Abstract
In this manuscript, we describe the generation of a gene library for the expression of HSP110/HSPH, HSP70/HSPA and HSP40/DNAJ members. First, the heat shock protein (HSP) genes were collected from the gene databases and the gene families were analyzed for expression patterns, heat inducibility, subcellular localization, and protein homology using several bioinformatics approaches. These results can be used as a working draft model until data are confirmed by experimental approaches. In addition, we describe the generation of a HSPA/DNAJ overexpression library and tested the effect of different fusion tags on HSPA and DNAJ members using different techniques for measuring chaperone activity. These results show that we have cloned a high-quality heat shock protein expression library containing most members from the HSPH, HSPA, DNAJA and DNAJB families which will be useful for the chaperone community to unravel the function of the highly diverse family of human molecular chaperones.
Keywords: HspH, HspA, DnaJ, Hsp110, Hsp70, Hsp40, Human chaperones, Bio-informatics
Introduction
All organisms, except some hyperthermophilic archaea, contain the family of HSP110/HSPH, HSP70/HSPA, and HSP40/DNAJ chaperones (Gribaldo et al. 1999). HSPA and DNAJ proteins function as molecular chaperones to assist in processes such as translation and transport of proteins across membranes.
The HSPA machine consists of the core HSPA protein along with a transient array of different co-factors such as DNAJ, HSPH, BAG-1, Hip, CHIP, and HSPBP1 (Kampinga 2006). The HSPH/HSPA and DNAJ families are protein families consisting of many members and as a whole, the HSPH/HSPA/DNAJ gene family is the largest chaperone gene family found in humans.
It is thought that many of its members are specialized (Sahi and Craig 2007). For instance, members of the HSPA, DNAJ, and HSPH family exist that are only expressed under stress conditions suggesting that these are specialized to function in the proteotoxic stress response (Albanese et al. 2006). Constitutively expressed members are found as well and such members are found in several cellular compartments such as the cytosol, mitochondria and the endoplasmic reticulum (ER) suggesting that cellular compartmentalization has driven some of the HSPA/DNAJ gene expansion (Brocchieri et al. 2008). In addition, some members have only been found at specific developmental stages or in specific cell types indicating the need for specialized members for specific substrates expressed only during specific developmental stages or in certain specialized cell types.
In contrast to the gene expansion as a result of compartmentalization, the gene expansion as a result of cellular specialization or organism development is poorly understood. It has been suggested that HSPA and DNAJ proteins bind small hydrophobic regions (Rudiger et al. 1997; Rudiger et al. 2001); yet there is great diversity and multiplicity within these families of which most members have not yet been studied in detail.
Although the various chaperone genes are now relatively well annotated, the molecular function for most of its members is currently unknown. For each of the families, only a single or restricted number of proteins has been studied in detail. In this paper, we used bioinformatics approaches to study the different HSPH, HSPA, and DNAJ members (Brocchieri et al. 2007). Thereafter, we describe the construction of a human HSPH/HSPA/DNAJ expression library.
Materials and methods
Bioinformatics
HSP gene retrieval HSPH, HSPA and DNAJ genes were collected from National Center for Biotechnology Information (NCBI) Gene (Maglott et al. 2007). Mouse orthologs were identified using NCBI Homologene (Wheeler et al. 2007). Protein molecular weights were calculated using the clone manager 7 suite (Sci-Ed Software).
EST count analysis Expression data based on tissue-specific and developmental stage specific expressed sequence tag (EST) numbers were collected from the NCBI UniGene database (Wheeler et al. 2007). EST numbers are displayed as counts per million.
Affymetrix gene array Investigation of genome-wide heat-induced transcriptional activation was described previously (Page et al. 2006). These experiments were performed in Hela cells using a 1.5-h heat shock at 43°C. Recovery times were 0.5, 2, and 4 h at 37°C. Affymetrix gene array data were downloaded (Page et al. 2006) and linear induction was calculated from the 2log fold change. Affymetrix uses different annotations for its probe sets. _at suffix designates a unique probe set, whereas the _s_at and _x_at suffixes designate probe sets that can cross hybridize with multiple genes. In the case of redundant probe sets, _at suffix were selected by default. In the case of no available _at suffix, the first probe set was selected routinely.
