Abstract
HIF transcription factors (HIF-1 and HIF-2) are central mediators of cellular adaptation to hypoxia. Because the resting partial pressure of oxygen is low in the intestinal lumen, epithelial cells are believed to be mildly hypoxic. Having recently established a link between HIF and the iron-regulatory hormone hepcidin, we hypothesized that HIFs, stabilized in the hypoxic intestinal epithelium, may also play critical roles in regulating intestinal iron absorption. To explore this idea, we first established that the mouse duodenum, the site of iron absorption in the intestine, is hypoxic and generated conditional knockout mice that lacked either Hif1a or Hif2a specifically in the intestinal epithelium. Using these mice, we found that HIF-1α was not necessary for iron absorption, whereas HIF-2α played a crucial role in maintaining iron balance in the organism by directly regulating the transcription of the gene encoding divalent metal transporter 1 (DMT1), the principal intestinal iron transporter. Specific deletion of Hif2a led to a decrease in serum and liver iron levels and a marked decrease in liver hepcidin expression, indicating the involvement of an induced systemic response to counteract the iron deficiency. This finding may provide a basis for the development of new strategies, specifically in targeting HIF-2α, to improve iron homeostasis in patients with iron disorders.
Introduction
Iron is an essential nutrient and is critical for a multitude of biological processes at the cellular level (including catalyzing essential enzymatic reactions and electron transport) and at the systemic level for oxygen transport in hemoglobin and myoglobin. Intestinal iron absorption is the only way for iron to be taken up by the body. Since humans do not have a regulated iron excretion pathway, systemic cues from either iron stores (liver) or sites of iron utilization (erythron or muscle) tightly regulate this process. Any disturbances in systemic regulators of iron transport machinery or inadequate nutrition and bleeding can disturb the fine balance of iron distribution and result in anemia (iron deficit) or iron accumulation in the parenchyma (1). Excess iron accumulation is observed in hereditary hemochromatosis, the most common genetic disorder in humans, while iron deficiency is one of the most frequently observed diseases in the world today, affecting as many as 2 billion people.
Non-heme dietary iron exists largely as ferric salts and is insoluble and thus bio-unavailable. Ferric iron (Fe3+) is rendered soluble in the proximal intestine, where it is reduced to ferrous iron (Fe2+), possibly by duodenal cytochrome b (DCYTB), an ascorbate-dependent ferric reductase. Ferrous iron is transported across the apical membrane by divalent metal transporter 1 (DMT1), the principal iron importer known to date. Depending on body iron requirements, iron can be either stored bound to ferritin or exported across the basolateral enterocyte membrane into the plasma by the sole iron exporter — ferroportin (FPN). Once in the plasma, iron is reoxidized by a copper ferroxidase, hephaestin, and, bound to the plasma iron transport molecule transferrin, delivered to all cell types that express transferrin receptors at their surface (1). At the cellular level, a central role of the RNA-binding proteins iron-regulatory protein 1 (IRP1) and IRP2 has been described in controlling the expression of critical iron metabolism proteins by a posttranscriptional mechanism based on cis-regulatory RNA motifs called iron-responsive elements (IREs) (2). At the systemic level, iron absorption is thought to be regulated by the liver-expressed peptide hepcidin, which is secreted into the serum. Hepcidin limits the amount of iron exported by the enterocyte into the serum by directly interacting with its cognate receptor FPN, resulting in its degradation (3). In addition, hepcidin transcription is upregulated by iron repletion and downregulated by iron deficiency, ineffective erythropoiesis, and hypoxia (4). More importantly, liver hepcidin levels are inversely correlated with the expression of iron absorption genes (5) and the rates of dietary iron absorption (6).
The gastrointestinal tract has a unique steady-state tissue oxygenation profile. Because of its juxtaposition with the anoxic lumen, the gastrointestinal mucosa has a uniquely steep oxygen gradient, ranging from the high level of oxygen in the richly vascularized subepithelial mucosa to the relative hypoxic state of the epithelium (reviewed in ref. 7). Adaptive transcriptional responses to oxygen deprivation are mediated by the HIFs, which function as heterodimeric transcription factors. The HIF heterodimers consists of 2 helix-loop-helix proteins: a regulatory subunit, which is the oxygen-responsive component, and HIF-1β, also known as the aryl hydrocarbon receptor nuclear translocator (ARNT), which is constitutively expressed. Three regulatory HIF subunits have been characterized: HIF-1α, HIF-2α, and HIF-3α. HIF-1α is expressed ubiquitously, whereas HIF-2α expression appears to be restricted to certain tissues (8). The expression patterns and functional properties of HIF-3 remain to be elucidated.
In normoxia, the regulatory subunit α is hydroxylated by the prolyl-hydroxylases (PHDs) and degraded through the ubiquitin-proteasome pathway via its interaction with the von Hippel–Lindau (vHL) tumor suppressor protein. Conversely, under hypoxia or iron depletion, hydroxylation is inhibited, resulting in stabilization of the α-subunit, heterodimerization with ARNT, nuclear translocation, and trans-activation of HIF target genes. The HIF heterodimer binds to hypoxia-responsive elements (HREs) of target gene regulatory sequences. Germline deletion of either the Hif1a or Hif2a subunit leads to unique phenotypes, suggesting essential roles for both HIF-α isoforms. Germline deletion of Hif1a, the most extensively studied subunit so far, is embryonic lethal (9, 10) and has been involved in the transcription of genes implicated in the control of angiogenesis, glycolytic metabolism, apoptosis, cellular stress, and other critical processes. Global deletion of Hif2a results in distinct phenotypes depending on the mouse strain (11–13), making it difficult to elucidate the specific roles of HIF-2α. However, recent evidence has shown that HIF-2α plays a critical role in adult erythropoiesis (14) and that it is the dominant HIF-α isoform in regulating erythropoietin (EPO) expression under physiological conditions (15). We recently showed in vivo that, through coordinate downregulation of hepcidin and upregulation of EPO and FPN in the liver, the vHL/HIF pathway mobilizes iron to support erythrocyte production (16).
