Abstract
The small heat shock proteins (sHSPs) are a diverse family of molecular chaperones. It is well established that these proteins are crucial components of the plant heat shock response. They also have important roles in other stress responses and in normal development. We have conducted a comparative sequence analysis of the sHSPs in three complete angiosperms genomes: Arabidopsis thaliana, Populus trichocarpa, and Oryza sativa. Our phylogenetic analysis has identified four additional plant sHSP subfamilies and thus has increased the number of plant sHSP subfamilies from 7 to 11. We have also identified a number of novel sHSP genes in each genome that lack close homologs in other genomes. Using publicly available gene expression data and predicted secondary structures, we have determined that the sHSPs in plants are far more diverse in sequence, expression profile, and in structure than had been previously known. Some of the newly identified subfamilies are not stress regulated, may not posses the highly conserved large oligomer structure, and may not even function as molecular chaperones. We found no consistent evolutionary patterns across the three species studied. For example, gene conversion was found among the sHSPs in O. sativa but not in A. thaliana or P. trichocarpa. Among the three species, P. trichocarpa had the most sHSPs. This was due to an expansion of the cytosolic I sHSPs that was not seen in the other two species. Our analysis indicates that the sHSPs are a dynamic protein family in angiosperms with unexpected levels of diversity.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-008-0023-7) contains supplementary material, which is available to authorized users.
Introduction
The small heat shock proteins (sHSPs) are a ubiquitous family of proteins found in all domains of life (Caspers et al. 1995; de Jong et al. 1998; Fu et al. 2006). Numerous studies have shown that the sHSPs, like most other HSPs, are molecular chaperones (Narberhaus 2002; van Montfort et al. 2002; Wang et al. 2004; Sun and MacRae 2005; Nakamoto and Vigh 2007). Small HSPs can bind to denatured proteins and thereby prevent their irreversible aggregation (Lee et al. 1995; Lee et al. 1997; van Montfort et al. 2002; Nakamoto and Vigh 2007). By binding to these denatured proteins, the sHSPs can create a reservoir of proteins that can be refolded by other parts of the chaperone system (Lee and Vierling 2000; Haslbeck et al. 2005). The importance of the sHSPs is illustrated by the finding that homologs of sHSPs have been found in almost all organismal genomes even in highly reduced genomes that have lost numerous other genes (Gil et al. 2002, 2003; Waters et al. 2003).
Most heat shock protein families (i.e., the HSP60s, HSP70s, HSP90s, and HSP100s) are highly conserved across great organismal distances; in fact, some of these families are among the most highly conserved protein families known (Boorstein et al. 1994; Gupta 1995; Stechmann and Cavalier-Smith 2003). In contrast, the amino acid sequences of the sHSPs are highly variable (Caspers et al. 1995; Waters 1995; de Jong et al. 1998). However, there are a number of conserved sHSP features. First, although these proteins get their name from the size of the monomers (usually less than 30 kDa), they function as large oligomers with 12–24 subunits (van Montfort et al. 2002). Second, all sHSPs share the conserved α-crystallin domain of approximately 100 residues (Caspers et al. 1995; de Jong et al. 1998; Fu et al. 2006). Third, the sHSPs share a compact β-sheet sandwich structure. These sheets can dimerize forming the building block of the large oligomers. Much of our knowledge of the sHSP structure is based on analysis of the crystal structures of two sHSPs: one from wheat Triticum aestivum HSP16.9 (a dodecamer; van Montfort et al. 2001) and the other from the archaeabacterium Methanococcus jannaschii, HSP 16.5 (a 24-mer; Kim et al. 1998). Comparisons of these structures find that even most secondary structural features are conserved (van Montfort et al. 2002). This is despite the fact that these proteins share less than 25% amino acid sequence identity. From this analysis, we can then assume that despite large levels of sequence diversity among the sHSPs, structural features are most likely conserved within this diverse and important family.
The sHSPs are part of both the plant stress response and normal development (Waters et al. 1996; Wang et al. 2004; Sun and MacRae 2005). Most of the sHSPs are highly upregulated during heat stress. This upregulation of the sHSPs can confer organismal thermal tolerance by protecting other proteins from irreversible denaturation. The sHSPs also play a role in plant responses to other stresses. At least some of the plant sHSPs are expressed in response to heavy metals, drought, UV, salinity, cold, osmotic, and oxidative stress. In addition, some plant sHSPs are a part of normal development during embryogenesis, seed germination, pollen development, and fruit maturation (Waters et al. 1996; Wang et al. 2004; Sun and MacRae 2005).
Gene duplication has been important in generating functional diversity within the sHSP family (Caspers et al. 1995; de Jong et al. 1998; Waters et al. 2003; Franck et al. 2004). For example, the sHSP family includes the α-crystallin proteins of the vertebrate eye lens. However, by far, one of the most interesting aspects of the functional diversity and evolution of the entire sHSP superfamily is the abundance, diversity, and importance of sHSPs in plants (Vierling 1991; Waters 1995; Waters et al. 1996; Waters 2003). In plants, in contrast to other organisms, the most abundant proteins during heat shock are the sHSPs (Vierling 1991). In addition, as well as being highly abundant, the plant sHSPs are also highly diverse in sequence and in cellular location.
There are at least seven different plant sHSP subfamilies (Waters 1995; Scharf et al. 2001; Siddique et al. 2003; Ma et al. 2006). A minimum of three plant sHSP subfamilies localize to the cytosol. In addition, there are subfamilies that localize to the chloroplast (CP), mitochondria (MT), endoplasmic reticulum (ER), and peroxisome (PX), respectively. The exact timing of the origin of the plant sHSP subfamilies is not yet known. However, at least some subfamilies originated very early in land plant evolution as two of the cytosolic subfamilies and the CP subfamily have been identified in bryophytes (Waters and Vierling 1999a; Waters and Vierling 1999b). It is interesting to note that none of the organelle-localized plant sHSP subfamilies have homologs in animals, fungi, or even in green algae (Waters and Vierling 1999a; Waters and Rioflorido 2007). There is still much to be learned about how and when the plant sHSP subfamilies evolved.
Previous analysis of the complete Arabidopsis thaliana genome revealed that this species has 19 sHSP genes (Scharf et al. 2001). Some of these sHSPs had not been identified previously despite extensive studies by a number of researchers of the A. thaliana heat shock response. The results of this genome analysis raised a number of important unanswered questions: How many of the previously unknown sHSPs are also present in other plants? Do these newly identified sHSPs have expression patterns that are highly distinct from the well-characterized heat-inducible sHSPs? In addition, what evolutionary forces are acting on the sHSPs? Further, are there sHSP-specific evolutionary patterns that are seen across species, or do genome-specific processes dominate sHSP evolution? Now that two additional complete flowering plant genomes are available, the genomes of Oryza sativa or rice (Goff et al. 2002; Yu et al. 2002), a member of the grass family, and Populus trichocarpa (Tuskan et al. 2006), a member of the Salicaeae family, these and additional questions can be addressed.
To answer these questions, we performed a comparative analysis of the sHSP sequences in A. thaliana, O. sativa, and P. trichocarpa. We found that in contrast to the 19 sHSPs in A. thaliana, O. sativa has 23 sHSPs, and P. trichocarpa has 36. Phylogenetic analysis of all of these sHSPs allowed us to identify new plant sHSP subfamilies and novel sHSPs within each genome. In addition, we have determined that the evolutionary forces acting on this protein family varies across genomes. Among the sHSPs, gene conversion is absent in A. thaliana and in P. trichocarpa and much more common in O. sativa. This analysis finds that the sHSPs are a dynamic protein family and that despite the long evolutionary histories of many sHSP subfamilies, this family is continuing to expand within different angiosperm lineages.
Materials and methods
Gene and protein identification
The programs Blastp, Blastn, and TBlastn were used at each of the individual genome websites for A. thaliana (http://www.arabidopsis.org/Blast/), O. sativa (http://tigrblast.tigr.org), and P. trichocarpa (http://genome.jgi-psf.org/Poptr1_1/ Poptr1_1.home.htmland) at National Center for Biotechnology Information. All sequences with at least an e value of less than 10−4 were downloaded and examined further. Gene models were confirmed by comparisons to available EST sequences. The chromosomal location and orientation were noted for each gene. Genes present in the duplicated region of each genome (Vision et al. 2000; Blanc et al. 2003; Blanc and Wolfe 2004; Wang et al. 2006) were identified using The Institute for Genomic Research and Plant Genome Duplication databases.
The sHSP sequences identified were analyzed using a variety of sequence analysis tools. The corresponding amino acid sequences were generated from each deoxyribonucleic acid (DNA) gene sequence using Vector NTI (v.7). Protein size in kilodaltons was calculated with Vector NTI (v.7). Amino acid sequence alignments were then generated with ClustalW and optimized by hand with BioEdit (v.7.0.5). The amino acid alignments were used to align the DNA sequences using the program protal2dna (http://bioweb.pasteur.fr/seqanal/interfaces/protal2dna.html). The complete amino acid alignment sHSPs can be found in the supplementary materials (Supplementary Fig. 1).
