Abstract
Rapamycin inhibits the activity of the target of rapamycin (TOR)-dependent signaling pathway, which has been characterized as one dedicated to translational regulation through modulating cap-dependent translation, involving eIF4E binding protein (eIF4E-BP) or 4E-BP. Results show that rapamycin strongly inhibits global translation in Drosophila cells. However, Hsp70 mRNA translation is virtually unaffected by rapamycin treatment, whereas Hsp90 mRNA translation is strongly inhibited, at normal growth temperature. Intriguingly, during heat shock Hsp90 mRNA becomes significantly less sensitive to rapamycin-mediated inhibition, suggesting the pathway for Hsp90 mRNA translation is altered during heat shock. Reporter mRNAs containing the Hsp90 or Hsp70 mRNAs’ 5′ untranslated region recapitulate these rapamycin-dependent translational characteristics, indicating this region regulates rapamycin-dependent translational sensitivity as well as heat shock preferential translation. Surprisingly, rapamycin-mediated inhibition of Hsp90 mRNA translation at normal growth temperature is not caused by 4E-BP-mediated inhibition of cap-dependent translation. Indeed, no evidence for rapamycin-mediated impaired eIF4E function is observed. These results support the proposal that preferential translation of different Hsp mRNA utilizes distinct translation mechanisms, even within a single species.
Keywords: Translation, Heat shock, Rapamycin, Hsp90, Hsp70
Introduction
Exposure of cells to stress leads to protein inactivation and ultimately cell death, unless protective responses occur (Parsell and Lindquist 1993; Gabai and Sherman 2002). A highly conserved response, termed the heat shock response or stress response, occurs in virtually all living organisms from bacteria to humans (Lindquist 1986). The characteristic features of the response were initially described and detailed in Drosophila cells exposed to elevated temperatures (Ritossa 1962; Tissieres et al. 1974), among others, although subsequent studies have shown that a wide array of stressful circumstances, including oxidative stress, abnormal protein accumulation, and endoplasmic reticulum perturbations, induce a similar molecular response (Lindquist 1986).
The heat shock response provides for the massive, rapid synthesis of a small group of proteins termed the heat shock proteins (Hsps), or stress proteins, that are required to protect, maintain, and restore protein activity in the face of stressful insults (Lindquist 1986). The heat shock proteins are typically present in cells at low to undetectable levels before stress. Following stress, a latent transcription factor, heat shock factor, is activated leading to the abundant synthesis of Hsp mRNAs. In Drosophila, significant amounts of the major Hsp, Hsp70, are detected within 30 min of heat shock, and by 3 h Hsp70 is one of the most abundant proteins in the cytoplasm (Cotto and Morimoto 1999; Lindquist 1980a).
Stress characteristically inhibits numerous facets of metabolic function. At the level of translation, the overall rate of protein biosynthesis is inhibited >90% (Lindquist 1986; Tissieres et al. 1974), yet concurrently Hsp mRNAs are efficiently translated (Lindquist 1980b; Li and Duncan 1995) due to primary structure features in their 5′ untranslated regions (5′UTRs) (Lindquist and Petersen 1990). Neither the specific translation-promoting features of the 5′UTRs nor the specific lesion in the translational machinery that can be circumvented by 5′UTR elements have been clearly defined. These two facets are likely mechanistically linked to provide preferential Hsp mRNA translation during heat shock. This study was developed to investigate how Hsp mRNA structural features allow evasion of translational machinery lesions, using the translational inhibitor rapamycin as a probe.
The macrolide antibiotic rapamycin interacts with cytosolic FKBP to inactivate the target of rapamycin (TOR) protein kinase (Gingras et al. 2001a). Several downstream TOR targets including the ribosomal protein S6 kinase (S6K) and eIF4E binding protein (eIF4E-BP), or 4E-BP, are prominent regulators of protein synthesis. Indeed, the TOR-dependent pathway has been described as a signal transduction pathway dedicated to “translational control” (Brown and Schreiber 1996). More specifically, rapamycin, through its target, the TOR-dependent pathway and 4E-BP, is characterized as specifically inhibiting eIF4E-dependent or cap-dependent translation (Gingras et al. 2001a). 4E-BP is multiply phosphorylated by TOR and other kinases (Gingras et al. 1999, 2001a, b), which prevents its association with eIF4E and maintains high translation rates. Rapamycin, by inactivating TOR, causes the rapid dephosphorylation of 4E-BP, its subsequent association with eIF4E, and the inhibition of translation (Gingras et al. 2001a), according to the prevailing current view.
In this study, we have used rapamycin as a probe to investigate the basis for Hsp mRNAs’ preferential translation. A prevalent hypothesis posits that Hsp mRNAs are translated by a cap-independent mechanism (Rhoads and Lamphear 1995). Heat shock causes an inhibition of molecular components of the cap-dependent translational machinery, according to this theory, leading to the repression of virtually all non-heat shock, cap-dependent mRNAs while Hsp mRNAs are spared. Substantial evidence indicates that eIF4F, the principal molecular complex in cap-dependent translation, is partially or wholly inactivated by heat shock (Duncan and Hershey 1984a; Duncan et al. 1987; Panniers et al. 1985; Lamphear and Panniers 1991; Zapata et al. 1991; Zapata et al. 1994; Hess and Duncan 1996). Furthermore, Sierra, Zapata, and colleagues have elegantly demonstrated that most Drosophila Hsp mRNAs are highly resistant to translational inhibition due to suppression of eIF4F activity in vitro (Zapata et al. 1991; Zapata et al. 1994). However, Hsp90 mRNA is a fly in the ointment; its translation is significantly inhibited in the Zapata et al. assays, yet numerous reports document its efficient translation during heat shock in intact cells. This suggests the cap-independence model for Hsp mRNA translation is inaccurate, or at least incomplete. Our results using rapamycin have provided a unique insight into this distinction between Hsp70 and Hsp90 mRNAs, suggest a resolution for the apparent contradictory results cited above, and lead us to an intriguing new model for Hsp90 mRNA translation in Drosophila.
Materials and methods
Chemicals Chemicals were purchased from Sigma Chemical Company unless otherwise indicated.
Transfection of Drosophila S2 tissue culture cells Schneider S2 cells were cultured at 22–23°C in Schneider’s Drosophila Medium (SM; Invitrogen, Inc) containing 10% fetal bovine serum (FBS), 200 μM l-glutamine, 100 units/ml penicillin, 0.1 mg/ml streptomycin, 0.25 mg/ml amphotericin B (Invitrogen). Twenty-four hours before transfection, the cells were seeded at a density of 1–1.5 × 106 cells/ml in a T25 flask (Corning). For transfection, 10 μg plasmid DNA was mixed with 250 μl 0.5 M CaCl2 in a total volume of 500 μl. The CaCl2–DNA complex was then added dropwise to 500 μl of 2X HEPES-buffered saline (280 mM NaCl, 10 mM KCL, 1.5 mM Na2PO4, and 50 mM HEPES) with concurrent bubbling. After 15 min incubation, the CaCl2–DNA–HEPES mixture was added slowly to the cells with gentle concurrent rocking. 16–18 h post transfection; the transfection media was removed and replaced with fresh SM/10% FBS. Cells were used for analysis 48–72 h post transfection, at which time the transgene(s) was induced for 3 h with 500 μM CuSO4.The two plasmids used in this study are termed here p70UTR and p90UTR. The construction of both has been described (Ahmed and Duncan 2004). Both contain a metallothionein promoter precisely appended to the transcribed region (Ahmed and Duncan 2004). The coding sequence is an internally deleted version of Hsp70 with Mr ∼44 kDa (Hsp70ΔCd) (McGarry and Lindquist 1985). The coding sequence/3UTR cannot be translated during heat shock unless a 5′UTR conferring preferential heat shock translation is present (McGarry and Lindquist 1985; Hess and Duncan 1996; Ahmed and Duncan 2004).
