Abstract
The cellular heat stress response is well studied in Drosophila in respect to the role of heat shock proteins (Hsp). Hsps are molecular chaperones, highly expressed during and after exposure to numerous stress types. Hsps are all regulated by a common transcription factor, the heat shock factor (HSF), and it is known that HSF is controlling other, so far uncharacterised, heat-responsive genes. In this study, we investigate whether novel candidate genes for heat resistance, identified by microarray experiments, are regulated by HSF. The microarray experiments recently identified several strongly upregulated genes in response to a short, non-lethal heat treatment in Drosophila melanogaster. To test whether or not a subset of these genes are HSF-induced, we studied 11 currently unannotated genes using quantitative polymerase chain reaction on HSF mutant flies with a non-functional HSF at elevated temperatures. We found indication of HSF regulation in most of the studied genes, suggesting a role of these unknown genes in heat tolerance. Surprisingly, some of the genes seemed to be upregulated independent of HSF function. The high induction in response to heat, which mimics the expression profile of Hsps, implies a role in the cellular heat response of these genes as well.
Introduction
All organisms have a universal cellular stress response that protects the cells from the damaging effects of temperature and other stress factors. Heat shock proteins (Hsp) are an important part of this response and are considered prime candidates for heat resistance in most life forms, including the fruit fly Drosophila. The heat shock genes (hsp) are induced as a response to stress factors and protect the cells by stabilising and refolding proteins whose structure is disturbed by stress exposure (reviewed by Feder and Hofmann 1999; Sørensen et al. 2003).
Undoubtedly, Hsps are principle components of heat tolerance; however, Hsps cannot explain all aspects of this trait. Several reports indicate other genes to be involved in heat resistance. In Drosophila melanogaster, the level of the major Hsp contributing to heat resistance, Hsp70, drops much earlier than acquired thermotolerance (Dahlgaard et al. 1998), pointing towards the action of other, and so far unknown, protective mechanisms. Moreover, not all heat-resistance traits are correlated (Hoffmann et al. 1997; Bubliy and Loeschcke 2005) or can be associated with Hsp in Drosophila (Sørensen et al. 2001), indicating that heat resistance is a complex trait likely to involve multiple factors and genes.
All known hsps are regulated by the same transcription factor, the heat shock factor (HSF). HSF recognises and binds to specific sequences in the promoters of hsp and promotes expression of Hsps in response to stress (Parker and Topol 1984; Wu et al. 1987). Furthermore, HSF has been proposed as regulator of other heat-induced genes. Using immunoflourescence staining of Drosophila chromosomes from heat-shocked salivary gland squashes, Westwood et al. (1991) found that HSF, besides being associated with hsps at the heat-shock puff sites, also was associated with many other chromosomal loci scattered all over the chromosomes. Birch-Machin et al. (2005) later identified 188 HSF targets in D. melanogaster embryos, many of which corresponded to the cytological locations identified by Westwood et al. (1991). These genes are annotated to several different biological processes such as metabolism, gene expression, protein modification and degradation and, as expected, chaperone activity (Birch-Machin et al. 2005). Similarly in yeast, several gene targets are bound to HSF during heat shock, and their corresponding proteins define a broad range of biological functions as well (Hahn et al. 2004). These studies show that HSF is apparently controlling other unknown heat-inducible genes potentially involved in heat resistance.
Microarray studies are a high throughput method for identifying new candidate genes involved in protective responses to different stimuli as heat stress. A microarray study on D. melanogaster describing the heat shock response in temporal detail identified several genes not previously associated with heat stress (Sørensen et al. 2005). These genes were highly upregulated immediately after a short, non-lethal heat shock, mimicking the response profile of hsps. The high upregulation of these genes after heat shock suggests that these genes might be new candidate genes involved in heat resistance as well as potentially regulated by HSF.