We hypothesized that the HIF transcription factors, stabilized in the hypoxic epithelial layers of the intestine, could play a central role in maintaining iron balance in the body through the regulation of intestinal iron absorption genes and thus iron absorption. We specifically deleted Hif1a or Hif2a in the intestinal epithelium in mice including the proximal part of the intestine, which is the main site of dietary iron uptake. The major genes involved in iron uptake and export (DMT1, DCYTB, FPN) have only recently been characterized, and the molecular mechanisms of their basal, iron-dependant, or hypoxic regulation are still largely unknown. In this study, we show by the use of specific conditional knockouts of Hif1a and Hif2a in the intestine that HIF-2α, but not HIF-1α, directly regulates DMT1 transcription and modulates the entry of iron into the organism even at basal level. Deletion of Hif2a in the intestine was correlated to a decrease in serum iron and the triggering of a systemic response through a reduction in liver hepcidin expression to counteract the iron deficiency. While this work was in progress, Shah et al. reported that duodenal inactivation of Arnt but not Hif1a abolished the intestinal response to low iron diet and suggested that HIF-2α signaling was required for iron absorption (17). However, in our study, the use of a direct Hif2a-knockout mouse model allowed us to demonstrate that HIF-2 signaling was also required in maintenance of basal iron absorption rates.
Results
The duodenum is hypoxic at basal level.
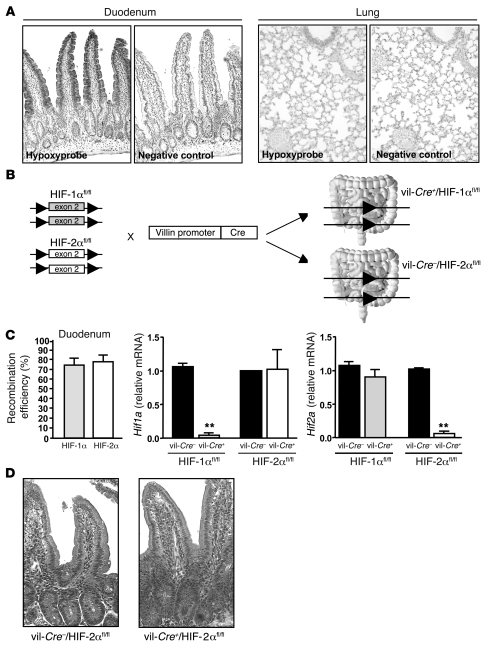
Several reports suggest that the intestinal epithelial cells that line the mucosa experience a uniquely steep physiological oxygen gradient in comparison with other cells of the body. To our knowledge, the status of the level of oxygen in the duodenum, the site of iron absorption, has not been previously investigated. We monitored in vivo hypoxia of the duodenum using nitroimidazole compound Hypoxyprobe, a marker for hypoxia. As shown in Figure 1A, Hypoxyprobe was retained in the superficial epithelial layers within the duodenum at the villus level, whereas it was weakly retained in a normoxic tissue, i.e., the lung. The hypoxic staining observed at the duodenum villi correlates with the described expression pattern of DMT1, beginning at the crypt-villus junction and visible along the entire length of the villi (18, 19).
Figure 1. Intestine-specific deletion of Hif1a and Hif2a.
(A) Immunostaining for the hypoxic marker Hypoxyprobe in WT mouse duodenum (original magnification, ×100) and lung (original magnification, ×200). The control corresponds to the omission of primary antibody. (B) HIF-1αfl/fl and HIF-2αfl/fl mice (both floxed at exon 2) were bred with a transgenic strain expressing Cre recombinase under the control of the murine villin promoter. (C) Recombination efficiency for HIF-1αfl/fl and HIF-2αfl/fl alleles quantified by real-time PCR of genomic DNA isolated from the duodenum as described previously (30). n = 4 in each group. Hif1a and Hif2a mRNA levels in duodenum scrapings of vil-Cre+/HIF-1αfl/fl and vil-Cre+/HIF-2αfl/fl mice and WT littermates as determined by real-time PCR. n = 4 in each group. **P < 0.01, unpaired Student’s t test. (D) H&E staining of duodenum of a vil-Cre+/HIF-2αfl/fl mouse and a WT littermate (original magnification, ×200).
Generation of conditional knockout of Hif1a and Hif2a in the intestine.
To examine the role of HIF transcription factors in iron uptake, we specifically deleted either Hif1a or Hif2a in the intestinal epithelium including the proximal part of the duodenum. HIF-1αfl/fl and HIF-2αfl/fl mice (both floxed at exon 2) were bred with a transgenic strain expressing Cre recombinase under the control of the murine villin promoter (Figure 1B).