Cellular location and protein secondary structure
All the sHSPs are nuclear encoded, and many are located in subcellular locations. To gain a better understanding of the possible cellular locations of the sHSPs identified here, we performed cellular location predictions using the following programs: PSORT (http://psort.nibb.ac.jp/form.html), Predotar (http://genoplanteinfo.infobiogen.fr/predotar/), and TargetP (http://www.cbs.dtu.dk/services/TargetP/). In cases where experimentally determined locations were known for some subfamily members, they agreed with our predicted locations (for example: cytosolic III, Siddique et al. 2003; peroxisome, Ma et al. 2006; MT, Lenne and Douce 1994; CP, Vierling et al. 1988).
The secondary structure of HSP16 from T. aestivum was obtained from the protein sequence database (1GME) and was compared to secondary structure predictions of the proteins identified here generated using the Predict Protein server (http://www.embl-heidelberg.de/predictprotein/predictprotein.html).
Phylogenetic analysis
To gain an understanding of how the sHSPs are related to each other, we generated phylogenetic trees using two different tree construction methods: neighbor-joining (NJ) with MEGA v. 3.1 (Kumar et al. 2004) and Bayesian with MrBayes v. 3.1 (Ronquist and Huelsenbeck 2003). We used both the DNA and amino acid sequences to estimate the phylogenetic relationships. Our alignments included only the highly conserved C-terminal domain (residues 170–300 in Supplementary Fig. 1). We performed phylogenetic analysis on the amino acid alignment and corresponding DNA alignment. Using the Bayesian method, we performed a combined analysis that included both the DNA and amino acid alignments. The tree topologies generated with each method and data type were highly congruent. Distance matrices were based on the Jones–Taylor–Thornton substitution matrix for the amino acid data and the Kimura 2 parameter for the DNA data. These analyses were performed in Mega v. 3.1 (Kumar et al. 2004). Pairwise deletions were excluded, and a gamma parameter was used to allow for variation across the sites. Estimates of support for the branches were generated from 1,000 bootstrap replicates. In the Bayesian analysis, four simultaneous chains of eight million generations each were run with trees saved every 100 generations. For the amino acid analysis, an initial analysis using the mixed model of amino acid evolution was used. This analysis determined that the WAG model was the best model for this data set, and this model was used in subsequent runs of MrBayes. The final analysis was stopped after the standard deviation of the split frequencies between the four independent tree searches was below the critical value of 0.001 (Ronquist et al. 2006). Trees from the first 4 million generations were discarded as burnin. The term “burnin” is defined by Ronquist and Huelsenbeck (2003) as those generations before convergence of the chains. By discarding the trees from these early generations, we keep only those trees generated after convergence of the chains. The remaining trees were used to generate the consensus tree and the posterior probabilities assigned to each branch. For the DNA analysis, we used MrModel Test (Posada and Buckley 2004) to choose the best model of DNA evolution. We then used the GTR+ gamma model for each codon position in the DNA analysis. For the combined analysis, the same models were used, but the tree was estimated using both amino acid and DNA data.
Sequence evolution and gene conversion
Estimates of nonsynonymous (Ka) and synonymous substitutions (Ks) were generated using the program MEGA v. 3.1 (Kumar et al. 2004) using the Kumar method. Our analysis of synonymous and nonsynonymous substitutions is based on the cytosolic I genes (Supplementary Figs. 2, 3, and 4). These genes are the most numerous in each genome, and in most cases, Ks has not yet become saturated. In comparisons within the same genome across subfamilies, almost all Ks values cannot be reliably estimated because they are so high. We detected gene conversion tracts using the program GENECONV v. 1.18 (Sawyer 1999). Alignments of sHSPs genes for each genome were generated and organized by chromosome, predicted subfamily, and levels of DNA sequence identity. Each of these alignments was analyzed separately with GENECOV. Following the recommendations of Sawyer and previous studies of gene conversion (Drouin 2002; Mondragon-Palomino and Gaut 2005), we evaluated the significance of each gene conversion tract based on the global p values rather than the pairwise p values. The global p values are more conservative and thus provide a better estimate of the statistical significance of the conversion tract. We considered values with a p value of less than 0.05 as statistically significant.
Gene expression
Publicly available gene chip data was analyzed. A. thaliana expression data were obtained from the GENEVESTIGATOR website (Zimmermann et al. 2004; http://www.genevestigator.ethz.ch/). This site has more than 2,000 A. thaliana Affymetrix gene chip experiments in its database. The programs Digital Northern, Meta Analyzer, Response Viewer, and Gene Atlas were used. All but one of the A. thaliana sHSPs are on the Affymetrix gene chip (At17.8I is not present). Expression profiles of the remaining 18 sHSPs are reported here. The website RED (Rice Expression Database http://red.dna.affrc.go. jp/RED/; Yazaki et al. 2004) has O. sativa gene chip experiments available for analysis. However, only a limited number of O. sativa sHSPs are on the gene chip. The following genes are on the O. sativa chip: Os22.3ER, Os21Cp, Os22MT, and Os22.4MT. Therefore, there is only limited information available for O. sativa sHSP gene expression. The poplar gene expression data was obtained from the Populus DB at htp://poppel.fysbot.umu.se/. Only some of the P. trichocarpa sHSPs were present on the gene chips, and very few stress experiments were conducted. As a result, comprehensive expression profiles are also not currently available for the P. trichocarpa sHSPs.
Results
The sHSPs of Arabidopsis thaliana, Oryza sativa, and Populus trichocarpa
Searches of the complete genomes of A. thaliana, O. sativa, and P. trichocarpa have identified 19 sHSP genes in A. thaliana (this work and Scharf et al. 2001), 36 sHSP genes in P. trichocarpa, and 23 sHSP genes in O. sativa. Each sHSP gene identified is listed along with chromosomal location and cellular location in Tables 1, 2, and 3. One of the most notable findings is the large expansion of the cytosolic I sHSPs in the P. trichocarpa genomes with 18 genes compared to 6 in A. thaliana and 7 in O. sativa.
Table 1.
A. thaliana small heat shock proteins
| Protein name | Subfamily | Gene ID | Orientation and chromosome location | Intron positiona | Cellular location | |
|---|---|---|---|---|---|---|
| 1 | At17.4 I | Cytosolic I | At3g46230 | 3 (−) 16994886–16995826 | None | Cytosol |
| 2 | At17.6A I | Cytosolic I | At1g59860 | 1 (+) 22035078–22035796 | None | Cytosol |
| 3 | At17.6B I | Cytosolic I | At2g29500 | 2 (−) 12640180–12640882 | None | Cytosol |
| 4 | At17.6C I | Cytosolic I | At1g53540 | 1 (+) 20045992–20046637 | None | Cytosol |
| 5 | At17.8 I | Cytosolic I | At1g07400 | 1 (+) 227490–2275755 | None | Cytosol |
| 6 | At18.1 I | Cytosolic I | At5g59720 | 5 (+) 24079803–24080499 | None | Cytosol |
| 7 | At17.6 II | Cytosolic II | At5g12020 | 5 (−) 3882238–3882939 | None | Cytosol |
| 8 | At17.7 II | Cytosolic II | At5g12030 | 5 (−) 3884108–3884738 | None | Cytosol |
| 9 | At17.4 III | Cytosolic III | At1g54050 | 1 (−) 20244952–20245809 | F | Cytosol |
| 10 | At15.4 IV | Cytosolic IV | At4g21870 | 4 (−) 11603582–11604343 | E | Cytosol |
| 11 | At21.7 V | Cytosolic V | At5g54660 | 5 (+) 22221131–22222378 | E | Cytosol |
| 12 | At18.5VI | Cytosolic VI | At2g19310 | 2 (−) 8376876–8377488 | None | Cytosol |
| 13 | At15.7 PX | Peroxisome | At5g37670 | 5 (+) 14986221–14986739 | None | Peroxisome |
| 14 | At22.0 ER | ER | At4g10250 | 4 (+) 6370339–6371286 | None | ER |
| 15 | At21 CP | CP | At4g27670 | 4 (−) 13818876–13819977 | B | Plastid |
| 16 | At23.5 MT | MT | AT5g1440 | 5 (+) 20908389–20909437 | B | Mitochondria |
| 17 | At23.6 MT | MT | At4g25200 | 4 (+) 12917027–12918063 | B | Mitochondria |
| 18 | At26.5 | MTII | At1g52560 | 1(−) 19578429–19579435 | C | Mitochondria |
| 19 | At14.2 | NAb | At5g47600 | 5 (−) 19317008–19317703 | D | Cytosol |
aIntron position in based on the complete amino acid alignment of sHSPs found in Supplementary Fig. 1.
bNA indicates that the protein is not a member of any of the subfamilies and is currently considered an “orphan” sHSP.
Table 2.