Rapamycin treatment, heat shock, [35S]methionine labeling, and protein extraction Sixty to 120 min before analysis Drosophila S2 cells were supplemented with fresh FBS, to 10% of the final medium volume. Previous studies have documented this serum boost results in optimal translation, measured as ribosomes in polysomes, for several hours (Duncan 1995). At the start of each experiment, cells were scraped from T25 flasks, pelleted by brief centrifugation (3 mi, 3,000 rpm) in a clinical centrifuge (IEC), and resuspended in Grace's Media lacking methionine (Invitrogen). The cells were then transferred to 20-ml glass scintillation vials, with a stir flea, and allowed to recover ≥15 min with stirring before analyses (at heat shock or normal growth temperature (22–24°C)). Following recovery cells were divided into equal-sized treatment conditions, with some being incubated with 40 ng/ml rapamycin (Calbiochem) for 60 min. For heat shock samples, a cell aliquot was transferred to a 35°C water bath with stirring for 15 min. Between 15 to 30 min (in some experiments 10–30 min), relative to the start of heat shock, all cell treatment conditions (∼5 × 106 cells each, or a 1-ml suspension) were labeled with 15–20 μCi [35S]methionine/cysteine (ICN Biochemicals) for 15 min. At the end of labeling, cells were rapidly pelleted by centrifugation, resuspended in and washed twice with 4°C wash buffer (Earle's buffered salt solution (EBSS): 50 mM KCL, 15 mM MgSO4, 4 mM CaCl2, 3 mM KH2PO4, 10 mM dextrose, 8.4 mM HEPES pH 7.2, and 20 μg/ml cycloheximide) by centrifugation. After the final centrifugal wash, the cell pellet was lysed with (for two-dimensional gel analyses) 100–150 μl of Ampholyse buffer (∼9.8 M Urea, 5% 3.5–10 Biolytes (Bio-Rad), 2% Nonidet P-40 (NP-40), 1% β-mercaptoethanol), microcentrifuged for 3 min at top speed (Eppendorf), and the supernatant recovered to give a final protein concentration of ∼2 mg/ml. Protein concentration was measured by Bradford Assay (Bio-Rad), and radioactivity per microgram protein was measured by trichloroacetic acid (TCA) precipitation. For samples to be analyzed by one-dimensional gel electrophoresis, washed cell pellets were lysed in hSDS (0.3% SDS, 50 mM Tris pH 8.0, 1% β-mercaptoethanol, at ∼98°C). The pellet was disrupted by pipeting. A volume of 1/20 of RNase/DNase solution {5 mg/ml DNase, 2.5 mg/ml RNase A, 500 mM Tris pH 7.0, 50 mM MgCl2} was added for ∼1 min, viscosity reduced by pipeting, then 1/4 volume of 4× Laemmli-formula sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer was added, and samples analyzed on 1D slab gels as described below for the second dimension of the 2D procedure.
Analysis of proteins by two-dimensional isoelectric focusing/SDS-PAGE Two dimensional IEF/SDS-PAGE was performed basically as described by O'Farrell (1975), with modifications as described by Duncan and Hershey (1984b) to promote spot focusing. Briefly, 15-cm 1st D tube gels were polymerized in standard lab supply glass tubing, ∼4 mm OD, to a height of 13.5 cm. The first dimension gel mix contained ∼9.8 M Urea, 4% 5–7, 1% 3.5–10 Biolytes (Bio-Rad), 2% NP-40, 3.5% acrylamide, (5%T bisacrylamide). The upper and lower electrode solutions were 50 mM NaOH and 25 mM phosphoric acid, respectively. Gels were pre-run for 400–600 V h. Approximately 100 μg sample was loaded, overlayed with 3:1 Ampholyse/H2O, and run for about 16 h at 800 V. Gels were extruded, equilibrated in 5 ml of equilibration buffer (60 mM Tris pH 6.8, 2.3% SDS, 5% β-mercaptoethanol, 10% glycerol) for 15 min, and resolved on a 10.5% slab gel, Laemmli discontinuous buffer system, at 20 mA for about 4 h, at which time the bromophenol blue tracking dye reached the lower buffer reservoir. Gels were fixed, dried, and exposed to Kodak X-OMAT film for 4–15 days. Protein bands/spots were quantitated by densitometry (LabWorks (UVP)). The Hsp70ΔCd reporter transgene used in this study is observed as two isoforms (e.g., Fig. 3). The relative abundance of each differs between experiments, such that in some transfections both forms are detected in approximately equal amounts, whereas in others the acidic or basic form predominates (e.g., Fig. 6).
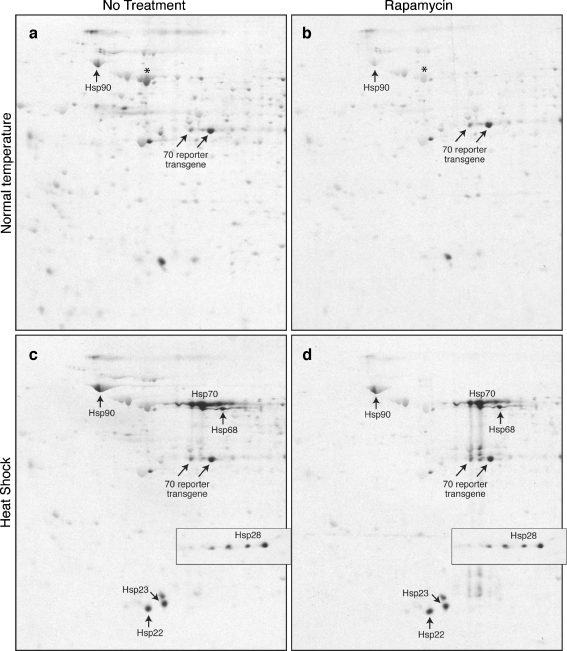
Fig. 3.
Rapamycin differentially affects heat shock protein synthesis. Drosophila S2 cells were treated and labeled as described in Fig. 1 legend. Protein samples were prepared as described for 2DGE (see “Materials and methods”) and analyzed by IEF/SDS-PAGE. Equal amounts of protein (equal cell numbers) were loaded into each first dimension gel (based on Bradford assays, and confirmed by Coomassie brilliant blue staining of the gels after electrophoresis). Dried gels were exposed to film and labeled proteins detected by autoradiography. For quantitation, films were scanned with a densitometer, and the intensity of spots determined using the Labworks software (UV Products). The basic region of the gel is to the right, the acidic end to the left. Only a sector of the entire gel is shown in the panels. a, b: Cells labeled at normal growth temperature, without (a) and with (b) rapamycin. c, d: Cells labeled at heat shock temperature, without (c) and with (d) rapamycin. The trio of vertical spots in panel d are degradation products of Hsp70 (whose main spot is located directly above), which are variably observed. Hsps were identified in accord with previous studies (Storti et al. 1980; Ireland et al. 1982; Li and Duncan 1995). The inset in the heat shock panels shows Hsp28 (four isoforms), which migrates at the basic perimeter of the gel (more basic than the sector of the gel shown in the panels). The location of Hsc70 is shown in panels a and b with asterisk
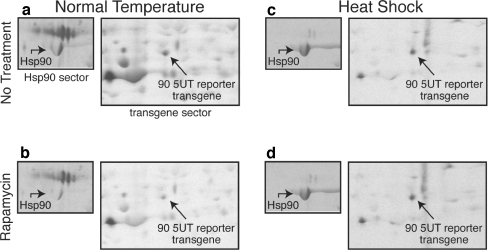
Fig. 6.