Deletion efficiency of Hif1a and Hif2a in duodenum was approximately 80%, as determined by quantitative PCR on mouse genomic DNA (Figure 1C). The levels of Hif1a and Hif2a mRNAs were markedly decreased in the duodenum of vil-Cre+/HIF-1αfl/fl mice and vil-Cre+/HIF-2αfl/fl mice, respectively compared with their WT littermates. No compensatory expression of the remaining HIF isoform was observed in either mouse strain.
The vil-Cre+/HIF-1αfl/fl mice and vil-Cre+/HIF-2αfl/fl mice present no macroscopic abnormalities, and histological duodenal crypt/villus units were indistinguishable from those of the WT mice (data not shown and Figure 1D), ruling out the possibility that intestinal iron malabsorption resulted from altered villus architecture.
HIF-2α but not HIF-1α regulates iron absorption–related genes in the intestine.
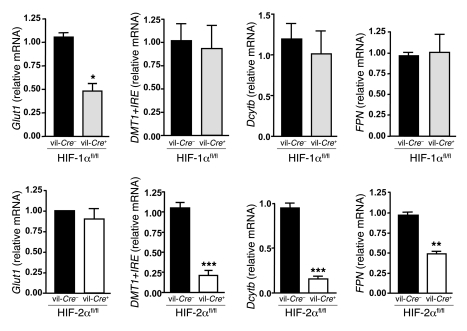
To specifically assess the respective roles of HIF-1α and HIF-2α in the regulation of iron absorption–related genes, we analyzed the expression of Dcytb, DMT1+IRE (the predominant isoform of DMT1 in intestine), and FPN in duodenal scrapings from vil-Cre/HIF-1αfl/fl and vil-Cre/HIF-2αfl/fl mice. In each experiment, matched littermate mice were used. Whereas deletion of Hif1a resulted in a 55% reduction in transcription of its well-characterized target, glucose transporter 1 (Glut1), it failed to affect the expression of iron-related genes (Figure 2). Interestingly, in contrast, the loss of HIF-2α strongly decreased the expression of DMT1+IRE (80%) and of Dcytb mRNA (85%), as well as the expression of FPN, albeit to a lesser extent (50%) (Figure 2). No change in Glut1 expression was observed in duodenum scrapings from vil-Cre/HIF-2αfl/fl mice. These data provide the first direct demonstration to our knowledge of a specific regulation of key iron absorption–related genes by HIF-2.
Figure 2. Deletion of Hif2a in the duodenum decreases the expression of genes involved in iron absorption.
Glut1, DMT1+IRE, Dcytb, and FPN mRNA levels in duodenum scrapings from vil-Cre+/HIF-1αfl/fl and vil-Cre+/HIF-2αfl/fl mice and WT littermates (n ≥ 6 in each group). *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test.
HIF-2α but not HIF-1α directly trans-activates the DMT1-1A promoter.
In contrast to Dcytb, which appears not to be necessary for iron absorption in the mouse (20), DMT1 is essential for non-heme dietary iron absorption (21). Specific deletion of DMT1 in mouse intestine (22) and a missense mutation in Belgrade rats (23) and mk/mk mice (24) are associated with severe iron deficiency anemia. Mutations of DMT1 in humans have also been associated with microcytic anemia (25, 26). We thus focused next on the regulation of DMT1, the only known iron importer in enterocytes. Four DMT1 transcript variants have been described. They are distinguished by the use of alternative exons 1A or 1B and alternative 3ι splicing giving rise to IRE- or non-IRE-containing (+IRE or –IRE) transcript variants (22, 27).
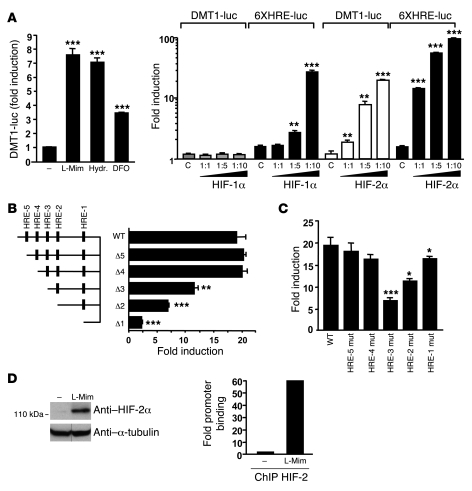
To assess whether the regulation of DMT1 by HIF-2α was direct, we first examined the 5ι flanking region of exon 1A for the presence of consensus HREs [A/G]CGTG. The DMT1-1A isoform is expressed mainly in the duodenum and shows the greatest increases in expression in response to hypoxia or iron deficiency (27, 28). We mapped 5 candidate HRE elements (2 in the sense and 3 in the antisense orientation) in the human DMT1-1A promoter (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI38499DS1). Unlike that of the exon 1B promoter, the activity of the 1A promoter has not been studied extensively. Hence, to examine whether the 5 putative HREs were involved in the HIF-dependent regulation of DMT1, we cloned the 616-bp 1A promoter region in front of a luciferase reporter. The activity of this DMT1-1A luciferase construct (DMT1-1A–luc) was measured in the human intestinal Caco-2 cell line (Caco-2/TC7), an intestinal model widely used to study iron absorption (29). An increase in 1A promoter activity was observed in Caco-2/TC7 cells incubated with l-mimosine (L-Mim), hydralazine, or desferrioxamine mesylate (DFO), 3 known pharmacological inducers of HIF-1α and HIF-2α (Figure 