An important first step in studying these new candidate genes in further detail is to elucidate how they are regulated in response to heat. We designed this study to investigate whether a subset of these new candidate genes is regulated in a manner similar to the Hsps, i.e. by HSF. We investigated unannotated genes identified by (Sørensen et al. 2005) as well as a cluster of upregulated genes all located at chromosome position 33B (2L), a site previously shown to be HSF associated (Westwood et al. 1991). To study the regulation of these genes, we used a temperature-sensitive HSF mutant line of D. melanogaster developed by Jedlicka et al. (1997) in which the HSF protein product is non-functional as a transcription factor at temperatures above 30°C due to a single amino acid change. Transcriptional activity of the genes following a short heat treatment was assayed in the mutant line and a rescued control line using real-time quantitative polymerase chain reaction (qPCR). Failure to induce the genes following heat shock by the HSF mutant implies HSF-mediated activation. If the genes are significantly induced in both lines, this will suggest heat-triggered regulation by an HSF-independent mechanism.
Materials and methods
Selection of genes
We selected unannotated genes from the microarray analysis by Sørensen et al. (2005) that were significantly upregulated more than twofold immediately after 1 h heat stress at 36°C and had a similar expression profile as known Hsps following a heat treatment: an immediate high induction following heat shock. Genes were denoted A–F (see Table 1 for gene symbols). Additionally, five significantly heat–induced genes, G–K; located at chromosome position 33B (2L) were chosen for analysis (Table 1). Of these, G, H and J were less than twofold induced in the microarray study.
Table 1.
Details on the investigated genes and the corresponding oligonucleotide primers used for qPCR
| Gene ID | Gene symbol | Primer sequence | Exon junction | Product size (bp) | FlyBase ID | |
|---|---|---|---|---|---|---|
| A | cg4269 | F | 5′-GAATGGCTAAGAATATGTTCA-3′ | 1–2 | 136 | FBgn0034741 |
| R | 5′-ACAGATTGCCGTGGTATTCGCTG-3′ | |||||
| B | cg6332 | F | 5′-TGCCCTTCGCTACAAGCCGAGT-3′ | 3–4 | 152 | FBgn0038921 |
| R | 5′-CGTGCTCGCCTTGTATTTGAGCG-3′ | |||||
| C | cg13227 | F | 5′-GAGAAGGAATACGACATCGAAATGGAT-3′ | – | 131 | FBgn0033589 |
| R | 5′-GAGCTTGGTTTCTCCTGCACCACT-3′ | |||||
| D | cg13664 | F | 5′-CGCCATTCTGGACTCACGTTGTT-3′ | – | 107 | FBgn0039294 |
| R | 5′-TGATCCGCAGCTTGCCGATTAT-3′ | |||||
| E | cg18331 | F | 5′-TCCACCACCTCATCTGACCCCG-3′ | – | 140 | FBgn0036181 |
| R | 5′-ACGACAGGAGCCGAAGAGGA-3′ | |||||
| F | cg32130 | F | 5′-GATGCTGACCCGCAACCTGCT-3′ | 3–4 | 106 | FBgn0086708 |
| R | 5′-ATCGAAGCCTGAATGACTTTGAT-3′ | |||||
| G | cg6734 | F | 5′-CTCATGTCCATTGCGGCTCTCTATG-3′ | 2–3 | 190 | FBgn0032395 |
| R | 5′-GTAGGTGCTCCATGATACTGGCGTC-3′ | |||||
| H | cg6766 | F | 5′-CGACAAGCCAGAGTTGTCTCCAATT-3′ | 2–3 | 189 | FBgn0032398 |
| R | 5′-TCATCGGTCCATTTGCCCAGGT-3′ | |||||
| I | cg6785 | F | 5′-TCTTTAGCTCCTAGCAGATCGCCTTA-3′ | 1–2 | 196 | FBgn0032399 |
| R | 5′-GATCCCAGATCTGCTTGAGGCTC-3′ | |||||
| J | cg6792 | F | 5′-CTTCAAGCTTAAGAACCCGCGAGCT-3′ | 1–2 | 194 | FBgn0032401 |
| R | 5′-GCCGTCGTAGGTGAGTGTCGTATTC-3′ | |||||
| K | cg14945 | F | 5′-CAAGGAGTTCCCAGTGGGTTTTGG-3′ | 1–2 | 215 | FBgn0032402 |
| R | 5′-CCACGGAACCAAACAGGACATCG-3′ | |||||
| – | Beta actin | F | 5′-ATGACGCACCGCGTGCAGTT-3′ | – | 109 | FBgn0000043 |
| R | 5′-CTCTGTGCCTCATCGCCGACAT-3′ |
F Forward primer, R reverse primer
Maintenance and origin of experimental flies