Hsp90 mRNA’s 5′ untranslated region confers rapamycin-dependent translational characteristics to a reporter gene. Drosophila S2 cell treatments, labeling, analysis, and methodology were as described in Figs. 1 and 3 legends. Cells were transfected with p90UTR [34]. Only sectors of the entire gel are shown in the panels, corresponding to the migration regions of endogenous Hsp90 and the reporter protein. a, b: Cells labeled at normal growth temperature, without (a) and with (b) treatment with 40 ng/ml rapamycin for 60 min before 15-min pulse labeling with [35S]met/cys. c, d: Cells labeled at heat shock temperature (35°C), without (c) and with (d) rapamycin. The locations of endogenous Hsp90 and the protein expressed by reporter transgene containing the Hsp90 mRNA 5′UTR are indicated by arrows. In this analysis, >90% of the transgene protein signal was detected in the acidic spot; see “Materials and methods” for additional description of transgene spot distribution. The analysis was repeated five times, with similar quantitative results obtained. In cells used in this series of experiments, both endogenous Hsp90 mRNA and transgene Hsp90 5′UTR reporter mRNA were translated during rapamycin/heat shock at 70–90% the no treatment/heat shock rate
Sepharose CL-6B column chromatography Lysates were prepared from a T75 flask (∼5 × 106 cells), untreated or treated. Cells were washed three times with 4°C EBSS by centrifugal pelleting and resuspension, using a clinical centrifuge (50 s, ∼3,500 rpm). The cell pellets were resuspended in 1000 μl of 4°C m7GTP column buffer (100 mM KCl, 20 mM HEPES pH 7.6, 7 mM β-mercaptoethanol, 0.2 mM EDTA, 10% glycerol) to which the following components were added for lysis: Triton ×-100 to 0.1% and Complete™ protease inhibitor mix (Roche). Cell suspensions were homogenized in a stainless steel homogenizer (7–10 strokes), then centrifuged for 3 min in a microcentrifuge. The supernatant was recovered and directly applied to the Sepharose CL-6B column (28 × 1 cm) at 4°C developed in the m7GTP-Sepharose buffer. Approximately 750-μl samples were collected using gravity flow; for analysis, the first seven fractions were discarded, based on pilot runs indicating that the excluded peak began to emerge in fraction 8–9. Thirty-microliter aliquots were removed from each fraction, mixed with 10 μl 4X SDS sample buffer, heated for 2 min at 98°C, and analyzed by 1DGE and immunoblotting. The amido schwartz staining pattern of the blot showed the predicted correlation between elution volume and protein molecular mass. The principal exception was that in the excluded material a range of protein sizes including some quite small species was detected.
Purification of eIF4E and associated proteins by m7GTP-Sepharose affinity chromatography Lysates were prepared as described above, for CL-6B analysis. The final supernatant was applied to a 1-ml (swollen) m7GTP-Sepharose column at 4°C, and allowed to drain by gravity flow. The flow-through was recovered and passed through the column a second time. This second eluate was saved as the “non-bound” fraction. The column was washed with ∼60 ml m7GTP-Sepharose buffer, at a flow rate of ∼ 1 ml/min, followed by a 10 ml wash with 100 μM guanosine diphosphate (GDP) in m7GTP-Sepharose buffer. Finally, the bound eIF4E and associated proteins were eluted with 100 μM m7GTP in m7GTP-Sepharose buffer. Ten 1-ml fractions were collected, and the content of proteins determined by mixing 30 μl of each fraction with 10 μl 4X SDS sample buffer, heat denaturation, 1D SDS-PAGE, and immunoblotting. In some analyses smaller, equal-volume aliquots from several column fractions were combined (identically for treated and untreated analyses) to provide a single bound fraction. For comparison of input and non-bound proteins, an equal volume aliquot of each fraction was analyzed.
Results
Rapamycin inhibits non-heat shock translation but not, to the same extent, heat shock translation
Protein synthesis rate in Drosophila tissue cells was analyzed by pulse-labeling with [35S]methionine. Cells were treated with rapamycin (40 ng/ml [low] or 120 ng/ml [high]) for 1 or 4 h, or left untreated (vehicle (0.1% EtOH) only). A portion of the cell culture from each treatment condition was then heat shocked for 30 min at 35°C (this represents a moderately severe heat shock; under these conditions protein synthesis is typically inhibited 60–80%, rather than >90% as is seen at 37°C). The lower temperature was selected because other investigations have shown that Hsp90 mRNA translation is optimal at 35°C(Ahmed and Duncan 2004). Proteins were labeled for the final 20 min of heat shock, or for a corresponding interval in the parallel non-heat shocked cultures.
Rapamycin proves to be a potent inhibitor of translation in Drosophila cells. TCA analysis of [35S]methionine incorporation into protein at normal growth temperature using cells pre-treated with rapamycin for 1 h showed 60–90% inhibition (Fig. 1a, dark bars; see also Fig. 2 for a PAGE depiction). The extent of inhibition observed was usually less after 4 h of incubation, indicating a modest translational adaptation to rapamycin occurs (data not shown). The significant inhibition observed in Drosophila cells contrasts with what is typically observed in mammalian cells, in which case rapamycin only inhibits translation by 10–30% within the first several hours (R. Duncan, unpublished results with HeLa and 293 cells, and, e.g., Beretta et al. (1996), Morley and Naegele (2002)). The results are similar to the strong translational repression observed in rapamycin-treated yeast cells (Barbet et al. 1996). The efficacy of rapamycin at inhibiting signaling downstream of TOR was verified by analyzing serum-stimulated lysates for S6 kinase activation using a phospho-T389 anti-active antibody and Western blotting (panel C).
Fig. 1.