P. trichocarpa small heat shock proteins
| Protein name | Subfamily | Gene ID | Chromosome location and orientation | Intron position a | Cellular location | |
|---|---|---|---|---|---|---|
| 1 | Pt17.7I | Cytosolic I | 203787 | IX(+): 8809749–8810195 | None | Cytosol |
| 2 | Pt17.6AI | Cytosolic I | 172186 | I(+): 16439861–16440320 | None | Cytosol |
| 3 | Pt17.5AI | Cytosolic I | 738820 | XIX(−): 9208574–9209317 | None | Cytosol |
| 4 | Pt17.8AI | Cytosolic I | 722968 | IX(+): 7423320–7424035 | None | Cytosol |
| 5 | Pt18.0I | Cytosolic I | 723183 | IX(+): 8168808–8169592 | None | Cytosol |
| 6 | Pt18.2AI | Cytosolic I | 679803 | 66(−): 191732–192600 | None | Cytosol |
| 7 | Pt17.5BI | Cytosolic I | 574257 | XIX(−): 9212097–9212555 | None | Cytosol |
| 8 | Pt17.4AI | Cytosolic I | 283987 | 2959(−): 6313–6762 | None | Cytosol |
| 9 | Pt18.3BI | Cytosolic I | 679801 | 66(−): 189735–190318 | None | Cytosol |
| 10 | Pt15.9I | Cytosolic I | 653054 | VI(+): 6360998–6361691 | None | Cytosol |
| 11 | Pt18.3AI | Cytosolic I | 579131 | 10195(+): 23–1502 | None | Cytosol |
| 12 | Pt18.5I | Cytosolic I | 659595 | X(+): 17309065–17309856 | None | Cytosol |
| 13 | Pt18.1I | Cytosolic I | 721475 | IX(+): 1290938–1293842 | G | Cytosol |
| 14 | Pt18.3CI | Cytosolic I | 836664 | 66(−): 186063–186792 | None | Cytosol |
| 15 | Pt17.4BI | Cytosolic I | 762435 | VI(−): 6358691–6359152 | None | Cytosol |
| 16 | Pt18.3DI | Cytosolic I | 563962 | VIII(−): 3534621–3535497 | None | Cytosol |
| 17 | Pt17.8BI | Cytosolic I | 549183 | I (+): 17839524–17840222 | None | Cytosol |
| 18 | Pt17.6BI | Cytosolic I | 650093 | IX(+): 7419936–7420544 | None | Cytosol |
| 19 | Pt17.5II | Cytosolic II | 832078 | VI(+): 14964013–14965409 | None | Cytosol |
| 20 | Pt17.5III | Cytosolic III | 712318 | III(+): 6588574–6589363 | F | Cytosol |
| 21 | Pt15.6IV | Cytosolic IV | 233215 | XI(−): 57306–57814 | E | Cytosol |
| 22 | Pt20.8V | Cytosolic V | 232356 | XI(+): 12969102–12969847 | E | Cytosol |
| 23 | Pt24.9V | Cytosolic V | 242489 | XIII(+): 7472125–7472978 | E | Cytosol |
| 24 | Pt16.9VI | Cytosolic VI | 826045 | XVIII(+):13045999–13046835 | None | Cytosol |
| 25 | Pt21.8ER | ER | 823806 | XIII(−):10336249–10337561 | None | ER |
| 26 | Pt15.9PX | PX | 830535 | II(+): 21093095–21094007 | E | Peroxisome |
| 27 | Pt23.1CP | CP | 574673 | XV(−): 393950–394768 | A, B | CP |
| 28 | Pt24.0CP | CP | 422073 | XII(−): 2456874–2457728 | B | CP |
| 29 | Pt24.0MT | MT | 799700 | III(−): 7110095–7111157 | B | MT |
| 30 | Pt23.9MT | MT | 817274 | III(+): 10505979–10507046 | None | Possible MT |
| 31 | Pt21.1 | MT II | 171902 | I (−): 1467389–14768209 | C | MT |
| 32 | Pt20.3 | NAb | 204055 | IX(−): 9528840–9529283 | None | Vacuole, SP |
| 33 | Pt16.2 | NAb | 459610 | I(+): 22799013–22799695 | None | Cytosol |
| 34 | Pt17.5 | NAb | 659379 | X(−): 5958515–15959331 | None | Cytosol |
| 35 | Pt20.4 | NAb | 560348 | VI(−): 2087984– 2088611 | B | CP/ MT |
| 36 | Pt22.1 | NAb | 769024 | X(+): 6355598–6356274 | B | CP/MT |
aIntron position in based on the complete amino acid alignment of sHSPs found in Supplementary Fig. 1.
bNA indicates that the protein is not a member of any of the subfamilies and is currently considered an “orphan” sHSP.
Table 3.
O. sativa small heat shock proteins
| Protein name | Subfamily | Gene ID | Chromosome location and orientation | Intron position a | Cellular location | |
|---|---|---|---|---|---|---|
| 1 | Os16.9 I | Cytosolic I | Os01g0136100 | 1 (−) 1948574–1947770 | None | Cytosol |
| 2 | Os16.91 I | Cytosolic 1 | Os01g0136000 | 1 (−) 1943922–1943473 | None | Cytosol |
| 3 | Os16.92 I | Cytosolic 1 | Os01g0136200 | 1 (+) 1950930–1951681 | None | Cytosol |
| 4 | Os16.93I | Cytosolic I | Os01g0135900 | 1 (+) 1940012–1940845 | None | Cytosol |
| 5 | Os17.4A I | Cytosolic 1 | Os03g0266900 | 3 (−) 8804578–8803829 | None | Cytosol |
| 6 | Os17.4B I | Cytosolic I | Os03g0267200 | 3 (−) 8808635–8807929 | None | Cytosol |
| 7 | Os17.4C I | Cytosolic 1 | Os03g0267000 | 3 (+) 8804977–8805684 | None | Cytosol |
| 8 | Os17.4D I | Cytosolic 1 | Os03g0266300 | 3 (+) 8775788–8776634 | None | Cytosol |
| 9 | Os17.6C II | Cytosolic II | Os02g0217900 | 2 (−) 6587165–6586638 | None | Cytosol |
| 10 | Os17.8II | Cytosolic II | Os01g0184100 | 1 (−) 4448876–4448070 | None | Cytosol |
| 11 | Os17.6B III | Cytosolic III | Os02g0782500 | 2 (+) 33197982–33198898 | F | Cytosol |
| 12 | Os18.8 IV | Cytosolic IV | Os07g0517100 | 7 (+) 19881971–19882841 | E | Cytosol |
| 13 | Os22.2 V | Cytosolic V | Os05g0500500 | 5 (+) 24482031–24483911 | E | Cytosol |
| 14 | Os17.6 PX | Peroxisome | Os06g0253100 | 6 (−) 7952497–7951773 | None | Peroxisome |
| 15 | Os22.3 ER | ER | Os04g0445100 | 4 (+) 22176650–22177580 | None | ER |
| 16 | Os26 CP | CP | Os03g0245800 | 3 (+) 7664447–7665459 | None | CP |
| 17 | Os22 MT | MT | Os02g0758000 | 2 (+) 31939347–31940561 | B | MT |
| 18 | Os22.4 MT | MT | Os06g0219500 | 6 (−) 6164038–6163155 | B | MT |
| 19 | Os16.9C | NAb | Os02g0711300 | 2 (+) 29486906–29487684 | None | Cytosol |
| 20 | Os17.6A | NAb | Os01g0135800 | 1 (+) 1933117–1933924 | None | Cytosol |
| 21 | Os18 | NAb | Os11g0244200 | 11 (−) 767–365–7669622 | None | ER |
| 22 | Os18.2 | NAb | Os02g128000 | 2 (−) 1450233–1450766 | None | Cytosol |
| 23 | Os21.2 | NAb | Os02g0107100 | 2 (+) 5640360–5639315 | None | MT |
aIntron position in based on the complete amino acid alignment of sHSPs found in Supplementary Fig. 1.
bNA indicates that the protein is not a member of any of the subfamilies and is currently considered an “orphan” sHSP.
The sHSP genes in A. thaliana are found dispersed on all five chromosomes (Table 1). Chromosome 3 has only one sHSP gene: At17.4C I. Chromosome 2 has two (At17.6BI and At18.5). Chromosomes 1 and 4 both have four genes each (see Table 1), and chromosome 5 has seven sHSP genes. Only one pair of genes (At17.6II and At17.7II) is found in a tandem repeat. There are three pairs of A. thaliana sHSP genes located in regions previously identified as being the product of segmental duplications: (1) At23.5 M and At 23.6M, (2) At17.4I and At18.1I, and (3) At17.8I and 17.6BI.