Two lines of D. melanogaster, created by Jedlicka et al. (1997) and obtained from the Bloomington Stock Center, were used. One line (HSF0; hsf4 in Jedlicka et al. 1997) is a heat-sensitive mutant line that has a mutation (V57M) in the hsf gene, resulting in a temperature-sensitive gene product which is non-functional as transcription factor above 30°C but functional at lower temperatures. The other line (HSF+) is a derived line with recovered HSF function. It has a P-element with a normal HSF gene inserted, and Hsps are induced in a normal manner (for details, see Jedlicka et al. 1997). On establishment, these lines were genetically identical except for the insertion of the rescuing hsf gene in the HSF+ line. Both lines were maintained on agar-sugar-yeast-oatmeal medium with a 12:12 h light/dark cycle at 25°C.
Induction of the stress response
Freshly emerged female flies of each line (HSF0 and HSF+) were collected under light CO2 anaesthesia and aged 2 days before onset of the experiment. Before the heat treatment, flies were transferred to 5-ml plastic vials with 18 flies per vial. The flies were exposed to 36°C for 1 h in a pre-heated water bath with the vials completely submerged in water (n = 3 vials for the HSF0 mutant line, n = 4 for the HSF+ line). One set of vials was not hardened but served as untreated controls (n = 5 for both lines). All flies were subsequently frozen in liquid nitrogen and stored at −55°C.
RNA extraction
Total RNA was extracted using RNeasy (QIAGEN) with on-column DNA digestion. Flies from each vial (18 females) were homogenised according to the manufacturer’s protocol using rotor-stator homogenisation. cDNA was synthesised using First-Strand cDNA synthesis Kit (Amersham Biosciences) with the first strand pd(N)6 primer provided by the kit. After synthesis, the cDNA was diluted 1:10 in H2O and stored at −20°C.
Primer design and lightcycler real-time PCR
Oligonucleotide primers for the target genes were designed to have annealing temperatures of 60°C (Table 1). When possible, primers were designed to span two exons to avoid amplification of genomic DNA. We used beta actin as reference gene. For the lightcycler reaction, a master mix was prepared as suggested by the manufacturer’s manual (Fast Start DNA Master SYBR Green I; Roche Diagnostics). Master mix (18 μl) was filled in the glass capillaries, and 2.0 μl cDNA added as PCR template. Capillaries were centrifuged and placed into the Lightcycler rotor (Roche, version 3.5). PCR products were quantified using the threshold cycle (Ct) calculated by the second derivate maximum values by the thermocycler analysis software.
Relative quantification and statistics
The ratio of target gene over the reference gene beta actin was calculated as the normalised expression to account for variability of initial concentrations of RNA and the efficiency of the cDNA synthesis. The ratio was calculated as 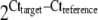 assuming primer efficiencies to be the same and optimal (=2) for both genes. The expression levels were
assuming primer efficiencies to be the same and optimal (=2) for both genes. The expression levels were 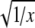 transformed to improve normality. t tests were performed to test for differences in expression levels using the statistical package PAST (Hammer et al. 2001). A sequential Bonferroni correction was applied to correct for multiple comparisons (Rice 1989).
transformed to improve normality. t tests were performed to test for differences in expression levels using the statistical package PAST (Hammer et al. 2001). A sequential Bonferroni correction was applied to correct for multiple comparisons (Rice 1989).