Rapamycin inhibits protein synthesis. Drosophila S2 cells were incubated with 40 ng/ml rapamycin for 1 h, or left untreated. Some cells were transfected with Hsp70 reporter mRNA expression plasmid p70UTR (Hess and Duncan 1996) 3 days before rapamycin treatment (subsequently used for the 1-h treatment condition). Cells were either heat shocked (35°C for 10 min) or not, then pulse-labeled with 30 μCi/ml [35S]methionine for 20 min. Protein samples were prepared as described (see “Materials and methods”). a: Quantified incorporation by TCA precipitation on GF/C filters and liquid scintillation counting. Rapamycin inhibition of protein synthesis at normal growth temperature is shown in dark bars at left, and at heat shock (35°C) in light bars at right. Parallel measurements of normal temperature translation, without and with rapamycin, were made by protein analysis by 2D IEF/SDS-PAGE, autoradiography, and densitometry of an array of ∼10 non-heat shock protein spots. b: Heat shock mRNA-directed protein synthesis was determined by 2D IEF/SDS-PAGE, autoradiography, and densitometry (see Fig. 3 for visual example) (dark bars, analysis at normal growth temperature; light bars, analysis at heat shock). Results represent the average and standard deviation of three to five replicates in most cases. c: Rapamycin treatment inhibits signaling downstream of TOR. S6 kinase activity/phosphorylation was analyzed using anti-active phospho-T389 antibody (Cell Signaling Technology #9205) and Western blotting after 1 h stimulation by 10% serum. S6K(P) (∼70 kDa; arrow) was reduced to undetectable levels following rapamycin treatment in several independent experiments
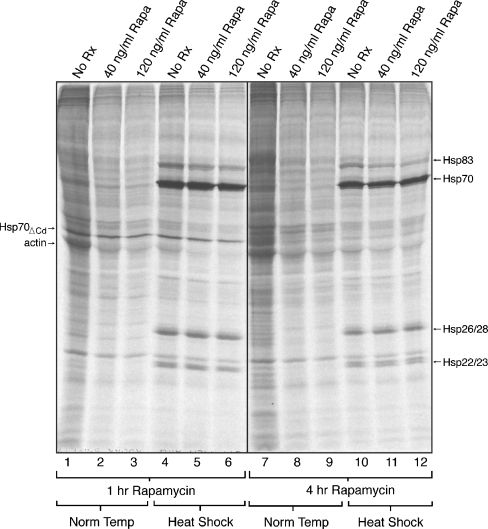
Fig. 2.
Analysis of rapamycin effects on protein synthesis by 1D SDS-PAGE. Drosophila S2 cells were incubated with or without 40 or 120 ng/ml rapamycin for 1 (lanes 1–6) or 4 h (lanes 7–12). Cells were heat shocked cells and pulse-labeled as described in Fig. 1 legend. Cells used for the 1-h treatment analysis were transfected with p70UTR. Equal amounts of protein (equal cell numbers) were loaded into each lane of an SDS-PAGE gel (based on Bradford assays, and confirmed by Coomassie brilliant blue staining of the gel after electrophoresis). The dried gel was exposed to film and labeled proteins detected by autoradiography. Norm temp, cells labeled at normal growth temperature. The location of bands corresponding to heat shock proteins Hsp83, Hsp70, Hsp26/28, and Hsp22/23 are depicted on the right. Reporter protein expression is indicated by the arrow on the left border, as is actin. This analysis has been repeated in part or completely more than three times; a representative analysis is shown
When the same TCA analysis was carried out in heat shocked cells, substantially less inhibition by rapamycin was observed (Fig. 1a, light bars); again there was evidence for modest translational adaptation in the longer (4 h) treated cells. These results suggest that Hsp mRNA translation may be resistant to rapamycin-mediated protein synthesis inhibition, as a significant fraction of translation during heat shock occurs using Hsp mRNAs. This was confirmed in the next series of analyses.
The translational effects observed by TCA incorporation were verified by SDS-PAGE analysis of labeled protein samples (Fig. 2). Comparison of lane 1 (untreated) to lanes 2, 3 (40 ng/ml and 120 ng/ml rapamycin, respectively) shows the inhibition of translation in non-heat shocked cells treated with rapamycin for 1 h. Similarly, lane 7 vs. lanes 8, 9 shows the inhibition observed after 4 h of rapamycin treatment. The lesser extent of inhibition at heat shock can be seen comparing lane 4 (untreated) to lanes 5 and 6 (low and high rapamycin). Particularly note that the major heat shock translation product Hsp70 shows little to no inhibition (arrow, Fig. 2). This point is more precisely characterized as described below. Likewise, the translation of the small Hsp mRNAs (producing Hsp 26/28 and Hsp 22/23) is not significantly inhibited by rapamycin treatment (arrows, Fig. 2). The relative rapamycin-resistance of Hsp mRNA translation is also seen in the 4-h rapamycin-treated heat-shocked samples (lanes 10–12).
Rapamycin inhibits Hsp90 but not Hsp70 mRNA translation at normal growth temperature
To specifically and precisely determine the effects of rapamycin treatment on Hsp mRNA translation, two additions to the basic protocol were incorporated. First, proteins were resolved by two-dimensional IEF/SDS-PAGE. This allows the quantitative analysis of Hsp90 mRNA translation. On one dimensional gels, especially under non-heat shock conditions, the Hsp90 band is faint and represents the summed intensity of numerous other ∼85-kDa proteins; 2D resolution allows its unambiguous detection. Second, cells used in the analyses were transfected with an inducible transgene expressing an mRNA containing the full-length Hsp70 mRNA 5′ untranslated region followed by a reporter sequence (Hess and Duncan 1996). The translational properties of the reporter transgene recapitulate endogenous Hsp70 mRNA in all analyses to date (McGarry and Lindquist 1985; Hess and Duncan 1996). Three hours before treatment the metallothionein promoter was induced with 500 μM Cu2SO4. The use of the inducible reporter system is required to investigate the effects of rapamycin on Hsp70 mRNA translation at non-heat shock temperatures, as no Hsp70 mRNA is normally expressed. The experimental goal is to compare and contrast the effects of rapamycin on the translation of Hsp90 and Hsp70 mRNA at non-heat shock and heat shock temperatures. As Hsp90 mRNA is present in non-heat shocked cells, no transfection analysis is required.
Rapamycin inhibits Hsp90 mRNA translation by up to 90% in non-heat shocked cells (Fig. 3; quantified in Fig. 1b); approximately the same extent of inhibition is seen at 1 and 4 h, using 40 or 120 ng/ml (data not shown). The extent of Hsp90 mRNA translation inhibition by rapamycin is equal to or greater than global (non-heat shock) mRNA translation inhibition (based on quantifying Hsp90 and >10 non-heat shock protein spots in ≥6 independent analyses), indicating that Hsp90 mRNA is, if anything, more sensitive to the inhibitory molecular events induced by rapamycin treatment in non-heat shocked cells.
In striking contrast, the translation of the Hsp70 reporter transgene is unaffected by rapamycin treatment in the non-heat shocked cells (Figs. 1b and 3). In contrast, the synthesis of the constitutive form of Hsp70 (Hsc70) is inhibited by rapamycin to the same extent as bulk non-heat shock proteins (quantification not shown, but see Fig. 3), and likewise is inhibited by heat shock. In conclusion, the Hsp90 and Hsp70 mRNAs respond quite distinctly to protein synthesis inhibition by rapamycin treatment under non-heat shock conditions.
Rapamycin has a reduced effect on Hsp90 mRNA translation during heat shock
The same 2D analysis of Hsp90 mRNA translation in rapamycin-treated cells was performed during heat shock. Surprisingly, its translation is significantly less inhibited (Figs. 1 and 3). This suggests that a heat-induced change in the mechanism of translation, based either in the mRNA itself or in the translational machinery, partially relieves the inhibitory effect of rapamycin. We can exclude the possibility that Hsp90 mRNA's enhanced translation during heat shock simply reflects the ability of a weakly initiating mRNA to translate more efficiently when competition is minimized due to the release of ribosomes from normal mRNA translation. In rapamycin-treated, non-heat shocked cells 70–80% of the translating ribosomes are released, thus relieving competition, yet there is no selective enhancement of Hsp90 mRNA translation; it is inhibited at least as much as the average non-heat mRNA.