Subcellular localization analysis Predictions on subcellular localizations were performed using pSort, pTarget, CELLO, Multiloc, and Proteome analyst (Szafron et al. 2004; Yu et al. 2006; Guda 2006; Hoglund et al. 2006; Horton et al. 2007). Sequences from complete gene families were uploaded as fasta files. In each case, only the first rank localization is displayed. For all predictors, the default settings for mammalian or animal proteins were used. The presence of prenelation motifs was determined using the PrePS webserver (Maurer-Stroh et al. 2007).
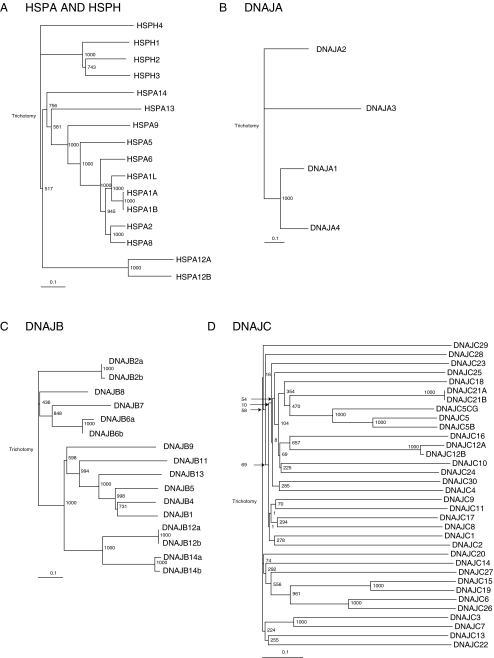
Protein Alignments Primary amino acid alignments were performed in ClustalX2 using the neighbor-joining algorithm and Blosum matrixes at the default settings (Larkin et al. 2007). Bootstrap analysis was performed using 1,000 random number generator seeds and 1,000 bootstrap trials. Phylograms were made by importing the homology tree output of ClustalX in TreeView (Page 1996). The distance is depicted in the scale bar of Fig. 1 as 0.1 amino acids substitutions per position.
Fig. 1.
Phylograms for the HSPH/A (A), DNAJA (B), DNAJB (C), and DNAJC (D) families. Primary amino acid alignments were performed using the Neighbor-joining algorithm using a Blosum scoring matrix in ClustalX (see “Methods” for details). Bootstrap values are indicated on the branch-points
Library cloning and validation
Gene Cloning
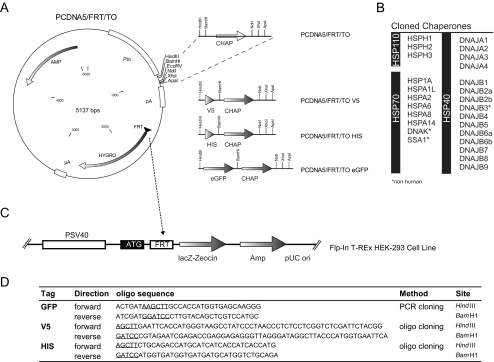
Detailed information about the plasmids used in this study can be found in Fig. 2. Briefly, tetracycline-inducible HSP expression plasmids were constructed as follows. First, the green fluorescent protein (GFP) and the v5 tag, harboring a Kozak consensus ATG initiation codon and lacking a stop codon, were cloned in the pCDNA5 FLP recombination target (FRT)/TO vector (Invitrogen). Subsequently, the coding sequence of the different chaperones was amplified using the primers listed in supplemental Table 8. As a template source, complementary DNA (cDNA) was made from total RNA as previously described (Hageman et al. 2005). As a source of total RNA, QPCR Human reference Total RNA (Stratagene) was used, which is a mixed source of RNA from the following cell line derivations: adenocarcinoma, mammary gland; hepatoblastoma, liver; adenocarcinoma, cervix; embryonal carcinoma, testis; glioblastoma, brain; melanoma, skin; liposarcoma, histiocytic lymphoma, macrophage, histocyte; lymphoblastic leukemia, T lymphoblast; plasmacytoma, myeloma, B lymphocyte. DNAJB4, DNAJB5, and DNAJB8 were amplified from cloned full-length cDNAs purchased from Open Biosystems (clone ID: DNAJB4: 4340658, DNAJB5: 4684829, and DNAJB8: 5296554). The fragments were cloned in pCDNA5 frt to GFP lacking a stop codon resulting in a N-GFP-cDNA-C protein. The presence of the correct gene was sequence verified. Protein expression was verified by Western blotting. Subsequently, fragments were subcloned to pCDNA5 frt to v5 and pCDNA5 frt to.