Results
Heat induction of genes
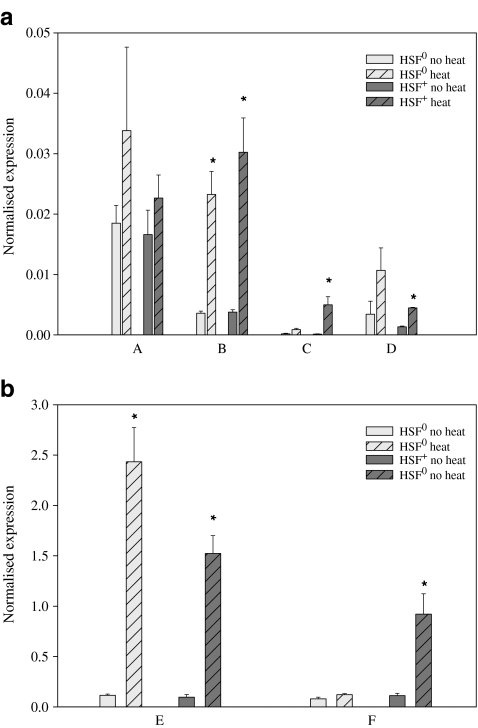
For the unannotated candidate genes, five out of the six investigated genes were significantly upregulated by heat exposure: B, C, D, E and F (Fig. 1a and b). Of these, C (P = 0.00810), D (P = 0.00007) and F (P = 0.00183) were only upregulated in the HSF+ control line and not in the HSF0 mutant line. B and E were significantly upregulated in both lines (BHSF0P = 0.00004, BHSF+P = 0.00017, EHSF0P = 0.00057, EHSF+P = 0.00002).
Fig. 1.
a and b The mean expression of genes A–F in HSF0 and HSF+ flies before and after a short heat treatment (±SE). An asterisk (*) denotes significant differences between heat-treated and untreated flies within that line after sequential Bonferroni correction. The genes are shown in two separate figures due to major differences in expression levels
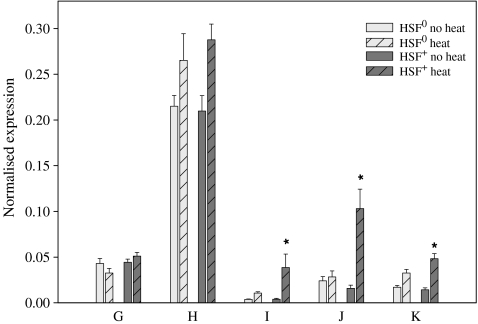
For the gene cluster at chromosome position 33B, three of the five genes were significantly upregulated by heat in the HSF+ control line and not in the HSF0 mutant line: I (P = 0.00148), J (P = 0.00087) and K (P = 0.00087; Fig. 2). Although not significant after Bonferroni correction, gene H shows a trend towards upregulation in both lines.
Fig. 2.
The mean expression of genes from chromosome location 33B in HSF0 and HSF+ flies before and after a short heat treatment (±SE). An asterisk (*) denotes significant differences between heat-treated and untreated flies within that line after sequential Bonferroni correction
Discussion
The results in this paper represent an important first step in the investigation of novel candidate genes of importance for heat resistance in D. melanogaster. We investigated the role of HSF in the heat induction of candidate genes using a temperature-sensitive HSF mutant of D. melanogaster. For 8 of 11 studied genes, results obtained from a microarray experiment by Sørensen et al. (2005) were verified, confirming a heat-induced upregulation of these genes. Despite the different genetic background of the fly strains used in the two studies, this verifies the strong correlation between Affymetrix gene chips and quantitative polymerase chain reaction (qPCR) shown by Park et al. (2004). It also demonstrates the usability of microarray studies in identifying candidate genes for further investigation.