Hsp70 mRNA translation is virtually unaffected by rapamycin treatment under heat shock labeling conditions (Fig. 3, quantified in Fig. 1b). This parallels its translation properties in non-heat shocked cells, and confirms the impression formed by 1D and TCA analyses. The same complete resistance was seen for the endogenous Hsp70 mRNA and the transgene reporter form. Hsp22, 23 and 28 mRNAs’ translation also was confirmed to be unaffected by rapamycin treatment using the 2D gel quantitation approach (Fig. 3; only heat shock translation could be examined). These results confirm the conjecture proposed above, that Hsp mRNA translation is resistant to rapamycin, and further show that rapamycin-resistance Hsp mRNA translation occurs under heat shock and non-heat shock conditions, with the exception of Hsp90 mRNA translation.
Rapamycin effects on 4E-BP and eIF4E in Drosophila
The idiosyncratic sensitivity of Hsp90 mRNA translation to rapamycin treatment is consistent with observations that it is the only Hsp mRNA that contains significant 5′UTR secondary structure (Hess and Duncan 1996), hence requiring eIF4E-dependent eIF4F-mediated RNA unwinding activity. Furthermore, its inhibition by rapamycin conforms to previous results showing that either 4E-BP overexpression or antisera-mediated eIF4E titration inhibits its translation (Zapata et al. 1991; Ahmed and Duncan 2004). To confirm that the mechanism of rapamycin repression of Hsp90 mRNA occurred through the well-established pathway in which 4E-BP dephosphorylation causes inhibition of eIF4E and hence eIF4F, several complementary assays were conducted.
Dephosphorylation of 4E-BP Rapamycin inhibits TOR leading to the dephosphorylation of 4E-BP, its increased association with eIF4E, and dissolution of the eIF4F complex (Gingras et al. 2001a). This well-established pathway is based on numerous studies in a wide range of cell types (Gingras et al. 1999). Our findings, described below, suggest the intriguing alternative conclusion that Hsp90 mRNA translation inhibition is mediated by a distinct, as yet unknown, series of rapamycin-induced molecular events.
To examine whether rapamycin causes dephosphorylation of dm4E-BP, cells were treated with 40 ng/ml for 1 h, protein extracts prepared, and analyzed by 1DGE/immunoblotting. In all species examined, including Drosophila (Miron et al. 2001, 2003), phosphorylated forms of 4E-BP can be identified as more slowly migrating (higher apparent molecular weight) forms. In our Drosophila cells under normal growth conditions (serum-replete; see “Materials and methods”) typically >70% of 4E-BP is detected in the lowest apparent Mr band (Fig. 4a; referred to as the α form by Miron et al. 2003). A minor portion of 4E-BP migrates more slowly in a higher molecular weight band (Fig. 4a; the β form (Miron et al. 2003)). Treatment with rapamycin reduces the β form to undetectable levels (Fig. 4a). Because the β form represents a minor fraction of the total, no detectable increase in the amount of α form is apparent in most analyses. Similar results have been reported by Miron et al. (2003) in their characterization of Drosophila 4E-BP.
Binding to eIF4E To examine whether rapamycin affected the extent of interaction of 4E-BP with eIF4E, rapamycin-treated and untreated cell extracts were applied to an m7GTP-Sepharose column, which quantitatively binds eIF4E and any associated proteins. In confirmation of the column analysis’ efficacy, virtually all of the eIF4E applied to the column was recovered in the bound fraction (Fig. 4b,c). 4E-BP is also exclusively detected in the bound fraction, without or with rapamycin treatment (Fig. 4b); there is no detectable 4E-BP in the non-bound fraction (Fig. 4c). This is not unexpected as the majority of dm4E-BP is found in the rapidly migrating, “unphosphorylated” form in untreated serum-replete cells (Fig. 4a). It mirrors our previous studies (Ahmed and Duncan 2004), although it differs from the results reported by Miron et al. (2003). eIF4G was primarily associated with eIF4E, based on retention of >75% of the input by the column either without or with rapamycin treatment. Hence, rapamycin causes no detectable dissociation of eIF4E-associated eIF4G in these cells (Fig. 4b). Another protein reported to be associated with eIF4E, and bound to m7GTP-Sepharose, is the poly(A)-binding protein (PABP) (Tarun Jr. and Sachs 1996; Borman et al. 2002); however, no detectable association of dmPABP with dmeIF4E was observed (Fig. 4b). As a control for non-specific retention, it was determined that the vast majority of proteins were quantitatively eluted in the non-bound fraction based on global comparison of protein abundance by PAGE/staining (1D and 2D analyses: data not shown).
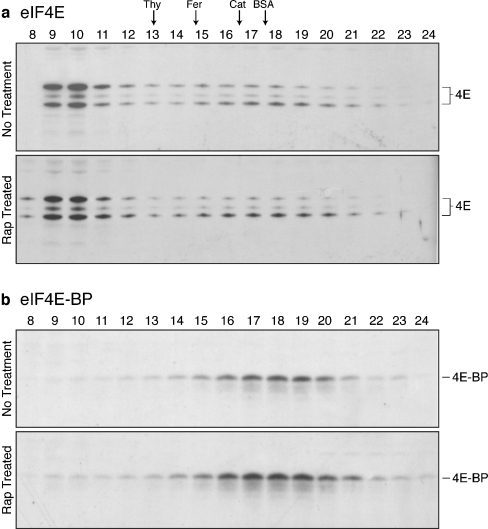
Fig. 4.
4E-BP phosphorylation and association with eIF4E by m7GTP-Sepharose column analysis. Drosophila S2 cells were treated (or not) with 40 ng/ml rapamycin for 60 min. Lysates were prepared for m7GTP-Sepharose analysis as described in “Materials and methods”. An aliquot from each cell culture was also lysed in hSDS buffer (see “Materials and methods”) and directly analyzed by SDS-PAGE and immunoblotting with anti-4E-BP antisera (panel a). Following rapamycin treatment, the prevalence of the 4E-BPα variant (lowest apparent Mr form) was unchanged to minimally increased (as shown in this panel). Note that the α band likely represents a mixture of phosphorylated and unphosphorylated 4E-BP. Results by ourselves and others (Fadden et al. 1997; Duncan and Song 1999; Gingras et al. 2001b) have determined that 4E-BP is phosphorylated on ≥4 sites; higher phosphorylation states produce apparently higher molecular weight forms, but the monophosphorylated form migrates at the same apparent molecular weight as “unphosphorylated” 4E-BP. b: After binding to the column and washing with column buffer and column buffer containing 100 μM GDP, proteins were eluted with 100 μM m7GTP. Bound fractions were analyzed by SDS-PAGE and immunoblotting using antisera to eIF4E, 4E-BP, PABP, and eIF4G. Binding to the column was also assessed by comparing the input to the non-bound flow-through fraction (see “Materials and methods”) (panel c) using the same panel of antibodies. The lower molecular weight band in panel c, PABP-sector, is a minimally degraded form of PABP (Lefrere and Duncan 1994). Highly labile DmPABP is not quantitatively recovered in the bound + unbound fractions, possibly reflecting weak association and loss or degradation during binding the analysis. The analysis was repeated ≥5 times; representative results are depicted
In conclusion, since dm4E-BP is primarily associated with dmeIF4E before and after rapamycin treatment, this mechanism does not appear to be the basis for Hsp90 mRNA-specific inhibition. Inquiry must be focused on other alternative pathways.