Fig. 2.
Schematic overview of the library construction. (A) The pCDNA5/FRT/TO vector system together with the cloned fusion tags. (B) List of the cloned molecular chaperones. (C) Schematic representation of the FRT locus within the Flp-In T-REx HEK-293 cell line. (D) Primer sequences for the construction of the indicated fusion tags
Table 8.
Summary of the library cloning
| Family | Gene | 5′-oligo | 3′-oligo | Site 1 | Site 2 |
|---|---|---|---|---|---|
| HSP110 | HSPH1 | CTCCCAGGGTTTCTTATCAG | GATTTTAATCACAGCCCTCTTG | NA | NA |
| HSPH1* | CACAGATATCACCATGTCGGTGGTGGGGTTG | CGCGATCCTCGAGCTAGTCCAAGTCCATATTAACAG | EcoRV | XhoI | |
| HSPH2 | ACCCACTGGAAGGACTTAGG | GAGCTCCTGCCATGTAAGTC | NA | NA | |
| HSPH2* | GACAGATATCACCATGTCGGTGGTGGGCATAGAC | GACTGCGGCCGCGGAATCAATCAATGTCCATTTCAG | EcoRV | NotI | |
| HSPH3 | GCAATAGCCCAGAAGAGGAC | GATGGACCCCGTGGTTACTTG | NA | NA | |
| HSPH3* | GACGGATATCACCATGTCTGTGGTTGGCATTGAC | GATCGCGGCCGCAGACTTAGTCCACTTCCATCTC | EcoRV | NotI | |
| HSP70 | HSPA1A | ACCAGAGGATCCACCATGGCCAAAGCCGCGGCGAT | ATCACTGCGGCCGCCTAATCTACCTCCTCAATGG | BamHI | NotI |
| HSPA1L | CACAGATATCACCATGGCTACTGCCAAGGGAAT | GACTGCGGCCGCTTAATCTACTTCTTCAATTGTGGGGC | EcoRV | NotI | |
| HSPA2 | CACACAGGATCCACCATGTCTGCCCGTGGCCCGGCT | GACTGCGGCCGCTTAGTCCACTTCTTCGATGGTGG | BamHI | NotI | |
| HSPA6 | GACAGATATCACCATGCAGGCCCCACGGGAGCT | GACTGCGGCCGCTCAATCAACCTCCTCAATGA | EcoRV | No I | |
| HSPA8 | CACACAGGATCCACCATGTCCAAGGGACCTGCAGTTG | GACTGCGGCCGCTTAATCAACCTCTTCAATGGTGGG | BamHI | NotI | |
| HSPA14 | CACACAGGATCCACCATGGCGGCCATCGGAGTTCA | GACTGCGGCCGCCTAAGATGCTATCTCAATAGAGATTG | BamHI | NotI | |
| DNAK | ACCAATGGATCCACCATGGGTAAAATAATTGGTATC | AATAATGCGGCCGCCCCGTGTCAGTATAATTACC | BamHI | NotI | |
| SSA1 | TACTAAGGATCCACCATGTCAAAAGCTGTCGGTATTG | TAGTATGCGGCCGCTTAATCAACTTCTTCAACGGTTGGACC | BamHI | NotI | |
| HSP40 | DNAJA1 | CACAATGGATCCACCATGGTGAAAGAAACAACTTACTACG | GACTGCGGCCGCTTAAGAGGTCTGACACTGAAC | BamHI | NotI |
| DNAJA2 | ATCCACGGATCCACCATGGCTAACGTGGCTGACACG | GACTGCGGCCGCTTACTGATGGGCACACTGCAC | BamHI | NotI | |
| DNAJA3 | ATTCGAGGATCCACCATGGCTGCGCGGTGCTCCACA | GACTGCGGCCGCGGCTGGGATATCATGAGGTA | BamHI | NotI | |
| DNAJA4 | ATAGCTGGATCCACCATGGTGAAGGAGACCCAGTA | GACTGCGGCCGCTCATGCCGTCTGGCACTGCAC | BamHI | NotI | |
| DNAJB1 | CACAATGGATCCACCATGGGTAAAGACTACTACCAGACG | GACTGCGGCCGCCTATATTGGAAGAACCTGCTCAAG | BamHI | NotI | |
| DNAJB2a | ATCGATGGATCCACCATGGCATCCTACTACGAGATC | TACGATGCGGCCGCTCAGAACACATCTGCGGGTTTC | BamHI | NotI | |
| DNAJB2b | ATCGATGGATCCACCATGGCATCCTACTACGAGATC | TACGATGCGGCCGCTCAGAGGATGAGGCAGCGAG | BamHI | NotI | |
| DNAJB3 | TACTACGGATCCACCATGGTGGACTACTACGAGGT | TACTGTGCGGCCGCTTACTGAGTATTGATGCGAAGCAG | BamHI | NotI | |
| DNAJB4 | TGCAAAGGATCCACCATGGGGAAAGACTATTATTGC | GACTGCGGCCGCCTATGAGGCAGGAAGATGTTTCC | BamHI | NotI | |
| DNAJB5 | GATCGCGGCCGCACCATGGGAAAAGATTATTACAAGATTCTTGGG | GATATCGGGCCCCTAGGAACAGGGTAGGTGCTGC | NotI | ApaI | |
| DNAJB6b | GATATAGGATCCGGAACCATGGTGGATTACTATGAAGTTCT | GATATTGCGGCCGCTTACTTGTTATCCAAGCGCAG | BamHI | NotI | |