3A, left panel). To distinguish between the effects of HIF-1α or HIF-2α on the DMT1-1A promoter, DMT1-1A–luc was transfected together with increasing concentrations of HIF-1α or HIF-2α expression vectors. In agreement with our in vivo observations, HIF-2α, but not HIF-1α, strongly induced, in a dose-dependent manner, the luciferase activity of DMT1-1A–luc (Figure 3A, right panel). Importantly, both HIF-1α and HIF-2α induced, in a dose-dependent manner, the activity of a positive control consisting of 6XHRE-driven luciferase reporter. Next, to determine the relative importance of the 5 HREs present in the promoter of DMT1-1A, we truncated the promoter to sequentially remove the HREs (Δ5 to Δ1). As shown in Figure 3B, the deletion of HRE-5– or HRE-4–containing regions (Δ5 and Δ4) had no effect on HIF-2α induction of the DMT1 promoter. However, DMT1-1A promoter activity decreased significantly upon removal of HRE-3 (Δ3) and further decreased with the deletion of the region containing HRE-2 (Δ2). Their combined deletion led to an additive decrease in HIF-2α induction (Δ1). Second, we mutated independently the 5 target HREs and analyzed their activity after transfection with HIF-2α (Figure 3C). In agreement with the deletion experiments, mutations in HRE-1, HRE-2 and HRE-3 significantly decreased the HIF-2α DMT1 promoter activation, with a particularly dramatic decrease (60%) in the context of the HRE-3 mutant. Interestingly, the degree of conservation of the HREs across species correlated with the importance of the HREs in DMT1 activation, with HRE-3 being the most highly conserved across species (Supplemental Figure 1B).
Figure 3. HIF-2α binds and trans-activates the human DMT1-1A promoter.
(A) Left: The HIF agonists L-Mim (500 μM), hydralazine (Hydr.; 150 μM), and desferrioxamine mesylate (DFO; 150 μM) induced DMT1-1A promoter–driven luciferase activity. Right: Caco-2/TC7 cells were transiently transfected with pGL3–DMT1-1A or pGL3-6XHRE vectors together with pcDNA3 empty vector as a control (C) or increasing concentrations of pcDNA3–HIF-1α and pcDNA3–HIF-2α. (B) Luciferase assay on Caco-2/TC7 cells transiently transfected with pGL3–DMT1-1A (WT) or serial truncated versions of DMT1-1A promoter (D5 to D1) in the presence of pcDNA3–HIF-2α at a 1:10 ratio. (C) Luciferase assay on Caco-2/TC7 cells transiently transfected with pGL3–DMT1-1A (WT) or pGL3–DMT1-1A mutated in each of the 5 HREs (HRE-1 mut to HRE-5 mut) in the presence of pcDNA3–HIF-2α at a 1:10 ratio. (D) Left: Western blot analysis of Caco-2/TC7 cells incubated or not (–) with L-Mim (500 μM). Lanes were run on the same gel but were noncontiguous, as indicated. Right: Quantification by real-time PCR of a ChIP experiment on Caco-2/TC7 cells incubated with or without L-Mim (500 μM). *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test.
Finally, to determine the direct binding of the DMT1-1A promoter by HIF-2, we performed a ChIP assay on Caco-2/TC7 cells incubated in the presence of the HIF agonist L-Mim, which strongly induced HIF-2 expression, as shown in Figure 3D, left panel. Primers flanking HRE-3 specifically amplified DNA sequences immunoprecipitated by an HIF-2α antibody, showing that HIF-2α strongly binds to the human DMT1-1A promoter, supporting the hypothesis that DMT1 is directly regulated by HIF-2α (Figure 3D, right panel). Together, these results correlate with our in vivo data and show that HIF-2α directly binds DMT1 and induces its transcription.
Decrease in serum and liver iron in vil-Cre+/HIF-2fl/fl mice.
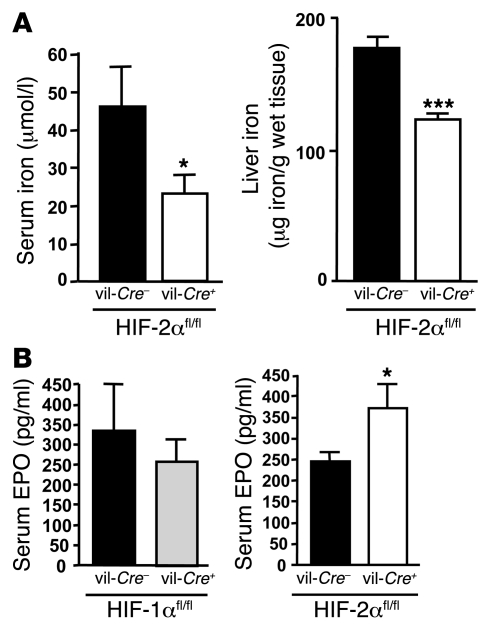
To determine the physiological consequence of Hif2a deletion in the duodenum on systemic iron, we measured iron-related parameters in vil-Cre/HIF-2αfl/fl mice. While no change in serum and liver iron was observed in vil-Cre+/HIF-1αfl/fl mice (data not shown), serum as well as liver iron levels were significantly reduced in vil-Cre+/HIF-2αfl/fl mice relative to WT littermates (Figure 4A). Concomitantly, EPO levels were significantly increased in the kidney (data not shown) and in the serum of vil-Cre+/HIF-2αfl/fl mice compared with control littermates (Figure 4B). The EPO levels in the serum of vil-Cre+/HIF-1αfl/fl mice were not altered.