In P. trichocarpa, the sHSP genes are found on at least 8 of the 19 chromosomes (Table 2). At this time, there are some sequencing scaffolds that have not yet been mapped to chromosomes. There are six sHSP genes on scaffold 66. Chromosome IX has seven sHSP genes, but of these, only two (Pt17.6BI and Pt17.8AI) are located near each other. Only one pair of sHSP genes is found in a tandem duplication: Pt17.5AI and Pt17.5BI on chromosome 9. The other cytosolic I genes are found dispersed on five chromosomes and one scaffold. There are four cytosolic I genes on chromosome IX, but only two are located near each other (Pt17.6BI and Pt17.8AI); they are separated by 3 kb. In addition, there are three pairs of genes found in duplicated regions within the P. trichocarpa genome: (1) Pt17.6AI and Pt17.4AI, (2) Pt18.5AI and Pt18.3AI, and (3) Pt20.8V and 24.9V.
The sHSP genes in O. sativa were found on 9 of the 12 chromosomes (Table 3). There are groups of sHSP genes on both chromosomes 1 (six sHSP genes) and 3 (five sHSP genes). Chromosome 2 also has six sHSP genes but none of these are located near each other. Only one pair of O. sativa sHSPs was found in segmentally duplicated regions Os22 M and Os22.4 M.
Phylogenetic analysis identified new sHSP subfamilies
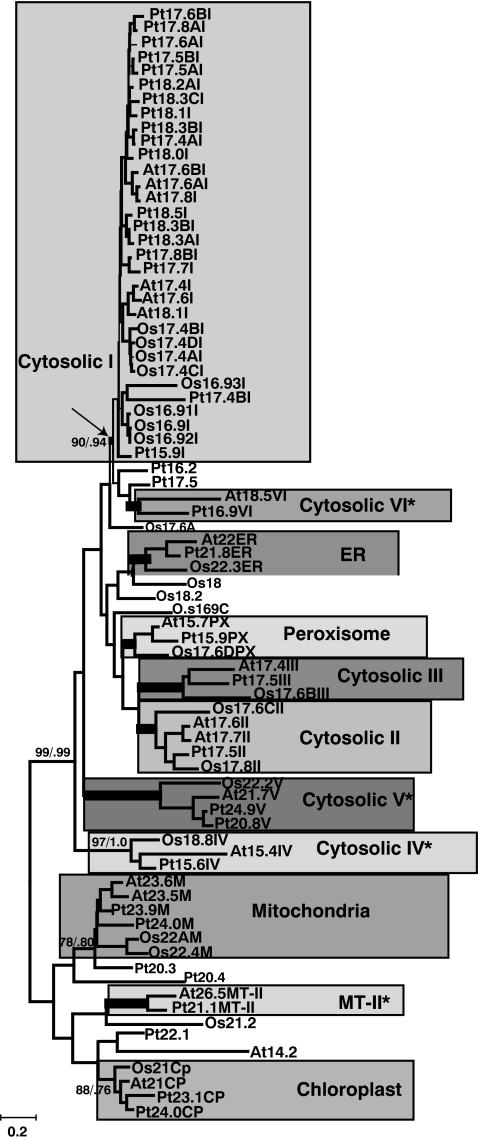
The previously identified sHSP subfamilies (cytosolic I, II, III, ER, PX, CP, and MT) can be easily identified in the phylogenetic tree of the sHSPs (Fig. 1). Each of these subfamilies contains A. thaliana, P. trichocarpa, and O. sativa sHSPs proteins indicating that the origin of these subfamilies at a minimum predates the divergence of the common ancestor of these three species. The individual subfamilies are well supported by bootstrap and posterior probabilities. However, there is little support for the branches that unite the different subfamilies, and thus the relationships of individual subfamilies to each other are not clear. However, in this analysis, as seen in previous analysis, the MT and CP subfamilies are closely related to each other. In addition, from the phylogenetic tree (Fig. 1), it is clear that the cytosolic II and III subfamilies are more closely related to each other than they are to the cytosolic I subfamily.
Fig. 1.
Phylogenetic tree of sHSP amino acid sequences. Only the residues and corresponding DNA sequence for the more conserved C-terminal domain (170–300 in Supplementary Fig. 1) were used to construct the phylogenetic tree and generate support values. Support for major branches is given in bootstrap values (based on 1,000 NJ bootstrap replicates) and Bayesian prior probabilities. The branches marked by the wide branches indicate the highest possible support of both 100% bootstrap values and 1.0 Bayesian prior probabilities. When provided, Bootstrap values are given first followed by prior probabilities. A different shaded background indicates each of the well-supported subfamilies. The subfamilies newly identified in this analysis are marked by an asterisk
The phylogenetic relationships within the cytosolic I subfamily deserves particular attention. The cytosolic I genes in both A. thaliana and O. sativa form two closely related groups (Fig. 1). In P. trichocarpa, there are three groups of cytosolic I genes. The phylogenetic groups in O. sativa reflect chromosomal location (all the Os16.9s are on chromosome 1, and the Os17.4s are on chromosome 3). In contrast, in both A. thaliana and P. trichocarpa, there is no correlation between chromosomal location and phylogenetic relationship among the cytosolic I genes.
In this analysis, we have identified four new plant sHSP subfamilies (cytosolic IV, V, VI, and MT-II). Two of these plant subfamilies (Cytosolic IV and V) have homologs in all three species examined. Cytosolic IV is composed of At15.4, Os18.8, and Pt15.6IV, and cytosolic V is composed of At21.7, Os22.2, Pt24.9V, and Pt20.8V. These proteins are predicted to localize to the cytosol and lack any recognizable organelle-targeting sequence. The two subfamilies have homologs in A. thaliana and P. trichocarpa but none in O. sativa (cytosolic VI and MT-II). Cytosolic VI includes At 18.5 and Pt16.9VI. The MT-II subfamily includes At26.5 and Pt21.1.
Analysis of sequence alignments of these new subfamilies reveals some interesting patterns of sequence conservation (Fig. 2). It is clear from the alignment of all the sHSPs that they do not share high levels amino acid identity within or between subfamilies (Supplementary Fig. 1). However, there are a number of secondary structural features that are conserved across subfamilies (Fig. 2). The structural features β3, β4, β5, β8, and β9 are the most highly conserved. It is important to note that both the cytosolic IV and VI families lack the β6 strand. The “GVL” sequence motif in β9 that is conserved across sHSPs from archaea to bacteria to eukaryotes is not present in any of the cytosolic V subfamily members and is also absent in At18.5VI (Fig. 2). However, the corresponding secondary structure β9 is conserved in these proteins. Finally, the cytosolic V family is missing the β10 strand (Fig. 2).
Fig. 2.
Amino acid alignment of newly identified sHSP subfamilies. The newly identified sHSP subfamilies are aligned to each other and to the cytosolic I protein from wheat whose crystal structure is known (van Montfort et al. 2001). The secondary structure for the A. thaliana, P. trichocarpa, and O. sativa proteins are based on structure predictions. The helical regions are denoted by italics and double underline. The β strands are denoted by single underlines. The Wheat 16.9 helical regions are represented by rectangles and the β strands by arrows. The structural numbering system is taken from van Montfort et al. (2001). The N-terminal region includes positions 1–139, and the C-terminal region containing the α-crystallin domain includes positions 140–279
Of particular interest are the “orphan” sHSPs because they may represent recent duplicates that are evolving new functions. We define an orphan protein as one that does not have a close homolog in the other genomes and does not belong to any subfamily. In the phylogenetic tree, some of these orphan genes appear to have close homologs; however, these relationships are not consistent across tree construction methods and are never highly supported. There is one orphan in A. thaliana: At14. This protein lacks an N-terminal domain. It does not have an organelle-targeting sequence and is predicted to localize to the cytosol. This is somewhat surprising because it appears to be closely related to the CP and MT sHSPs (Fig. 1). P. trichocarpa has four orphan sHSPs: Pt17.5, Pt22.1, Pt20.4, and Pt24.0MT. One of these sHSPs, Pt20.3, is predicted to be targeted to the vacuole. The predicted locations of two other P. trichocarpa orphan genes (Pt20.4 and Pt22.1) are equivocal with both CP and MT localizations possible. The remaining two P. trichocarpa proteins are clearly cytosolic proteins. There are five orphan sHSP genes in the O. sativa genome: Os16.9C, Os17.6A, Os18, Os18.2, and Os21.2. Three of these, Os16.9C, Os17.6A, and Os18.2, are most likely cytosolic proteins. One orphan gene, Os18, is predicted to be an ER protein, and this protein is closely related to the ER subfamily. Finally, Os21.2 is predicted to localize to the MT.