Protein complexes including eIF4E and 4E-BP To further examine whether eIF4E was targeted during rapamycin-mediated translation inhibition, an alternative approach was investigated. The most active form of eIF4E occurs when it associates with eIF4A and eIF4G in the eIF4F complex (Haghihat and Sonenberg 1997; Gross et al. 2003), which likely contains many other translational components in the cellular context (Holz et al. 2005). m7GTP-Sepharose analysis provides little information about the portion of eIF4E present in high molecular mass, high activity complexes. 4E-BP is generally considered to exist as a free protein when inactive, and in a dimeric complex with eIF4E when functioning as a translational repressor (Gingras et al. 2001b), although neither of these aspects has been rigorously addressed. Size-exclusion column chromatography can be used to characterize the macromolecular complexes in which eIF4E and 4E-BP occur (Duncan and Song 1999).
Extracts from rapamycin-treated and untreated cells were applied to a Sepharose CL-6B column. Fractions from the elution profile were analyzed by immunoblotting using dmeIF4E and dm4E-BP antisera. In untreated cells eIF4E is detected in (1) very large complexes eluting in the void volume and in (2) a broad, small molecular-mass peak (Fig. 5a). This broad peak approximates the elution profile of purified eIF4E. Treatment of cells with rapamycin has no effect on eIF4E's distribution profile (compare panels a, b, in Fig. 5). While this assay cannot establish which molecules interact with eIF4E to form high molecular mass complexes, numerous previous reports implicate the scaffolding protein eIF4G as bridging initiation factor and other translational regulatory proteins (Holz et al. 2005).
Fig. 5.
Association of eIF4E and 4E-BP with high molecular weight complexes. Drosophila S2 cells were treated (or not) with 40 ng/ml rapamycin for 60 min, lysates were prepared as described, and analyzed by Sepharose CL-6B column chromatography. Aliquots from fractions were analyzed by SDS-PAGE/immunoblotting using antisera to eIF4E (panel a) and 4E-BP (panel b). The excluded volume (Blue Dextran) is observed beginning in fraction ∼9 and an included volume marker (phenol red) elutes in fraction ∼26. The elution positions of several molecular weight standards (bovine serum albumin [BSA], 68 kDa; catalase [Cat], 232 kDa; ferritin [Fer], 440; thyroglobulin [Thy], 669 kDa) are shown above panel a
4E-BP is principally detected in a broad small Mr peak that roughly parallels the distribution of eIF4E (Fig. 5). Rapamycin treatment has little effect on its distribution (compare panels in Fig. 5b). These results indicate that rapamycin-induced translational inhibition, which reduces activity by >80%, does not cause any significant changes in the macromolecular states of either eIF4E or 4E-BP: eIF4E is not released from its high molecular mass complexes reflecting an induced association with 4E-BP, nor is increased 4E-BP/eIF4E association inferred from the 4E-BP distribution.
In sum, results in this section suggest that the rapamycin-induced inhibition of protein synthesis cannot be simply explained by eIF4E/4E-BP interactions, and that the differential effects of rapamycin on heat shock mRNA protein synthesis may result from its targeting a heretofore uncharacterized step in mRNA translation (see Discussion section) to which Hsp90 mRNA is idiosyncratically sensitive under non-heat shock conditions.
The Hsp90 mRNA’s 5′ untranslated region confers the rapamycin-dependent translational characteristics
To begin to probe the RNA basis for Hsp90 mRNA's translation inhibition by rapamycin treatment, chimeric mRNAs were analyzed. The ability of Hsp22 and Hsp70 mRNA to be preferentially translated during heat shock can be recapitulated by placing their respective 5′ untranslated regions before a reporter coding sequence (McGarry and Lindquist 1985; Hultmark et al. 1986; Lindquist and Petersen 1990; Hess and Duncan 1996). Similarly, we have recently determined that Hsp90 mRNA’s 5′UTR likewise contains all the requisite signals for heat shock translation (Ahmed and Duncan 2004). Experiments reported above document that the Hsp70 mRNA 5′UTR also confers rapamycin-resistant translation at both normal and heat shock temperatures. In this section, the influence of Hsp90 mRNA’s 5′UTR on rapamycin-perturbed translation is addressed.
An inducible reporter transgene containing the full-length Hsp90 mRNA 5′UTR (Ahmed and Duncan 2004) was transfected into cells, and rapamycin treatment and pulse-labeling analysis performed 72 h later. The analysis shows (Fig. 6) that: 1) the transgene mRNA’s translation was inhibited by rapamycin to the same extent as endogenous Hsp90 mRNA’s translation under non-heat shock conditions (compare panels a and b in Fig. 6); and 2) during heat shock, the transgene's Hsp90 5′UTR conferred significant resistant to rapamycin-mediated translational repression (compare panels c and d in Fig. 6). Again, this parallels the translational characteristics of the complete, endogenous Hsp90 mRNA. In conclusion, the Hsp90 mRNA 5′UTR is sufficient to confer the unique heat-regulated translational response to rapamycin comparing normal and heat shock temperatures.
Discussion
Rapamycin treatment strongly inhibits overall protein synthesis in Drosophila-cultured cells, but has little effect on Hsp70 mRNA translation at either normal growth temperature or during heat shock. In contrast, rapamycin strongly inhibits Hsp90 mRNA translation at normal growth temperature but only moderately represses it during heat shock. The analysis of the molecular mechanism for rapamycin-dependent inhibition of general protein synthesis points to a novel basis for repression.
Hsp mRNAs translate via a cap-dependent mechanism that may, however, remain quite efficient when levels of functional eIF4E/eIF4F are reduced. Evidence supporting this assertion includes: (1) translation is blocked by eIF4G cleavage or m7GTP cap analogue treatment (Song et al. 1995); (2) translation of in vitro synthesized uncapped Hsp mRNA is poor in vitro and in vivo (ibid.); and (3) overexpression of Dm4E-BP inhibits Hsp mRNA translation (Ahmed and Duncan 2004). However, the sensitivity of Hsp70 mRNA to these inhibitors of cap-dependent translation is substantially less than most non-heat shock mRNAs, as we (Ahmed and Duncan 2004; Song et al. 1995) and others reported, showing that antibodies to eIF4E or eIF4G fail to significantly reduce translation of most Hsp mRNAs (Zapata et al. 1991, 1994) and that eIF4E antisense RNA accentuates Hsp mRNA translation (De Benedetti et al. 1991). Furthermore and notably, Zapata and colleagues observed that Hsp90 mRNA translation was significantly repressed by antibody treatment of Drosophila cell free translation systems using amounts that had little to no effect on other Hsp mRNAs (Zapata et al. 1991, 1994). Hsp90 mRNA translation is also significantly more sensitive than Hsp70 mRNA to inhibition by overexpression of 4E-BP at normal growth temperature (Ahmed and Duncan 2004). Hsp90 mRNA is irrefutably efficiently translated during heat shock.