Although all genes except cg6734 (G, Fig. 2) appear to be upregulated in both lines, significant upregulation was observed for the HSF+ control line only in six cases. This strongly suggests HSF-dependent regulation of these genes during heat shock. The heat induction of these genes coupled with HSF regulation points to a role for these genes in heat resistance. Further studies of these genes and their protein products are required to confirm such a role.
Due to the method of heat treatment, it is possible that a small time frame exists in which the rising temperature is sufficient to activate HSF and induce transcription yet insufficient to impair the function of HSF in the mutant line. Is not clear whether such conditions apply to the temperature-sensitive mutant HSF. However, if so, this could explain the observed non-significant increase in the mutant flies. Alternatively, additional factors acting independent of HSF and resulting in a more modest regulation could be responsible for the effect.
Three of the five genes clustering at chromosome position 33B seemed to be HSF controlled: I (cg6785), J (cg6792) and K (cg14945). This is in agreement with previous observations of HSF staining at this location following heat shock of polytene chromosomes (Westwood et al. 1991) and further supports that some of these genes are under transcriptional control by HSF and thus may have important functions contributing to heat resistance. Transcriptional start points of these three genes lie within close proximity (5 kb) which may suggest the presence of a common HSF-driven promoter region.
Especially interesting was the observation that two of the heat induced genes were induced equally in both HSF0 and HSF+ flies, implying heat-responsive regulation independent of the HSF system. The mechanism behind this could be induced by heat alone or by several stressors, like Hsps. Putative candidates for the latter are members of the mitogen-activated protein kinase (MAPK) family. In D. melanogaster, the MAPKs are activated in response to stress factors such as heat, oxidative stress and UV light, and thus seem to be part of a general stress response like Hsps (Botella et al. 2001; Inoue et al. 2001; Craig et al. 2004). Another candidate could be Menin, a recently identified regulator of the general stress response in D. melanogaster (Papaconstantinou et al. 2005). Menin itself is induced by stress and appears to be (part of) a regulator system independent of HSF (Papaconstantinou et al. 2005).
Since this study was initiated, gene F (cg32130, starvin) has been annotated for molecular function, revealing that this protein is indeed involved in protein binding and can be associated with apoptosis, proteolysis and peptidolysis (http://flybase.bio.indiana.edu/). The protein product contains a Bcl-2-associated athanogene (BAG) domain, also present in Hsp70 regulators, where it regulates chaperone activity through binding to the ATPase domain of Hsp70 (Doong et al. 2002). This protein function correlates well with the heat induction we observe here, and it is not surprising that the gene is regulated by the same transcription factor as its putative target gene. In addition, gene J (cg6792) has been annotated and is involved in transcription regulation activity and protein binding. The function of this gene in heat resistance remains to be investigated.
In the present study, we show that several heat-responsive genes appear to be HSF regulated, indicating these genes to be directly involved in the protective roles of the heat-shock response. Much less expected was the discovery that two of these heat-induced genes seem regulated by factors outside the HSF system. Computer analysis of the promoter regions using the MAPPER software (Marinescu et al. 2005) revealed HSF binding sites in five of the seven HSF-dependent genes but was not found in the two non-dependent genes, supporting our empirical data. Our results support other studies pointing towards a complicated heat-stress response consisting of numerous genes besides the well-characterised hsps. Future studies of these heat-responsive candidate genes will help broaden our picture of the universal stress response.
Acknowledgements
We thank Jesper Sørensen for assistance with the fly strains and Kamilla Pedersen, Torsten Kristensen, Cino Pertoldi and Pernille Sarup for helpful suggestions to the manuscript. The study was supported by a center grant from the Natural Sciences Research Council and a grant from the Novo Nordisk Foundation to VL. LTJ was supported by a stipend from the Faculty of Science, University of Aarhus.