Intron position
Some but not all sHSP subfamilies contain introns. The ER, peroxisome, cytosolic I (except for one Populus cytosolic I gene) and II subfamilies all lack introns (Tables 1, 2, and 3). However, when present, many of the intron positions are shared by subfamilies or groups of subfamilies. The cytosolic IV and V subfamilies share a unique intron position, suggesting a close phylogenetic relationship. In addition, most of the CP and MT sHSPs share an intron. This is consistent with the close relationships seen in the tree (Fig. 1). However, the MT-II subfamily (At26.5 and Pt21.1) that is within the larger CP/MT family does not share this intron but instead has an intron in a different position. It is noteworthy that there are two CP sHSPs in the P. trichocarpa genome. Both posses the conserved B intron position, but Pt23.1 has another unique intron. Pt23.1 also lacks the highly conserved methionine-rich region found in all other known angiosperm sHSPs (Waters 2003). The cytosolic III subfamily has an intron that is not shared with any other sHSP gene. Two of the P. trichocarpa orphan sHSPs have introns. Both Pt20.4 and Pt22.1 share an intron position with the CP and MT subfamilies. Finally, the orphan At14.2 that has no close homolog in any other genome is a member of the larger CP/MT lineage and has an intron that is not shared with any of the other sHSPs. The presence of shared introns indicates close relationships among some subfamilies. However, intron position, presence, or absence does not shed light on the order of gene duplications that gave rise to this large family of proteins.
Gene conversion among sHSPs genes
Using the program GeneConv, we found no evidence of any statistically significant (at the 0.05 probability level) gene conversion events among the P. trichocarpa sHSP genes. In addition, no evidence for significant gene conversion was found between any of the A. thaliana sHSP genes, not even between the two cytosolic II genes (At17.6II and At17.7II) that are found in a tandem duplication. Upon close inspection of the DNA alignment, it was clear that the substitutions between At17.6II and At17.7II are evenly distributed across the genes (data not shown). The longest stretch of identical sequence was only 34 bp long. The nonsynonymous (Ka) substitution rate between these two genes is estimated to be 0.06 per site, and the Ks (or synonymous subs) is 0.452 per site. This high level of Ks and relatively low Ka were also found among the cytosolic I A. thaliana genes. This pattern of high Ks and lower Ka is also seen among the P. trichocarpa cytosolic I genes (Supplementary Table 1).
The chromosomal organization and rates of gene conversion are considerably different in O. sativa. Chromosomes 1 and 3 in O. sativa each have a number of sHSPs that are found in close proximity (Table 2). Evidence of gene conversion occurring among sHSP genes located on chromosomes 1 and chromosomes 3 is presented in Table 4. No other gene conversion events were detected. On chromosome 1 of O. sativa, there is a tandem array of five sHSP genes (Os17.6A, Os16.93I, Os16.91I, Os16.9I, and Os16.92I). Analysis of possible gene conversion events found statistically significant gene conversion tracts among three of the cytosolic I genes (Os16.91I, Os16.9I, and Os16.92I) but not between these genes and the other sHSP genes on this chromosome.
Table 4.
Gene conversion tracts in the O. sativa cytosolic I genes
| Chromosome | Sequences | Global p valuea | BC/KA P valueb | Lengthc | |
|---|---|---|---|---|---|
| 1 | Cr 1 | 16.92 vs. 16.9 | 0.0009 | 0.0135 | 311 |
| 2 | Cr 1 | 16.91 Ivs. 16.9 | 0.0021 | 0.0315 | 251 |
| 3 | Chr 3 | 17.4A vs. 17.4 C | 0.0001 | 0.0001 | 353 |
| 4 | Chr 3 | 17.4 D vs. 17.4 C | 0.0001 | 0.0001 | 391 |
| 5 | Chr 3 | 17.4B vs. 17.4 C | 0.0001 | 0.0001 | 353 |
| 6 | Chr 3 | 17.4D vs 17.4A | 0.0001 | 0.0001 | 241 |
| 7 | Chr 3 | 17.4D vs. 1.4 A | 0.0001 | 0.0001 | 99 |
| 8 | Chr 3 | 17.4 B VS. 17.4D | 0.0001 | 0.0001 | 289 |
| 9 | Chr 3 | 17.4 BVS. 17.4A | 0.0001 | 0.00045 | 203 |
| 10 | Chr 3 | 17.4AVS 17.4C | 0.001 | 0.00056 | 85 |
ap value obtained from 10,000 resampling replicates.
bBC/KA p values are based Bonferroni-corrected Karlin–Altshul estimates. Both Global and BC/KA values are corrected for multiple comparisons.
cLength of the predicted gene conversion tract
Five O. sativa sHSP genes are found on chromosome 3. Four of these are closely related cytosolic I genes (17.4A, B, C, and D). Os26CP is also located on this chromosome, but it is at least 1 MB from the 17.4 cytosolic I genes. Despite the distance and presence of other genes between the cytosolic I genes, there is considerable evidence of gene conversion among all the cytosolic I Os17.4 genes (Table 4). Many of the predicted gene conversion tracts are at least 200 bp in length. It is important to note that it can be difficult to distinguish between recent duplications and continuing gene conversion. However, when the gene sequences for Os17.4 and Os17.4C are compared, it is clear that there are islands of complete sequence identity surrounded by regions of sequence divergence. This pattern is more consistent with gene conversion than it is with recent duplication or selection. Further, the presence of other genes in this region other than the cytosolic 17.4 genes argues against recent duplication. There was no evidence of any gene conversion across chromosomes or across subfamilies.
Gene expression of sHSP subfamilies in Arabidopsis thaliana
Based on the very large dataset of A. thaliana Affymetrix GeneChip experiments available at Genevestigator (Zimmermann et al. 2004), there is a clear sHSP stress-induced expression pattern (see Table 5). This pattern is shared by all the previously identified sHSP subfamilies: cytosolic I, II and II, PX, ER, CP, and MT. These sHSP genes are all induced during heat stress. In most cases, there is a more than 100-fold increase in expression during heat shock when compared to control levels. However, some sHSP genes have even higher heat stress expression levels. For example, the gene for the CP-targeted sHSP At21 has a 400-fold increase in expression during heat shock. In addition to being expressed during high temperature stress, many of the sHSP genes are also highly induced in response to hypoxia, anoxia, osmotic stress, salt, and wounding. Further, none of the sHSP genes are upregulated by cold, drought, or genotoxic conditions (Table 5).
Table 5.
A. thaliana Stress-Induced Expression Patterns
| Abiotic stress | Biotic stress | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| An | Ht | Hy | Os | Ox | Sa | Wo | H2O2 | Uv | Oz | A.b. | A.t. | B.c. | E.c. | M.p. | Nm | P.s. | |
| Gene | |||||||||||||||||
| 17.4 I | ++ | +++ | ++ | + | + | ++ | + | ++ | + | ++ | ++ | 0 | ++ | + | 0 | + | ++ |
| 17.6A I | + | ++ | ++ | ++ | + | + | ++ | + | ++ | + | 0 | + | + | 0 | + | ++ | |
| 17.6B I | + | +++ | ++ | + | + | + | + | ++ | + | ++ | ++ | 0 | ++ | 0 | 0 | + | ++ |
| 17.6C I | + | +++ | ++ | ++ | + | + | + | ++ | 0 | + | + | 0 | 0 | 0 | 0 | 0 | + |
| 18.1 I | + | +++ | ++ | ++ | + | + | + | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | + | 0 |
| 17.6 II | + | +++ | ++ | + | + | + | + | +++ | 0 | + | + | ++ | 0 | 0 | + | 0 | ++ |
| 17.7 II | + | +++ | ++ | ++ | + | ++ | + | + | 0 | ++ | + | 0 | ++ | 0 | 0 | + | ++ |
| 17.4 III | ++ | ++ | ++ | ++ | + | + | 0 | ++ | + | ++ | ++ | 0 | ++ | + | 0 | + | ++ |
| 15.4 IV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ++ | ++ | 0 |
| 21.7 V | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18.5VI | 0 | + | 0 | 0 | 0 | 0 | 0 | 0 | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15.7 PX | + | ++ | ++ | + | + | + | + | ++ | 0 | ++ | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 ER | 0 | +++ | ++ | ++ | ++ | ++ | + | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 CP | 0 | +++ | ++ | + | + | + | + | ++ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | + | 0 |
| 23.5 MT | 0 | ++ | + | + | + | + | 0 | ++ | + | + | + | 0 | ++ | 0 | 0 | + | 0 |
| 23.6 MT | 0 | +++ | ++ | + | + | ++ | + | + | + | ++ | ++ | 0 | + | 0 | 0 | ++ | + |
| 26.5 | + | ++ | ++ | + | + | ++ | + | + | + | 0 | 0 | 0 | + | 0 | 0 | 0 | 0 |
| 14.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
0 No measurable expression, + expression increase from two- to tenfold over control levels, ++ expression increase from 11- to 99-fold over control levels, +++ expression increase greater than 100-fold over control levels, An anoxia, Ht heat, Os osmotic, Ox oxidative, Sa salt, Wo wounding, Oz Ozone, A.b. Alternaria brassicola, A.t. Agrobacterium tumefaciens, B.c. Botrytis cinera, E.e. Erysiphe cichoracearum, M.p. Myzus persicae, Nm nematode, P.s. Pseudomonas syringae
There are considerable differences among the members of the conserved subfamilies in their developmental and tissue-specific expression levels. Some but not all sHSPs are expressed in dry seeds, in flowers, or flower parts (Table 6). The genes with the highest expression in seeds are At17.4I and At17.7II (Table 6). At17.7II also has high expression in petals (as does the orphan gene At18.5; Table 6).