Rapamycin treatment causing potent inhibition of Hsp90 mRNA translation but with virtually no effect on Hsp70 mRNA translation is consistent with its primarily targeting cap-dependent translation, paralleling results reported by Zapata et al. (1991, 1994) (normal temperature analyses). Numerous previous reports implicate 4E-BP dephosphorylation as the mechanism of rapamycin-mediated repression. Indeed, we predicted our investigation of the mechanistic basis for rapamycin-mediated Hsp90 mRNA translational repression would confirm 4E-BP dephosphorylation, and release of eIF4E from eIF4G and high molecular mass high activity complexes, with attendant inhibition of cap-dependent translation. Surprisingly, careful characterization of each of these aspects provided no evidence for this mechanism of inhibition. While our analyses do not support a causal role for 4E-BP in inhibiting cap-dependent translation, it remains likely that other rapamycin-sensitive target(s) also exert their inhibitory effect on cap binding/5′UTR unfolding/ribosome subunit scanning. Several attractive candidate targets, including eIF4G and eIF4B (Gingras et al. 2001a; Holz et al. 2005; Harris et al. 2006), are dephosphorylated after rapamycin treatment (Raught et al. 2000, 2004), but assays to characterize phosphorylation state-dependent functional alterations are not available for these despite extensive efforts, especially with regards to Drosophila factors. Whereas eIF4G cleavage, m7GTP cap analogue inhibition, or overexpression of 4E-BP all block Hsp70 mRNA translation, high concentrations of rapamycin do not. This provides further evidence that the mechanism of rapamycin inhibition in Drosophila cells is not occurring through 4E-BP.
Intriguingly, under heat shock conditions the rapamycin-induced inhibition of Hsp90 mRNA is significantly less pronounced. Hsp90 mRNA translation also becomes less sensitive to the inhibitory effects of overexpressed 4E-BP during heat shock (Ahmed and Duncan 2004). Hsp70 mRNA, as well as Hsp22, 23, and 28 mRNAs, remain resistant to rapamycin treatment under heat shock conditions (Figs. 1 and 3). A global consequence of these observations is that overall heat shock protein synthesis, which is predominantly accounted for by Hsp mRNA translation, is only modestly inhibited by rapamycin (Fig. 1). The sequence elements that facilitate heat shock-dependent escape from rapamycin-induced inhibition of translation occur in Hsp90 mRNA's 5′UTR, as shown in Fig. 6. This region also confers heat shock preferential translation on a reporter (Ahmed and Duncan 2004). All evidence accumulated to date suggests that the 5′UTR region of the heat shock mRNAs confers their unique translational characteristics, to heat shock as shown previously by ourselves and others (McGarry and Lindquist 1985; Hultmark et al. 1986; Hess and Duncan 1996; Ahmed and Duncan 2004), and to rapamycin as shown in this report.
Rapamycin treatment's differential effects on Hsp90 vs. Hsp70 mRNA translation support the proposal (Ahmed and Duncan 2004) of distinct “pathways” for preferential translation. A corollary is that the mechanism of Hsp90 mRNA translation is altered by heat shock. We suggest that heat reprograms Hsp90 mRNA translation from being highly cap-dependent to relatively resistant to cap-dependent function inhibition. Heat may destabilize secondary structure elements resulting in enhanced access of translation factors and/or exposure of primary sequence elements that enhance translation during heat shock. Our deletion analyses of the Hsp90 5′UTR indicate that an ∼40 nucleotide segment preceding the initiator AUG is critical for Hsp90 mRNA’s translation during heat shock (Ahmed and Duncan 2004). The proposal of two distinct temperature-dependent modes of Hsp90 mRNA translation is consistent with the results of Zapata et al. (1991, 1994) who only documented Hsp90 mRNA inhibition by antibody treatments in normal temperature translation analyses. It provides a resolution to the enigma of how Hsp90 mRNA can both be translated efficiently during heat shock when eIF4F is inhibited, which is a well-documented consequence of heat shock (Duncan 1996), and yet exquisitely sensitive to eIF4F inhibition at normal growth temperature.
Future experiments will provide evidence to buttress this proposal of a translational pathway shift for Hsp90 mRNA translation in Drosophila, and to elucidate its mechanism. Additional work will also be required to determine the generality of heat shock translation mechanisms elucidated in Drosophila to other organisms, including mammals, which contain a significantly more diverse repertoire of heat shock protein mRNAs. For example, mammals possess distinct Hsp90 isoforms whose mRNAs may show distinct heat shock translation responses. Furthermore, mammalian Hsp mRNAs (including Hsp70 and Hsp90 isoforms) characteristically possess more extensive secondary structure than the average non-heat shock mRNA (Duncan, unpublished results) precluding the types of “structure-less bypass” models proposed for Drosophila Hsp70 mRNA. Indeed, Schneider and colleagues have provided evidence for an Hsp70 mRNA translation pathway based on ribosome shunting in mammals (Yueh and Schneider 2000). It will be intriguing to determine if the same mechanism applies to Hsp90 mRNA translation in mammals, or if, as in Drosophila, different Hsp mRNAs have evolved distinct mechanisms to achieve preferential translation during heat shock.
Acknowledgements
This research was supported by NSF grant MCB-9728753. We thank Drs. N. Sonenberg, M. Miron, and J. Sierra for providing antisera to Drosophila eIF4E, 4E-BP, and eIF4G. We thank Dr. A. Vincent for providing antisera to Drosophila PABP. We thank Dr. R. Ahmed for the preparation of the plasmid vectors.