| DNAJB6a | TATATAGGATCCACCATGGTGGATTACTATGAAGTTCT | TATATAGCGGCCGCCTAGTGATTGCCTTTGGTCG | BamHI | NotI | |
| DNAJB7 | GATTACGATATCACCATGGTGGATTACTATGAAGT | GATTACGCGGCCGCTTAACAATTCCTTTTGGTAGACTTC | EcoRV | NotI | |
| DNAJB8 | AAGTAAGGATCCACCATGGCTAACTACTACGAAGTG | GATATAGCGGCCGCCTACTTGCTGTCCATCCATTTG | BamHI | NotI | |
| DNAJB9 | GATCGCGGCCGCACCATGGCTACTCCCCAGTCAAT | GATATCGGGCCCCTACTGTCCTGAACAGTCAG | NotI | ApaI | |
| DNAJB10 | ATCGATGGATCCACCATGGCATCCTACTACGAGATTC | TACGATGCGGCCGCTCAGAACACATCTGCTGGCTTC | BamHI | NotI |
Luciferase refolding Assay Cell lysis and luciferase activity measurements were done as previously described (Michels et al. 1995). Luciferase activity was plotted relative to the percentage of activity in an unheated control. Error bars on plots represent standard deviations.
Filter trap assay To determine protein aggregates, the filter trap assay was performed as previously described (Carra et al. 2005). Briefly, 10, 2, and 0.4 µg of protein extracts were applied onto 0.2-µm pore cellulose acetate membrane pre-washed with FTA + 0.1% sodium dodecyl sulfate (SDS). Mild suction was applied and the membrane was washed three times. Aggregated proteins trapped in the membrane were probed with a mouse anti-GFP antibody JL-8 (Clontech) at a 1:5,000 dilution or a mouse anti-V5 antibody (Invitrogen) at a 1:5,000 dilution followed by horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (Amersham) at 1:5,000 dilution. Visualisation was performed using enhanced chemiluminescence and Hyperfilm (ECL, Amersham).
Results
Bioinformatic analysis on the HSPH, HSPA, and DNAJ gene family
Collecting the HSPH, HSPA, and DNAJ gene family
In order to get a comprehensive overview of the gene family, we first extracted all human HSPH, HSPA, and DNAJ family members from the NCBI gene database. It should be noted that beside these protein encoding genes, we found many pseudogenes scattered throughout the human genome. Typically, pseudogenes show types of decay such as frame-shifts, stop-codons or gaps. For the HSPA family alone, already 30 pseudogenes have been documented (Brocchieri et al. 2008). For gene selection, we extracted the annotated non-pseudo genes from the NCBI gene bank (Maglott et al. 2007). We found 4 HSPH chaperones, 13 HSPA chaperones, and 49 DNAJ chaperones. The genes including the protein name, the old and alternative names, the NCBI human gene ID, the mouse ortholog gene ID, the human gene locus, the protein length, and the calculated molecular mass are listed in Table 1. Classically, human chaperones were ranked according to molecular mass. However, as can be seen from Table 1, many HSP genes deviate from this and contain only a HSP-like domain such as the HSP70 ATPase domain or the HSP40 DNAJ domain. For instance, while many HSP40/DNAJ proteins are around 40 kD, the sizes of proteins within this family range from 16 kD (DNAJC15) to 254 kD (DNAJC13). For this reason, a revised nomenclature was recently suggested (this issue of Cell Stress and Chaperones).