Figure 4. Decrease in serum and liver iron and increase in serum EPO levels in intestine-specific Hif2a-knockout mice.
(A) Quantification of serum and liver iron levels in vil-Cre+/HIF-1αfl/fl and vil-Cre+/HIF-2αfl/fl mice and WT littermates (n = 6 in each group). (B) Serum EPO levels in vil-Cre+/HIF-1αfl/fl and vil-Cre+/HIF-2αfl/fl mice and WT littermates. *P < 0.05, ***P < 0.001, unpaired Student’s t test.
Deletion of Hif2a in the intestine induces a systemic iron response by the organism.
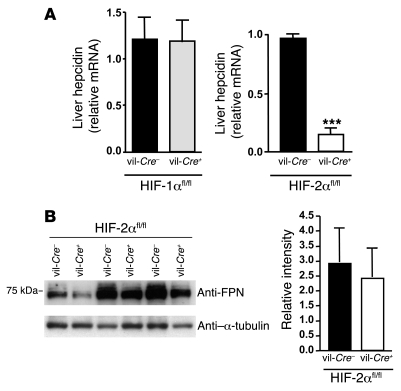
We next examined whether deletion of Hif2a in the intestine could trigger a systemic response by the organism by analyzing the expression of hepcidin, the iron-regulatory hormone. As expected, deletion of Hif1a in the intestine had no effects on hepatic hepcidin mRNA levels (Figure 5A). In contrast, hepcidin expression was drastically reduced in the liver of vil-Cre+/HIF-2αfl/fl mice relative to WT littermates. Therefore, a decrease in duodenal HIF-2α leads to iron deficiency and triggers a systemic iron response through downregulation of hepatic hepcidin. Despite the drastic decrease in hepcidin, duodenal FPN protein expression was not significantly altered in vil-Cre+/HIF-2αfl/fl mice compared with control littermates (Figure 5B). The observed lack of FPN protein increase, expected because of the downregulation of hepcidin in these mice, may be due to the diminished FPN mRNA levels observed in vil-Cre+/HIF-2αfl/fl mice (Figure 2), as well as to other posttranscriptional mechanisms due to local iron deficiency.
Figure 5. Decrease in hepatic hepcidin levels in intestine-specific Hif2a-knockout mice without any significant changes in duodenal FPN protein levels.
(A) Hepcidin mRNA expression in livers of vil-Cre+/HIF-1αfl/fl and vil-Cre+/HIF-2αfl/fl mice and WT littermates by real-time RT-PCR (n = 7). ***P < 0.001, unpaired Student’s t test. (B) FPN expression in duodenum scrapings of WT and vil-Cre+/HIF-2αfl/fl mice from 3 independent littermates, by Western blotting. Results were quantified and normalized (FPN/α-tubulin).
Duodenal Hif2a deletion results in decreased levels of hematocrit, red blood cell size, and hemoglobin content in response to low-iron diet.
We next examined whether the effects of HIF-2α observed at basal levels could be further exacerbated by iron deficiency. After a 2-month iron-deficient diet, WT mice showed strong increase in duodenal DMT1+IRE (~23-fold) and Dcytb (~45-fold) mRNA levels (Supplemental Figure 2A). Although an upregulation of DMT1 and Dcytb was detected in vil-Cre+/HIF-2αfl/fl mice, the absolute level of these mRNAs were lower in vil-Cre+/HIF-2αfl/fl as compared with WT littermates.
As expected, both WT and vil-Cre+/HIF-2αfl/fl mice demonstrated a repression of hepcidin expression (Supplemental Figure 2B). While no change in hematological parameters was observed between vil-Cre+/HIF-2αfl/fl mice and WT littermates fed a standard diet (data not shown), red blood cell, hemoglobin, and hematocrit levels were all decreased in vil-Cre+/HIF-2αfl/fl compared with WT mice fed the iron-deficient diet (Supplemental Figure 2C).
Discussion
Direct trans-activation of iron-related genes by HIFs, including the transferrin receptor, ceruloplasmin, heme oxygenase, and heme exporter BCRP (reviewed in ref. 30), has been demonstrated in in vitro studies. Here, we report a specific regulation of murine DMT1, Dcytb, and FPN in vivo by HIF-2α, but not by HIF-1α. Our detailed analyses of direct trans-activation of human DMT1 promoter by HIF-2α in Caco-2 cells extend the recently published work of Shah et al. (17). Although, we found an HIF-2α–dependent regulation of duodenal FPN mRNA levels at the basal level, no HREs were found to be clearly conserved between mice and other species, suggesting a possible indirect regulation of FPN by HIF-2α. The direct transcriptional regulation of DMT1 by HIF-2α suggests that HIF-2α regulation of DMT1 isoforms that contain IREs in their transcripts operates upstream of the IRP/IRE system. It has recently been suggested that IRPs are essential in intestinal function and coordinate the synthesis of DMT1 and FPN in the duodenum (31). However, deletion of IRP genes in the intestine does not correlate with a change in serum iron or iron stores, and the extent of their role in vivo remains to be fully elucidated.