Abbreviations
- TOR
Target of rapamycin
- S6K
ribosomal protein S6 kinase
- PI3K
phosphatidyl inositol 3-kinase
- eIF
eukaryotic initiation factor
- 4E-BP
eIF4E binding protein (eIF4E-BP)
- 5′UTR
5′ untranslated region of mRNA
- FBS
fetal bovine serum
- SM
Schneider's medium
- EBSS
Earle's buffered salt solution
- PABP or dmPABP
poly(A)-binding protein (from Drosophila melanogaster)
- “d” prefix precedingmolecular designations
Drosophila forms of the enzymes
References
- Ahmed R, Duncan RF (2004) Translational Regulation of Hsp90 mRNA: AUG-proximal 5’¢ untranslated region elements essential for preferential heart shock translation. J Biol Chem 279:49919–49930 [DOI] [PubMed]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN (1996) TOR controls translation initiation and early G1 progression in yeast. Mole Biol Cell 7:25–42 [DOI] [PMC free article] [PubMed]
- Beretta L, Gingras A-C, Svitkin YV, Hall MN, Sonenberg N (1996) Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J 15:658–664 [PMC free article] [PubMed]
- Borman AM, Michel YM, Malnou CE, Kean KM (2002) Free poly(A) stimulates capped mRNA translation in vitro through the eIF4G-poly(A)-binding protein interaction. J Biol Chem 277:36818–36824 [DOI] [PubMed]
- Brown EJ, Schreiber SL (1996) A signaling pathway to translational control. Cell 86:517–520 [DOI] [PubMed]
- Cotto JJ, Morimoto RI (1999) Stress-induced activation of the heat-shock response: cell and molecular biology of heat-shock factors. Biochem Soc Symp 64:105–118 [PubMed]
- De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads RE (1991) Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol Cell Biol 11:5435–5445 [DOI] [PMC free article] [PubMed]
- Duncan RF (1995) Cordycepin blocks recovery of non-heat shock mRNA translation following heat shock in Drosophila. Eur J Biochem 233:784–792 [DOI] [PubMed]
- Duncan RF (1996) Translational control during heat shock. In: Hershey JWB, Mathews M, Sonenberg N (eds) Translational control. Cold Spring Harbor Laboratory Press, New York
- Duncan R, Hershey JWB (1984a) Heat-shock induced translational alterations in HeLa cells: initiation factor modifications and the inhibition of translation. J Biol Chem 259:11882–11889 [PubMed]
- Duncan R, Hershey JWB (1984b) Evaluation of isoelectric focusing running conditions during two-dimensional isoelectric focusing/sodium dodecyl sulfate-polyacrylamide gel electrophoresis: variation of gel patterns with changing conditions and optimized isoelectric focusing conditions. Anal Biochem 138:144–155 [DOI] [PubMed]
- Duncan RF, Song HJ (1999) Striking multiplicity of eIF4E-BP1 phosphorylated isoforms identified by 2D gel electrophoresis regulation by heat shock. Eur J Biochem 265:728–743 [DOI] [PubMed]
- Duncan R, Milburn SC, Hershey JWB (1987) Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. J Biol Chem 262:380–388 [PubMed]
- Fadden P, Haystead TA, Lawrence JC Jr (1997) Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. J Biol Chem 272:10240–10247 [DOI] [PubMed]
- Gabai VL, Sherman MY (2002) Interplay between molecular chaperones and signaling pathways in survival of heat shock. J Appl Physiol 92:1743–1748 [DOI] [PubMed]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N (1999) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 13:1422–1437 [DOI] [PMC free article] [PubMed]
- Gingras AC, Raught B, Sonenberg N (2001a) Regulation of translation initiation by FRAP/mTOR. Genes Dev 15:807–826 [DOI] [PubMed]
- Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N (2001b) Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 15:2852–2864 [DOI] [PMC free article] [PubMed]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G (2003) Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115:739–750 [DOI] [PubMed]
- Haghihat A, Sonenberg N (1997) eIF4G dramatically enhances the binding of eIF4E to the mRNA 5’¢-cap structure. J Biol Chem 272:21677–21680 [DOI] [PubMed]
- Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC Jr (2006) mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J 25:1659–1668 [DOI] [PMC free article] [PubMed]
- Hess MA, Duncan RF (1996) Sequence and structure determinants of Drosophila Hsp70 mRNA translation: 5’¢-UTR secondary structure specifically inhibits heat shock protein mRNA translation. Nucleic Acids Res 24:2441–2449 [DOI] [PMC free article] [PubMed]
- Holz MK, Ballif BA, Gygi SP, Blenis J (2005) mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123:569–580 [DOI] [PubMed]
- Hultmark D, Klemenz R, Gehring WJ (1986) Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell 44:429–438 [DOI] [PubMed]
- Ireland RC, Berger E, Sirotkin K, Yund MA, Osterbur D, Fristrom J (1982) Ecdysterone induces the transcription of four heat-shock genes in Drosophila S3 cells and imaginal discs. Dev Biol 93:498–507 [DOI] [PubMed]
- Lamphear BJ, Panniers R (1991) Heat shock impairs the interaction of cap-binding protein complex with 5’¢ mRNA cap. J Biol Chem 266:2789–2794 [PubMed]
- Lefrere V, Duncan RF (1994) Heat shock-induced repression of proteolysis:poly(A)-binding protein degradation patterns can illusorily suggest its specific loss during heat shock. Nucleic Acids Res 22:1640–1642 [DOI] [PMC free article] [PubMed]
- Li D, Duncan RF (1995) Rapid turnover of heat shock protein 70 in Drosophila: stabilization during heat shock, and consequent effects on accumulation. Eur J Biochem 231:454–465 [DOI] [PubMed]
- Lindquist S (1980a) Varying patterns of protein synthesis in Drosophila during heat shock: implications for regulation. Dev Biol 77:463–479 [DOI] [PubMed]
- Lindquist S (1980b) Translational efficiency of heat-induced messages in Drosophila melanogaster cells. J Mol Biol 137:151–158 [DOI] [PubMed]
- Lindquist S (1986) The heat -shock response. Ann Rev Biochem 55:1151–1191 [DOI] [PubMed]
- Lindquist S, Petersen R (1990) Selective translation and degradation of heat-shock messenger RNAs in Drosophila. Enzyme 44:147–166 [DOI] [PubMed]
- McGarry TJ, Lindquist S (1985) The preferential translation of Drosophila hsp70 mRNA requires sequences in the untranslated leader. Cell 42:903–911 [DOI] [PubMed]
- Miron M, Verdu J, Lachance PE, Birnbaum MJ, Lasko PF, Sonenberg N (2001) The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat Cell Biol 3:596–601 [DOI] [PubMed]
- Miron M, Lasko PF, Sonenberg N (2003) Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol Cell Biol 23:9117–9126 [DOI] [PMC free article] [PubMed]
- Morley SJ, Naegele S (2002) Phosphorylation of initiation factor (eIF) 4E is not required for de novo protein synthesis following recovery from hypertonic stress in human kidney cells. J Biol Chem 277:32855–32859 [DOI] [PubMed]
- O’Farrell PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021 [PMC free article] [PubMed]
- Panniers R, Stewart EB, Merrick WC, Henshaw EC (1985) Mechanism of inhibition of polypeptide chain initiation in heat-shocked Ehrlich cells involves reduction of eukaryotic initiation factor 4F activity. J Biol Chem 260:9648–9653 [PubMed]
- Parsell DA, Lindquist S (1993) The function of heat -shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27:437–496 [DOI] [PubMed]
- Raught B, Gingras AC, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N (2000) Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J 19:434–444 [DOI] [PMC free article] [PubMed]
- Raught B, Peiretti F, Gingras A-C, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JWB (2004) Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 23:1761–1769 [DOI] [PMC free article] [PubMed]
- Rhoads RE, Lamphear BJ (1995) Cap-independent translation of heat shock messenger RNAs. Curr Top Microbiol Immunol 203:131–153 [DOI] [PubMed]
- Ritossa FM (1962) A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 18:571–573 [DOI]
- Song H-J, Gallie DR, Duncan RF (1995) m7GpppG cap dependence for efficient translation of Drosophila HSP70 mRNA. Eur J Biochem 232:778–788 [DOI] [PubMed]
- Storti RV, Scott MP, Rich A, Pardue ML (1980) Translational control of protein synthesis in response to heat shock in D. melanogaster cells. Cell 22:825–834 [DOI] [PubMed]
- Tarun SZ Jr., Sachs AB (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15:7168–7177 [PMC free article] [PubMed]
- Tissieres A, Mitchell HK, Tracy WM (1974) Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol 84:389–398 [DOI] [PubMed]
- Yueh A, Schneider RJ (2000) Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev 14:414–421 [PMC free article] [PubMed]
- Zapata JM, Maroto FG, Sierra JM (1991) Inactivation of mRNA cap-binding protein complex in Drosophila melanogaster embryos under heat shock. J Biol Chem 266:16007–16014 [PubMed]
- Zapata JM, Martinez MA, Sierra JM (1994) Purification and characterization of eukaryotic polypeptide chain initiation factor 4F from Drosophila melanogaster embryos. J Biol Chem 269:18047–18052 [PubMed]