References
- Birch-Machin I, Gao S, Huen D, McGirr R, White RAH, Russell S (2005) Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol 6:R63 [DOI] [PMC free article] [PubMed]
- Botella JA, Baines IA, Williams DD, Goberdhan DC, Proud CG, Wilson C (2001) The Drosophila cell shape regulator c-Jun N-terminal kinase also functions as stress-activated protein kinase. Insect Biochem Mol Biol 1:839–847 [DOI] [PubMed]
- Bubliy OA, Loeschcke V (2005) Correlated responses to selection for stress resistance and longevity in a laboratory population of Drosophila melanogaster. J Evol Biol 18:789–803 [DOI] [PubMed]
- Craig CR, Fink JL, Yagi Y, IP YT, Cagan RL (2004) Drosophila p38 orthologue is required for environmental stress responses. EMBO Rep 5:1058–1063 [DOI] [PMC free article] [PubMed]
- Dahlgaard J, Loeschcke V, Michalak P, Justesen J (1998) Induced thermotolerance and associated expression of the heat-shock protein Hsp70 in adult Drosophila melanogaster. Funct Ecol 12:786–793 [DOI]
- Doong H, Vrailas A, Kohn EC (2002) What’s in the ‘BAG’? – a functional domain analysis of the BAG-family proteins. Canc Lett 188:25–32 [DOI] [PubMed]
- Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu Rev Physiol 61:243–282 [DOI] [PubMed]
- Hahn J, Hu Z, Thiele DJ, Iyer VR (2004) Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24:5249–5256 [DOI] [PMC free article] [PubMed]
- Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electronica 4:9
- Hoffmann AA, Dagher H, Hercus M, Berrigan D (1997) Comparing different measures of heat resistance in selected lines of Drosophila melanogaster. J Insect Physiol 43:393–405 [DOI] [PubMed]
- Inoue H, Tateno M (2001) A Drosophila MAPKKK, D-MEKK1, mediates stress responses through activation of p38 MAPK. EMBO J 20:5421–5430 [DOI] [PMC free article] [PubMed]
- Jedlicka P, Mortin MA, Wu C (1997) Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J 16:2452–2462 [DOI] [PMC free article] [PubMed]
- Marinescu VD, Kohane IS, Riva A (2005) The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res 33:D91–97 (Database issue) [DOI] [PMC free article] [PubMed]
- Papaconstantinou M, Wu Y, Pretorius HN, Singh N, Gianfelice G, Tanguay RM, Campos AR, Bédard A (2005) Menin is a regulator of the stress response in Drosophila melanogaster. Mol Cell Biol 25:9960–9972 [DOI] [PMC free article] [PubMed]
- Park PJ, Cao YA, Lee SY, Kim J, Chang MS, Hart R, Choi S (2004) Current issues for DNA microarrays: platform comparison, double linear amplification, and universal RNA reference. J Biotechnol 112:225–245 [DOI] [PubMed]
- Parker C, Topol J (1984) A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp70 gene. Cell 37:273–283 [DOI] [PubMed]
- Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225 [DOI] [PubMed]
- Sørensen JG, Dahlgaard J, Loeschcke V (2001) Genetic variation in thermal tolerance among natural populations of Drosophila buzzatii: Down regulation of Hsp70 expression and variation in heat stress resistance traits. Funct Ecol 15:289–296 [DOI]
- Sørensen JG, Kristensen TN, Loeschcke V (2003) The evolutionary and ecological role of heat shock proteins. Ecol Lett 6:1025–1037 [DOI]
- Sørensen JG, Nielsen MM, Kruhøffer M, Justesen J, Loeschcke V (2005) Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones 10:312–328 [DOI] [PMC free article] [PubMed]
- Westwood JT, Clos J, Wu C (1991) Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature 353:822–827 [DOI] [PubMed]
- Wu C, Wilson S, Walker B, Dawid I, Paisley T, Zimarino V, Ueda H (1987) Purification and properties of Drosophila heat shock activator protein. Science 238:1247–1253 [DOI] [PubMed]