Several studies indicated that HIF-1α and HIF-2α modulate the transcription of an overlapping but distinct set of target genes (32, 33). The N-terminal transactivation domain is thought to confer HIF target gene specificity by interacting with additional transcriptional cofactors. It is therefore plausible that other auxiliary transcription factors may be required in order for HIF-2α to specifically induce DMT1 transcription.
A decrease in hepcidin expression by the body in response to iron deficiency (Figure 5A) is expected to increase iron absorption and expression of the iron absorption genes DMT1 and DCYTB. Basolaterally expressed FPN responds directly to the humoral regulator hepcidin, which triggers FPN internalization and degradation, although this mechanism has not yet been formally proven in vivo in the intestine. Interestingly, our data show that a decrease in hepcidin fails to promote iron absorption when Hif2a is deleted in the intestine. According to our model, iron absorption genes at the apical membrane would be regulated in a cell-autonomous manner by HIF-2, whereas hepcidin would exert its systemic effect at the basolateral membrane of the enterocyte.
HIF-2α, stabilized in the hypoxic epithelial layers of the intestine, could be important in maintaining the iron balance in the body by counteracting any transient changes in tissue oxygenation or iron deficiency that could occur daily under a regular diet. Indeed, the intestinal mucosa experiences multiple daily dynamic fluctuating rates of perfusion in the physiologic state. During fasting, the blood volume in the gut is relatively low; however, after the ingestion of a meal, perfusion rises significantly, resulting in large daily pO2 fluctuations. Importantly, even if hypoxic states are relatively constant in the intestine, both the barrier and absorptive functions of the intestinal epithelium can be further physiologically regulated by oxygen (reviewed in ref. 7). Interestingly, Raja et al. demonstrated in mice an increase in duodenal iron uptake by hypoxia as early as 6 hours after induction. This was independent of erythropoietic stimulation, as either mice were nephrectomized or their bone marrow was irradiated (34). This rapid change suggests direct involvement of HIF transcription factors in this process. Moreover, under a chronic iron-deficient diet, HIF-2α may be further stabilized through a likely decrease in PHD activity due to very low intracellular iron levels in enterocytes. Therefore, in conditions of systemic iron deficiency, not only would hepcidin be low to prevent FPN degradation, but HIF-2α, further stabilized by local iron deficiency in enterocytes, would boost iron absorption by increasing the expression of apical transporters.
Shah et al. recently reported that duodenal Arnt deletion prevents the increase in expression of iron absorption–related genes in response to low iron diet (17). However, they did not show any changes in iron absorption–related gene expression upon inactivation of duodenal ARNT, at basal levels. While we used a specific knockout of the Hif1a or Hif2a regulatory subunit in the duodenum, the above study used a combination of conditional knockout of VhL, Hif1a, and Arnt in the intestine. In the Arnt knockout, the oxygen-responsive components HIF-1α and HIF-2α, while unable to form a functional heterodimer with ARNT, are still present. This discrepancy may explain the lack of effect of the Arnt knockout on the maintenance of iron metabolism under a regular diet.
In conditions of iron-refractory iron deficiency anemia or anemia of chronic inflammation, high levels of hepcidin, by degrading duodenal FPN would increase intracellular iron in enterocytes. Supplementary ferrous iron has been shown in vitro to greatly enhance capture of HIF by vHL, even under hypoxia (35). Therefore, we can speculate that an increase in intracellular iron in enterocytes would contribute to HIF degradation and the subsequent decrease in the expression of apical transporters.
Together, these results suggest that monitoring levels of HIF-2α could benefit patients with iron disorders, considering its activation may allow iron mobilization and its reduction favors decreased iron absorption.
Methods
Animals.
Cell-specific inactivation of Hif1a and Hif2a in the intestine was achieved by cross-breeding villin-Cre transgenic mice (provided by Sylvie Robine, Institut Curie, Paris, France) with HIF-1αfl/fl (provided by Randall Johnson, UCSD, La Jolla, California, USA) and HIF-2αfl/fl mice, all on a C57BL/6 background. In all experiments, littermates from the same breeding pair were used as controls. Six- to 10-week-old mice were used and maintained on a standard rodent diet or an iron-deficient diet (3 ppm iron; Scientific Animal Food & Engineering). All mice used in the experiments were cared for according to criteria outlined in the European convention for the protection of vertebrate animals used for experimental and other scientific purposes (Council of Europe, ETS 123. 1991). Animal studies described here were reviewed and approved (agreement no. 75-1466) by the Directeur départemental des Services Vétérinaires of the Prefecture de Police de Paris.
Hypoxia staining.
Briefly, animals were administered Hypoxyprobe (NPI Inc.) via intraperitoneal injection (60 mg/100 g body weight). One hour after injection, small intestine and lungs were harvested, fixed overnight in 4% buffered formalin, and embedded in paraffin. Paraffin sections (5 μm) were processed according to the manufacturer’s instructions (Hypoxyprobe-1 Omni-Kit).
Iron measurements and hematological analysis.
Serum and tissue iron were quantified colorimetrically by a method described in ref. 4 using the IL test (Instrumentation Laboratory). Hematological parameters were determined using a Coulter MAXM automatic analyzer (Beckman Coulter).
Reverse transcription and real-time quantitative PCR.