Table 6.
A. thaliana expression Organ and Tissue Expression Patterns
| Cotyledons | Petal | Sepal | Stamen | Pollen | Silique | Seed | Stem | Leaf | Shoot apex | |
|---|---|---|---|---|---|---|---|---|---|---|
| 17.4 I | − | ++ | − | + | +++ | − | +++ | − | − | − |
| 17.6A I | − | +++ | + | + | + | − | − | − | − | − |
| 17.6B I | − | + | + | + | ++ | − | − | − | − | − |
| 17.6C I | − | − | − | − | − | − | − | − | − | − |
| 18.1 I | − | − | − | − | − | − | − | − | − | − |
| 17.6 II | − | ++ | + | + | ++ | − | − | − | − | − |
| 17.7 II | − | +++ | − | − | − | − | +++ | − | − | − |
| 17.4 III | − | − | − | − | − | − | − | − | − | − |
| 15.4 IV | +++ | ++ | + | + | + | + | ++ | + | +++ | + |
| 21.7 V | ++ | + | ++ | + | + | + | + | + | + | + |
| 18.5VI | + | +++ | ++ | ++ | − | − | ++ | + | + | |
| 15.7 PX | − | ++ | ++ | + | − | − | ++ | − | − | − |
| 22 ER | − | − | − | − | − | − | − | − | − | − |
| 21 CP | − | − | − | − | − | − | − | − | − | − |
| 23.5 MT | − | ++ | + | + | ++ | − | + | − | + | − |
| 23.6 MT | − | + | − | − | − | − | ++ | − | − | − |
| 26.5 | − | − | − | − | − | − | ++ | − | − | − |
| 14.2 | − | − | − | − | − | − | − | − | − | +++ |
The symbol − no detectable expression, + expression at least two times above background, ++ expression at least ten times above background, +++ expression at least 100 times over background
Newly identified sHSP subfamilies have distinct expression patterns
Notably, two of the newly identified plant subfamilies (Cytosolic IV and V) do not share the sHSP stress-induced expression pattern (Table 5). It is interesting that At21.7 (Cytosolic V) is constitutively expressed in all tissues examined and is not upregulated by heat or any other stress (Table 6). In fact, there were no conditions that dramatically increased the expression of this gene, but rather consistent levels were found in all tissues. This constitutive expression pattern is unusual for sHSPs. The A. thaliana member of the cytosolic IV subfamily, At15.4, is also expressed in many tissue types (Table 6). However, in contrast to At21.7, it is expressed in seeds and has very high expression levels in the cotyledon, petals, and adult leaves (Table 6). This gene is also not upregulated by any stress (Table 5). Our finding of seed expression based on the data available at the Genevestigator site is somewhat at odds with data presented in Kotak et al. (2007) that were based on data available at AtGenExpress. Kotak et al. (2007) reported that At15.4 is expressed only during early seed stages. The differences between our findings and that of Kotak et al. (2007) may reflect differences in the experimental data available at these two different sites, and differences in how seed stages are defined at these sites. Some sHSPs are induced by ABA exposure (Kotak et al. 2007) however in general we found little correlation between seed and ABA gene expression for the sHSPs (data not shown). For example, At18.5 is expressed in seeds but is not induced by ABA exposure.
Two other newly identified subfamilies, cytosolic VI and MT II, have very different expression patterns (Tables 5 and 6). At26.5, a member of the MT-II subfamily, has a pattern of stress-induced expression that is nearly identical to that of the previously identified sHSP subfamilies. However, the cytosolic VI subfamily represented by At18.5 has only a very mild increase in expression during heat stress (7X). In fact, there are very few conditions or tissues that have high levels of At18.5 expression. It is notable that At18.5VI is also expressed in petals, dry seeds, and in sepals. The only A. thaliana orphan gene, At14.2, also has very unusual expression patterns (Tables 5 and 6). It is highly expressed in the shoot apex and has very low or no expression in most other growth stages and in stress treatments. The only other growth stage where there is some measurable expression is in the inflorescence.
Expression patterns of segmental or tandem duplications
In Arabidopsis, there are no differences in the responses of the sHSP genes from segmental duplications to abiotic stress, and there are only minor differences in their developmental regulation (Tables 5 and 6). Previous analysis of the rice cytosolic I genes indicates that there are some differences in gene expression patterns (Guan et al. 2004). However, comparisons with the A. thaliana sHSP genes are limited because there is considerably less public microarray data for O. sativa and P. trichocarpa. The O. sativa gene expression data are also limited because of the small number of genes on the chip used in these experiments. The O. sativa array has only 9,000 genes, considerably less than the more than 20,000 genes represented on the A. thaliana chip. In fact, only five sHSP genes are on the Rice chip, and analysis of their gene expression patterns was conducted. Most have expression patterns consistent with their homologs in A. thaliana. While more P. trichocarpa sHSP genes are present on the poplar array, very few stress experiments are available in the public databases. Therefore, meaningful comparisons with the A. thaliana expression patterns are not possible now.
Discussion
Structural and functional features of newly identified sHSP subfamilies
A large number of studies have established that sHSPs form large oligomers and act as molecular chaperones (Narberhaus 2002; van Montfort et al. 2002; Nakamoto and Vigh 2007). Recent studies on the roles of particular residues and sHSP regions in the ability to both form large oligomers and interact with substrate proteins have established important structure–function relationships. It is now known that a lack of the N-terminal domain can prevent oligomerization and can eliminate molecular chaperone function (van Montfort et al. 2002; Haslbeck et al. 2004; Giese et al. 2005; Basha et al. 2006). Further, the β6 strand in the C-terminal domain has been associated with the ability to form oligomers (van Montfort et al. 2002). In addition, the usually highly conserved “GVL” residues in the β9 strand have been associated with chaperone function (van Montfort et al. 2002; Nakamoto and Vigh 2007). Given this information and the variation seen in the newly identified sHSP subfamilies and some of the orphan sHSPs, we can conclude that not all members of the plant sHSP family can form large oligomers or act as chaperones.
Both the cytosolic IV and VI subfamilies lack the β6 strand important in oligomerization. This indicates that these proteins most likely do not share the conserved sHSP oligomer structure and may not share chaperone function. It is important to note that both the A. thaliana and P. trichocarpa members of the cytosolic IV subfamily lack the β6 strand but the rice protein Os18.8IV retains a predicted strand in this region. In this analysis, homologs of the cytosolic VI subfamily were only found in A. thaliana and P. trichocarpa. However, there is important variation between these homologs. At18.5VI lacks the important “GVL” residues. However, the corresponding predicted β9 strand is still present. Pt16.9VI retains both the “GVL” residues and the predicted β9 strand. Thus, there may be a variation in structure and function within these subfamilies. It is important to note that within Arabidopsis, members of both the IV and VI families (At15.4IV and At18.5VI) lack any stress-induced gene expression. They also have overlapping but distinct developmental gene expression patterns. These subfamilies do not appear to be evolutionary related to each other as they do not share an intron position and are not closely related to each other in the plant sHSP phylogenetic tree.
The cytosolic V subfamily also lacks the “GVL” residues, but it is predicted to retain the β9 strand. In addition, this subfamily has retained the β6 strand known to be important in oligomerization but lacks the β10 strand. Some of the residues in the β10 strand are known to be involved in the intermolecular contacts needed for oligomer function (Sun and MacRae 2005), and so it is possible that the cytosolic V family may not form oligomers. Intron position would suggest a close relationship between cytosolic IV and V subfamilies. However, there is considerable sequence variation between these two subfamilies. In A. thaliana, both of these subfamilies lack any stress-induced expression, but the expression of At15.4IV varies across organs and tissues, while At21.7V is constitutively expressed. Taken together, this evidence clearly indicates that these subfamilies have divergent structures and functions and that further studies of these proteins are needed to understand their roles in plants. However, not all the newly identified sHSP subfamilies have diverged significantly from the other subfamilies. The MT-II subfamily that lacks a rice homolog retains all conserved secondary features, and the A. thaliana member, At26.5, has a stress-induced expression pattern.
It is very interesting to note that the A. thaliana orphan gene At14.2 lacks the N-terminal domain. Studies of C. elegans proteins, which also lack the N-terminal domain, show that these sHSPs do not form large oligomers and are not chaperones (Sun and MacRae 2005; Nakamoto and Vigh 2007). There is no evidence that the plant and C. elegans sHSPs are closely related (Waters and Vierling 1999a; Waters and Rioflorido 2007), and thus this lack of an N-terminal domain appears to be a convergent feature. A recent study in Synechocystis found that the N-terminal domain of sHSPs is essential for the ability to confer thermotolerance (Giese et al. 2005). At14.2 is not heat regulated but is highly expressed in the shoot apex. Taken together, all of this evidence strongly suggests that At14.2 is not a molecular chaperone. The evolutionary origin of this protein is also intriguing; At14.2 is a member of the larger CP-MT family of proteins and is most likely derived from a MT or CP localized protein, but its lack of a targeting sequence would indicate that it is a cytosolic protein. None of the sHSP orphan genes in P. trichocarpa and O. sativa have such extreme sequence variation, and the lack of detailed expression data for these proteins limits our understanding of their function in plants. However, identification of these proteins is important because they may represent newly arisen proteins that are evolving new functional roles and expression patterns. Thus, further study of their evolution and function may help us understand how proteins evolve new functions.
