl. J. Med. 2001;344:1979–1984. doi: 10.1056/NEJM200106283442604. [DOI] [PubMed] [Google Scholar]
- 18.Eggers P.W., Kimmel P.L. Is there an epidemic of HIV Infection in the US ESRD program? J. Am. Soc. Nephrol. 2004;15:2477–2485. doi: 10.1097/01.ASN.0000138546.53152.A7. [DOI] [PubMed] [Google Scholar]
- 19.Kopp J.B., et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao W.H., et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heath K.E., et al. Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am. J. Hum. Genet. 2001;69:1033–1045. doi: 10.1086/324267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelley M.J., Jawien W., Ortel T.L., Korczak J.F. Mutation of MYH9, encoding non-muscle myosin heavy chain A, in May-Hegglin anomaly. Nat. Genet. 2000;26:106–108. doi: 10.1038/79069. [DOI] [PubMed] [Google Scholar]
- 23.Seri M., et al. Mutations in MYH9 result in the May-Hegglin anomaly, and Fechtner and Sebastian syndromes. The May-Heggllin/Fechtner Syndrome Consortium. Nat. Genet. 2000;26:103–105. doi: 10.1038/79063. [DOI] [PubMed] [Google Scholar]
- 24.Papeta N., et al. Susceptibility loci for murine HIV-associated nephropathy encode trans-regulators of podocyte gene expression. J. Clin. Invest. 2009;119:1178–1188. doi: 10.1172/JCI37131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gharavi A.G., et al. Mapping a locus for susceptibility to HIV-1-associated nephropathy to mouse chromosome 3. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2488–2493. doi: 10.1073/pnas.0308649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schadt E.E., et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pravenec M., et al. Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat. Genet. 2008;40:952–954. doi: 10.1038/ng.164. [DOI] [PubMed] [Google Scholar]
- 29.Tsukaguchi H., et al. NPHS2 mutations in late-onset focal segmental glomerulosclerosis: R229Q is a common disease-associated allele. . J. Clin. Invest. 2002;110:1659–1666. doi: 10.1172/JCI16242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber T.B., et al. Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17079–17086. doi: 10.1073/pnas.0607465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber T.B., et al. Molecular basis of the functionalpodocin-nephrin complex: mutations in the NPHS2 gene disrupt nephrin targeting to lipid raft microdomains. Hum. Mol. Genet. 2003;12:3397–3405. doi: 10.1093/hmg/ddg360. [DOI] [PubMed] [Google Scholar]
- 32.Hubner N., et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 33.Ratelade J., et al. Maternal environment interacts with modifier genes to influence progression of nephrotic syndrome. J. Am. Soc. Nephrol. 2008;19:1491–1499. doi: 10.1681/ASN.2007111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quaggin S.E. Transcriptional regulation of podocyte specification and differentiation. Microsc. Res. Tech. 2002;57:208–211. doi: 10.1002/jemt.10076. [DOI] [PubMed] [Google Scholar]
- 35.Brem R.B., Yvert G., Clinton R., Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 36.Somlo S., Mundel P. Getting a foothold in nephrotic syndrome. Nat. Genet. 2000;24:333–335. doi: 10.1038/74139. [DOI] [PubMed] [Google Scholar]
- 37.Kopp J.B., et al. Progressive glomerulosclerosis and enhanced renal accumulation of basement membrane components in mice transgenic for human immunodeficiency virus type 1 genes. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1577–1581. doi: 10.1073/pnas.89.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuo Y., et al. HIV-1 genes vpr and nef synergistically damage podocytes, leading to glomerulosclerosis. J. Am. Soc. Nephrol. 2006;17:2832–2843. doi: 10.1681/ASN.2005080878. [DOI] [PubMed] [Google Scholar]
- 39.Zhong J., et al. Expression of HIV-1 genes in podocytes alone can lead to the full spectrum of HIV-1-associated nephropathy. Kidney Int. 2005;68:1048–1060. doi: 10.1111/j.1523-1755.2005.00497.x. [DOI] [PubMed] [Google Scholar]
- 40.Husain M., et al. HIV-1 Nef induces dedifferentiation of podocytes in vivo: a characteristic feature of HIVAN. AIDS. 2005;19:1975–1980. doi: 10.1097/01.aids.0000191918.42110.27. [DOI] [PubMed] [Google Scholar]
- 41.Chan K.T., et al. Accelerated development of collapsing glomerulopathy in mice congenic for the HIVAN1 locus. Kidney Int. 2009;75:366–372. doi: 10.1038/ki.2008.625. [DOI] [PMC free article] [PubMed] [Google Scholar]