Total RNA was extracted from tissues using RNA-PLUS reagent (Qbiogene) according to the instructions provided by the manufacturer. Reverse transcription was done with 2–3 μg of total RNA as described previously (36). Quantitative PCR was performed with 2 μl of a 1:10 dilution of reverse-transcribed total RNA, 10 μM of each primer, and 2 mM MgCl2 in 1× LightCycler DNA Master SYBR Green I mix using a LightCycler apparatus (Roche Applied Science). All samples were normalized to the threshold cycle value for 18S. Sequences of the primers are available in Supplemental Table 1.
Western blot studies.
Proximal duodenum was harvested and ground in a mortar, and tissue powder was resuspended in 1 ml of 0.25 M sucrose/0.03 M histidine (pH 7.2) supplemented with 1× complete inhibitor cocktail EDTA-free (Roche). The homogenate was centrifuged for 15 minutes at 6,000 g, and crude membrane fraction was subsequently isolated by ultracentrifugation of the supernatant at 75,779 g (TLA-45 rotor; Beckman-Coulter) for 1 hour. Protein concentration was determined using the Bradford assay; 50 μg of membrane extracts were loaded on a 10 % gel; and Western blot analysis was performed using standard methods. FPN primary antibody (gift of François Cannone-Hergaux, CNRS [ICSN], Gif-sur-Yvette, France) was diluted at 1:500. Immunoblots were quantified using ImageJ ( http://rsbweb.nih.gov/ij/).
EPO ELISA.
EPO protein levels in blood plasma were determined with the Quantikine Mouse EPO ELISA kit (R&D Systems).
Cloning procedures.
A 616-bp 5′ flanking region of the DMT1-1A exon was amplified from human genomic DNA and subcloned into the pGL3-basic luciferase reporter vector (Promega). All additional HRE mutant constructs were generated using Quick-Change Site-Directed Mutagenesis Kit (Stratagene) as per the manufacturer’s protocol. Each HRE element was mutated by insertion of an XhoI restriction site, which also enabled us to sequentially truncate the 5′ regions of the DMT1-1A promoter. All promoter constructs were confirmed by sequencing (see Supplemental data for the primers).
Transient transfection and luciferase assay.
Caco-2/TC7 cells (provided by Monique Rousset, Centre de Recherche des Cordeliers, Paris, France) were grown in 24-well plates and transfected using FuGENE 6 HD (Roche Applied Science) as per the manufacturer’s protocol. In all experiments, as a normalization control, the pRL-TK Renilla luciferase vector (Promega) was cotransfected with respective luciferase constructs at a 1:50 ratio. For the coexpression studies, pGL3-basic, pGL3–DMT1-1A, or 6XHRE-luc reporter vectors (40 ng) were cotransfected with pcDNA3, pcDNA3–HIF-1α, or pcDNA3–HIF-2α expression vectors at a 1:1 (40 ng), 1:5 (200 ng), or 1:10 (400 ng) ratio relative to the reporter vectors. For all mutant and deletion studies of the DMT1 reporter, pcDNA3–HIF-2α was cotransfected at a 1:10 ratio. pcDNA3–HIF-1α and pcDNA3–HIF-2α were provided, respectively, by Eric Clottes (Université de Toulouse/CNRS-IPBS, Toulouse, France) and David Russell (University of Texas Southwestern Medical Center, Dallas, Texas, USA); Fabrice Soncin (CNRS, Lille, France) provided the reagents produced by David Russell. The 6XHRE-luc construct, derived from the VEGF promoter, was provided by Randall Johnson. In some experiments, 24 hours after transfection, cells were incubated with DFO, hydralazine (both at 150 μM), or L-Mim (500 μM) in serum-free medium and incubated for an additional 24 hours. The luciferase assay was performed using a Dual-Glo Luciferase Reporter Kit (Promega) on a Berthold luminometer as indicated by the manufacturer, 48 hours after transfection.
ChIP assay.
Caco-2/TC7 cells, grown in 15-cm dishes, were incubated with or without 500 μM L-Mim (Sigma-Aldrich). Twenty-four hours later, the ChIP assay was performed according to the protocol described in Nelson et al. (37). Anti–HIF-2α (NB100-122; Novus Biologicals) was used for immunoprecipitation. The primer sequences, designed to amplify the HRE-containing region of DMT1 promoter, are shown in Supplemental Table 1. We quantified the amplification by quantitative PCR as the ratio: (2^(Ct input – Ct ChIP)DMT1)/(2^(Ct input – Ct ChIP)negative), where Ct ChIP is the Ct value corresponding to the immunoprecipitated DNA and Ct input is the Ct value of an aliquot of sheared chromatin sample before immunoprecipitation.
Statistics.
All values in the figures are expressed as mean ± SD. Student t test (unpaired, 2-tailed) was used for comparison between experimental groups. A P value of 0.05 or less was considered significant.
Supplementary Material
Acknowledgments
We are grateful to Randall Johnson for providing the HIF-1αfl/fl mice and Sylvie Robine for the villin-Cre mice. We thank François Cannone-Hergaux for the FPN antibody; Monique Rousset for the Caco-2/TC7 cells; and members of Sophie Vaulont and Christine Perret’s laboratory for helpful discussions, reading of the manuscript, and technical help with the ChIP procedure. This study was supported by funding from the Agence Nationale pour la Recherche and European Economic Community (EEC) Framework 6 (LSHM-CT-037296 Euroiron1). M. Mastrogiannaki is supported by a fellowship from the Ministère de l’Education Nationale de la Recherche et de la Technologie and P. Matak by the EEC.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: ARNT, aryl hydrocarbon receptor nuclear translocator; Dcytb, duodenal cytochrome b; DMT1, divalent metal transporter 1; EPO, erythropoietin; FPN, ferroportin; HRE, hypoxia-responsive element; IRE, iron-responsive element; IRP, iron-regulatory protein; L-Mim, l-mimosine; vHL, von Hippel–Lindau.