Plant sHSP family evolution
Previous research has shown that at least some of the sHSP subfamilies have long evolutionary histories (Waters and Vierling 1999a,b; Waters and Rioflorido 2007). The presence of the cytosolic I, II, and CP subfamilies in mosses indicate that these subfamilies originated at least 450 million years ago. The research presented here indicates that despite the long evolutionary history in plants of diverse sHSP subfamilies, new sHSP subfamilies and new sHSPs are continuing to arise and evolve new functions. In our analysis, we identified a major division between the CP/MT lineages of sHSPs and all the other sHSPs including all six cytosolic subfamilies, as well as the ER and peroxisome subfamilies. From this, we can conclude that the ER and peroxisome sHSPs evolved from the cytosolic sHSP homologs. However, there is little to no support for the branches uniting the different subfamilies, and at this time, it is not clear how they are related to each other. Based on intron position, we can conclude that the CP/MT proteins are all related and the cytosolic V and IV subfamilies are related. A better understanding of the origin of the 11 sHSP subfamilies will require considerable additional genome data from other angiosperm species as well as from plants representing seed and vascular plant lineages. The timing and mechanism of origin of the orphan sHSPs will require studies of homologs in close relatives of the species studied here. It is likely that analysis of complete genomes of other monocots may reduce the number of O. sativa orphan genes and may identify monocot-specific sHSP subfamilies.
It is then clear that the sHSPs are a dynamic protein family in plants and that comparisons of the evolutionary dynamics of the sHSPs in the three different angiosperm genomes may provide insights into gene family evolution. There are a number of models for gene family evolution; for a review, see Taylor and Raes (2004). These models differ in the importance of positive selection to change amino acid sequences (neofunctionalization) and gene expression changes (subfunctionalization) in the generation of gene family diversity (Lynch and Force 2000; Taylor and Raes 2004; Force et al. 2005; Hughes 2005). In addition, considerable attention has been paid to the importance of genome duplication (polyploidy and chromosomal duplication) as a driving force in plant gene family evolution (Zhang et al. 2002; Blanc and Wolfe 2004; Harberer et al. 2004; Casneuf et al. 2006; Duarte et al. 2006). Because our study examined one gene family in three different complete genomes, it provides an excellent opportunity to consider if evidence exists to support any one of these models in all or some of these plant genomes.
We found no evidence for recent positive selection and abundant evidence of purifying selection among the sHSPs in all three genomes. However, the signal of positive selection might be difficult to detect if the period of positive selection was short-lived and was then followed by strong purifying selection. However, evidence of positive selection in the Dof (DNA-binding proteins) family of proteins among the same species has been reported (Yang et al. 2006) suggesting that if there had been extensive positive selection among the sHSPs within these species, we should have detected it.
We have also found little evidence for a dominant role for segmental duplication in generating sHSP diversity with few sHSPs found in regions generated by genome duplication. This is in contrast to the important role of segmental duplication in the evolution of other protein families in the same species including the expansin family of proteins (Sampedro et al. 2006), the receptor-like kinases (Shiu et al. 2004), and the Aux/IAA, ARF genes (Remington et al. 2004).
It is also significant that we found no consistent evolutionary patterns among the sHSPs across genomes. For example, in each genome, the cytosolic I sHSPs are the most numerous, but the evolutionary forces that dominate this particular subfamily varies across genomes. The six A. thaliana cytosolic I sHSPs are found dispersed across the genome, and sequence similarity among these genes is maintained by strong purifying selection. In rice, the cytosolic I genes are found linked on two different chromosomes, and it is gene conversion that maintains sequence similarity. In P. trichocarpa, there has been an expansion of the cytosolic I genes, and there is also no evidence of gene conversion. As with A. thaliana, high synonymous and low nonsynonymous values suggest the action of purifying selection on the P. trichocarpa sHSPs.
The expansion of the cytosolic I genes in P. trichocarpa is interesting. It will be important to examine other angiosperm genomes as they become available to determine if the expansion of cytosolic I genes in P. trichocarpa is shared with related species or if it is species specific. It is unlikely that the expansion of the P. trichocarpa cytosolic I genes is due to genome-wide duplication events. First, there are only two pairs of P. trichocarpa cytosolic I genes present in duplicated regions of the genome, the same number seen in A. thaliana. In addition, a genome-wide event would have increased copy number of all the subfamilies equally. In order for the 18 cytosolic I genes to be due to past polyploidy events, we would have to propose preferential maintenance of only this subfamily. While it has been reported that cytosolic proteins are differentially maintained and that organelle-localized proteins are differentially lost after polyploidy (Blanc and Wolfe 2004), the other cytosolic subfamilies (II, III, IV, and V) do not show any Populus-specific expansions. Most likely, the expansion of the cytosolic I sHSPs in P. trichocarpa was driven by local events or a number of single duplication events followed by strong selection to maintain the duplicate cytosolic I copies. It is known that gene families associated with cell wall synthesis, meristem development, disease resistance, and metabolite transport are over-represented in P. trichocarpa (Tuskan et al. 2006). It will be important to determine if the cytosolic I genes are preferentially expressed in these tissues and/or under these conditions. Considerable future work on the expression and function of the cytosolic I genes in P. trichocarpa will be needed to understand the forces that generated and maintained the numerous cytosolic I genes in this genome.
Conclusions
Analysis of the sHSPs in three complete angiosperm genomes has identified four new plant sHSP subfamilies bringing the total number of sHSP subfamilies to 11. In addition, a number of orphan sHSP genes were also identified. These newly identified subfamilies and the orphan genes expand the previously known ranges for variation in plant sHSP sequence, structure, and expression. Evaluation of this data in light of past structural and functional studies of sHSPs indicates that there is also considerable functional variation among members of the plant sHSP family. Thus, these proteins are all related and share the α-crystallin domain but may vary in function. Members of the new subfamilies and the orphan genes should be the subjects of future detailed biochemical studies to determine exactly how their functions differ from established sHSP chaperone function. The evolutionary patterns of the sHSPs demonstrate that this is a dynamic family within flowering plants that continues to evolve and establish new members. For example, within P. trichocarpa, we see a selective expansion of the only the cytosolic I sHSP subfamily. It is interesting to note that we did not find a single pattern of sHSP evolution across all three genomes. We found that gene conversion among cytosolic I sHSPs is present in rice but absent in the other two genomes. In addition, we found no evidence for a significant role for positive selection or segmental evolution in generating the diverse sHSP subfamilies. In the future, examination of the sHSPs in additional angiosperm genomes, as they become available, and surveys of variation of sHSPs in species closely related to the species examined here should help provide a finer level of understanding of the evolution of this important and diverse protein family.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Alignment of sHSP amino acid sequences (DOC 67.5 KB)
A. thaliana cytosolic I DNA alignment (DOC 67.0 KB)
Synonymous (Ks) and Nonsynonymous substitutions for the Cytosolic I genes (DOC 118 KB)
Acknowledgements
We thank two anonymous reviewers for their careful reading of and helpful comments on an earlier version of this manuscript. This work was partially supported by award IBN:0313900 to ERW from the National Science Foundation. Z. Sanders-Reed was partially supported by the HCOP program at SDSU.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-008-0023-7) contains supplementary material, which is available to authorized users.