Citation for this article: J. Clin. Invest. 119:1159–1166 (2009). doi:10.1172/JCI38499
References
- 1.Andrews N.C. Forging a field: the golden age of iron biology. Blood. 2008;112:219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muckenthaler M.U., Galy B., Hentze M.W. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 3.Nemeth E., et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 4.Nicolas G., et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J. Clin. Invest. 2002;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viatte L., et al. Deregulation of proteins involved in iron metabolism in hepcidin-deficient mice. Blood. 2005;105:4861–4864. doi: 10.1182/blood-2004-12-4608. [DOI] [PubMed] [Google Scholar]
- 6.Leung P.S., Srai S.K., Mascarenhas M., Churchill L.J., Debnam E.S. Increased duodenal iron uptake and transfer in a rat model of chronic hypoxia is accompanied by reduced hepcidin expression. Gut. 2005;54:1391–1395. doi: 10.1136/gut.2004.062083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor C.T., Colgan S.P. Hypoxia and gastrointestinal disease. J. Mol. Med. 2007;85:1295–1300. doi: 10.1007/s00109-007-0277-z. [DOI] [PubMed] [Google Scholar]
- 8.Wiesener M.S., et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J. 2003;17:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 9.Iyer N.V., et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan H.E., Lo J., Johnson R.S. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Compernolle V., et al. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat. Med. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- 12.Peng J., Zhang L., Drysdale L., Fong G.H. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian H., et al. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber M., et al. Acute postnatal ablation of Hif-2alpha results in anemia. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rankin E.B., et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J. Clin. Invest. 2007;117:1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peyssonnaux C., et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J. Clin. Invest. 2007;117:1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah Y.M., Matsubara T., Ito S., Yim S.H., Gonzalez F.J. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152–164. doi: 10.1016/j.cmet.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canonne-Hergaux F., Gruenheid S., Ponka P., Gros P. Cellular and subcellular localization of the Nramp2 iron transporter in the intestinal brush border and regulation by dietary iron. Blood. 1999;93:4406–4417. [PubMed] [Google Scholar]
- 19.McKie A.T., et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 20.Gunshin H., et al. Cybrd1 (duodenal cytochrome b) is not necessary for dietary iron absorption in mice. Blood. 2005;106:2879–2883. doi: 10.1182/blood-2005-02-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunshin H., et al. Slc11a2 is required for intestinal iron absorption and erythropoiesis but dispensable in placenta and liver. J. Clin. Invest. 2005;115:1258–1266. doi: 10.1172/JCI24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunshin H., et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 23.Fleming M.D., et al. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming M.D., et al. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat. Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 25.Beaumont C., et al. Two new human DMT1 gene mutations in a patient with microcytic anemia, low ferritinemia, and liver iron overload. Blood. 2006;107:4168–4170. doi: 10.1182/blood-2005-10-4269. [DOI] [PubMed] [Google Scholar]
- 26.Mims M.P., et al. Identification of a human mutation of DMT1 in a patient with microcytic anemia and iron overload. Blood. 2005;105:1337–1342. doi: 10.1182/blood-2004-07-2966. [DOI] [PubMed] [Google Scholar]
- 27.Hubert N., Hentze M.W. Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: implications for regulation and cellular function. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12345–12350. doi: 10.1073/pnas.192423399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lis A., et al. Hypoxia induces changes in expression of isoforms of the divalent metal transporter (DMT1) in rat pheochromocytoma (PC12) cells. Biochem. Pharmacol. 2005;69:1647–1655. doi: 10.1016/j.bcp.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 29.Sharp P. Methods and options for estimating iron and zinc bioavailability using Caco-2 cell models: benefits and limitations. Int. J. Vitam. Nutr. Res. 2005;75:413–421. doi: 10.1024/0300-9831.75.6.413. [DOI] [PubMed] [Google Scholar]
- 30.Peyssonnaux C., Nizet V., Johnson R.S. Role of the hypoxia inducible factors HIF in iron metabolism. Cell Cycle. 2008;7:28–32. doi: 10.4161/cc.7.1.5145. [DOI] [PubMed] [Google Scholar]
- 31.Galy B., Ferring-Appel D., Kaden S., Grone H.J., Hentze M.W. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Hu C.J., et al. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol. Cell. Biol. 2006;26:3514–3526. doi: 10.1128/MCB.26.9.3514-3526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu C.J., Sataur A., Wang L., Chen H., Simon M.C. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Mol. Biol. Cell. 2007;18:4528–4542. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raja K.B., Simpson R.J., Pippard M.J., Peters T.J. In vivo studies on the relationship between intestinal iron (Fe3+) absorption, hypoxia and erythropoiesis in the mouse. Br. J. Haematol. 1988;68:373–378. doi: 10.1111/j.1365-2141.1988.tb04217.x. [DOI] [PubMed] [Google Scholar]
- 35.Jaakkola P., et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 36.Nicolas G., et al. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4596–4601. doi: 10.1073/pnas.072632499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson J.D., Denisenko O., Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.