References
- Basha E, Friedrich KL, Vierling E (2006) The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J Biol Chem 281:39943–39952 [DOI] [PubMed]
- Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13:137–144 [DOI] [PMC free article] [PubMed]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691 [DOI] [PMC free article] [PubMed]
- Boorstein WR, Ziegelhoffer T, Craig EA (1994) Molecular evolution of the HSP70 multigene family. J Mol Evol 38:1–17 [DOI] [PubMed]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y (2006) Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol 7:R13 [DOI] [PMC free article] [PubMed]
- Caspers GJ, Leunissen JA, de Jong WW (1995) The expanding small heat-shock protein family, and structure predictions of the conserved “alpha-crystallin domain”. J Mol Evol 40:238–248 [DOI] [PubMed]
- de Jong WW, Caspers GJ, Leunissen JA (1998) Genealogy of the alpha-crystallin-small heat-shock protein superfamily. Int J Biol Macromol 22:151–162 [DOI] [PubMed]
- Drouin G (2002) Characterization of the gene conversion between the multigene family members of the yeast genome. J Mol Evol 55:14–23 [DOI] [PubMed]
- Duarte JM, Cui L, Wall PK et al (2006) Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23:469–478 [DOI] [PubMed]
- Force A, Cresko WA, Pickett FB, Proulx SR, Amemiya C, Lynch M (2005) The origin of subfunctions and modular gene regulation. Genetics 170:433–446 [DOI] [PMC free article] [PubMed]
- Franck E, Madsen O, van Rheede T, Ricard G, Huynen MA, de Jong WW (2004) Evolutionary diversity of vertebrate small heat shock proteins. J Mol Evol 59:792–805 [DOI] [PubMed]
- Fu X, Jiao W, Chang Z (2006) Phylogenetic and biochemical studies reveal a potential evolutionary origin of small heat shock proteins of animals from bacterial class A. J Mol Evol 62:257–266 [DOI] [PubMed]
- Giese KC, Basha E, Catague BY, Vierling E (2005) Evidence for an essential function of the N terminus of a small heat shock protein in vivo, independent of in vitro chaperone activity. Proc Natl Acad Sci USA 102:18896–18901 [DOI] [PMC free article] [PubMed]
- Gil R, Sabater-Munoz B, Latorre A, Silva FJ, Moya A (2002) Extreme genome reduction in Buchnera spp.: toward the minimal genome needed for symbiotic life. Proc Natl Acad Sci USA 99:4454–4458 [DOI] [PMC free article] [PubMed]
- Gil R, Silva FJ, Zientz E et al (2003) The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc Natl Acad Sci USA 100:9388–9393 [DOI] [PMC free article] [PubMed]
- Goff SA, Ricke D, Lan TH et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100 [DOI] [PubMed]
- Guan J, Jinn T, Yeh C, Feng S, Chen Y, Lin C (2004) Characterization of the genomic structures and selective expression profiles of nine class I small heat shock genes clustered on two chromosomes in rice (Oryza sativa L.). Plant Mol Biol 56:795–809 [DOI] [PubMed]
- Gupta RS (1995) Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationships among animals, plants and fungi species. Mol Biol Evol 12:1063–1073 [DOI] [PubMed]
- Harberer G, Hindemitt T, Meyers BC, Mayer K (2004) Transcriptional similarities: dissimilarities and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136:3009–3022 [DOI] [PMC free article] [PubMed]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J (2005) Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol 12:842–846 [DOI] [PubMed]
- Haslbeck M, Ignatiou A, Saibil H, Helmich S, Frenzl E, Stromer T, Buchner J (2004) A domain in the N-terminal part of Hsp26 is essential for chaperone function and oligomerization. J Mol Biol 343:445–455 [DOI] [PubMed]
- Hughes A (2005) Gene duplication and the origin of novel proteins. Proc Natl Acad Sci USA 102:8791–8792 [DOI] [PMC free article] [PubMed]
- Kim R, Kim KK, Yokota H, Kim S-H (1998) Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc Natl Acad Sci USA 95:9129–9133 [DOI] [PMC free article] [PubMed]
- Kotak S, Vierling E, Baumlein H, von Koskull-Doring P (2007) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19:182–195 [DOI] [PMC free article] [PubMed]
- Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163 [DOI] [PubMed]
- Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122:189–198 [DOI] [PMC free article] [PubMed]
- Lee GJ, Pokala N, Vierling E (1995) Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem 270:10432–10438 [DOI] [PubMed]
- Lee GJ, Roseman AM, Saibil HR, Vierling E (1997) A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J 16:659–671 [DOI] [PMC free article] [PubMed]
- Lenne C, Douce R (1994) A low molecular mass heat-shock protein is localized to higher plant mitochondria. Plant Physiol 105:1255–1261 [DOI] [PMC free article] [PubMed]
- Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154:459–473 [DOI] [PMC free article] [PubMed]
- Ma C, Haslbeck M, Babujee L, Jahn O, Reumann S (2006) Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol 141:47–60 [DOI] [PMC free article] [PubMed]
- Mondragon-Palomino M, Gaut BS (2005) Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol Biol Evol 22:2444–2456 [DOI] [PubMed]
- Nakamoto H, Vigh L (2007) The small heat shock proteins and their clients. Cell Mol Life Sci 64:294–306 [DOI] [PMC free article] [PubMed]
- Narberhaus F (2002) Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev 66:64–93 [DOI] [PMC free article] [PubMed]
- Posada D, Buckley T (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808 [DOI] [PubMed]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135:1738–1752 [DOI] [PMC free article] [PubMed]
- Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed]
- Ronquist F, Larget B, Huelsenbeck JP, Kadane JB, Simon D, van der Mark P (2006) Comment on “Phylogenetic MCMC algorithms are misleading on mixtures of trees”. Science 312:367 (author reply 367) [DOI] [PubMed]
- Sampedro J, Carey RE, Cosgrove DJ (2006) Genome histories clarify evolution of the expansin superfamily: new insights from the poplar genome and pine ESTs. J Plant Res 119:11–21 [DOI] [PubMed]
- Sawyer SA (1999) GENECONV: a computer package for the statistical detection of gene conversion. Distributed by the author. Department of Mathematics, Washington University, St. Louis
- Scharf KD, Siddique M, Vierling E (2001) The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones 6:225–237 [DOI] [PMC free article] [PubMed]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16:1220–1234 [DOI] [PMC free article] [PubMed]
- Siddique M, Port M, Tripp J, Weber C, Zielinski D, Calligaris R, Winkelhaus S, Scharf KD (2003) Tomato heat stress protein Hsp16.1-CIII represents a member of a new class of nucleocytoplasmic small heat stress proteins in plants. Cell Stress Chaperones 8:381–394 [DOI] [PMC free article] [PubMed]
- Stechmann A, Cavalier-Smith T (2003) Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J Mol Evol 57:408–419 [DOI] [PubMed]
- Sun Y, MacRae TH (2005) Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci 62:2460–2476 [DOI] [PMC free article] [PubMed]
- Taylor JS, Raes J (2004) Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38:615–643 [DOI] [PubMed]
- Tuskan GA, Difazio S, Jansson S et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604 [DOI] [PubMed]
- van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E (2001) Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol 8:1025–1030 [DOI] [PubMed]
- van Montfort RL, Slingsby C, Vierling E (2002) Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Adv Protein Chem 59:105–156 [DOI] [PubMed]
- Vierling E (1991) The heat shock response in plants. Annu Rev Plant Physiol Plant Mol Biol 42:579–620 [DOI]
- Vierling E, Nagao RT, DeRocher AE, Harris LM (1988) A heat shock protein localized to chloroplasts is a member of a eukaryotic superfamily of heat shock proteins. EMBO J 7:575–581 [DOI] [PMC free article] [PubMed]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290:2114–2117 [DOI] [PubMed]
- Wang W, Vinocur B, Shoseyov O, Altman A (2004) Roles of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252 [DOI] [PubMed]
- Wang X, Shi X, Li Z, Zhu Q, Kong L, Tang W, Ge S, Luo J (2006) Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Bioinformatics 7:447 [DOI] [PMC free article] [PubMed]
- Waters ER (1995) The molecular evolution of the small heat shock proteins in plants. Genetics 141:785–795 [DOI] [PMC free article] [PubMed]
- Waters ER (2003) Molecular adaptation and the origin of land plants. Mol Phyl Evol 29:456–463 [DOI] [PubMed]
- Waters ER, Rioflorido I (2007) Evolutionary analysis of the small heat shock proteins in five complete algal genomes. J Mol Evol 65:162–174 [DOI] [PubMed]
- Waters ER, Vierling E (1999a) Chloroplast small heat shock proteins: Evidence for atypical evolution of an organelle-localized protein. Proc Natl Acad Sci USA 96:14394–14399 [DOI] [PMC free article] [PubMed]
- Waters ER, Vierling E (1999b) The diversification of plant cytosolic small heat shock proteins preceded the divergence of mosses. Mol Biol Evol 16:127–139 [DOI] [PubMed]
- Waters ER, Lee G, Vierling E (1996) Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot 47:325–338 [DOI]
- Waters ER, Hohn MH, Ahel I et al (2003) The genome of Nanoarchaeum equitans: insights into early archaeal evolution, parasitism and the minimal genome. Proc Natl Acad Sci USA 100:12984–12988 [DOI] [PMC free article] [PubMed]
- Yang X, Tuskan GA, Cheng MZ (2006) Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol 142:820–830 [DOI] [PMC free article] [PubMed]
- Yazaki J, Kojima K, Suzuki K, Kishimoto N, Kikuchi S (2004) The Rice PIPELINE: a unification tool for plant functional genomics. Nucleic Acids Res 32:D383–D387 [DOI] [PMC free article] [PubMed]
- Yu J, Hu S, Wang J et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92 [DOI] [PubMed]
- Zhang L, Vision TJ, Gaut BS (2002) Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol Biol Evol 19:1464–1473 [DOI] [PubMed]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol 136:2621–2632 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Alignment of sHSP amino acid sequences (DOC 67.5 KB)
A. thaliana cytosolic I DNA alignment (DOC 67.0 KB)
Synonymous (Ks) and Nonsynonymous substitutions for the Cytosolic I genes (DOC 118 KB)