Table 1.
Overview of the human HSP70/HSP40 gene family
| Gene Name | Protein Name | Alternative Name | Human GeneID | Mouse ortholog ID | Locus (human) | Protein length (aa) | Calculated Mass (kD) | |
|---|---|---|---|---|---|---|---|---|
| HSPH | HSPH1 | HSPH1 | HSP105 | 10808 | 15505 | 13q12.3 | 858 | 96.9 |
| HSPH2 | HSPA4 | HSPA4, APG-2; HSP110 | 3308 | 15525 | 5q31.1–q31.2 | 840 | 94.3 | |
| HSPH3 | HSPA4L | HSPA4L, APG-1 | 22824 | 18415 | 4q28 | 839 | 94.5 | |
| HSPH4 | HSPH4 | HYOU1; GRP170 | 10525 | 12282 | 11q23.1 | 999 | 111.3 | |
| HSPA | HSPA1A | HSPA1A | HSP70–1, HSP72, HSPA1 | 3303 | 193740 | 6p21.3 | 641 | 70.0 |
| HSPA1B | HSPA1B | HSP70–2 | 3304 | 15511 | 6p21.3 | 641 | 70.0 | |
| HSPA1L | HSPA1L | hum70t, hum70t | 3305 | 15482 | 6p21.3 | 641 | 70.4 | |
| HSPA2 | HSPA2 | Heat-shock 70kD protein-2 | 3306 | 15512 | 14q24.1 | 639 | 70.0 | |
| HSPA5 | HSPA5 | BIP, GRP78, MIF2 | 3309 | 14828 | 9q33-q34.1 | 654 | 71.0 | |
| HSPA6 | HSPA6 | heat shock 70kD protein 6 (HSP70B’) | 3310 | X | 1q23 | 643 | 71.0 | |
| HSPA7 | HSPA7 | 3311 | X | 1q23.3 | ? | ? | ||
| HSPA8 | HSPA8 | HSC70, HSC71, HSP71, HSP73 | 3312 | 15481 | 11q24.1 | 646/493 | 70.9/53.5 | |
| HSPA9 | HSPA9 | GRP75, HSPA9B, MOT, MOT2, PBP74, mot-2 | 3313 | 15526 | 5q31.1 | 679 | 73.7 | |
| HSPA12A | HSPA12A | FLJ13874, KIAA0417 | 259217 | 73442 | 10q26.12 | 1296 | 141.0 | |
| HSPA12B | HSPA12B | RP23–32L15.1, 2700081N06Rik | 116835 | 72630 | 20p13 | 686 | 75.7 | |
| HSPA13 | HSPA13 | Stch | 6782 | 110920 | 21q11 | 471 | 51.9 | |
| HSPA14 | HSPA14 | HSP70–4, HSP70L1, MGC131990 | 51182 | 50497 | 10p14 | 509 | 54.8 | |
| DNAJA | DNAJA1 | DNAJA1 | DJ-2; DjA1; HDJ2; HSDJ; HSJ2; HSPF4; hDJ-2 | 3301 | 15502 | 9p13–p12 | 397 | 44.9 |
| DNAJA2 | DNAJA2 | DNJ3; mDj3; DNAJ3; HIRIP4 | 10294 | 56445 | 16q11.1–q11.2 | 412 | 45.7 | |
| DNAJA3 | DNAJA3 | Tid-1; Tid1l | 9093 | 83945 | 16p13.3 | 480 | 52.5 | |
| DNAJA4 | DNAJA4 | Dj4; Hsj4 | 55466 | 58233 | 15q24.1 | 397 | 44.7 | |
| DNAJB | DNAJB1 | DNAJB1 | HSPF1; HSP40 | 3337 | 81489 | 19p13.2 | 340 | 38.2 |
| DNAJB2 | DNAJB2 | HSJ1; HSPF3; DNAJB10 | 3300 | 56812 | 2q32–q34 | 324/277 | 35,6/30,6 | |
| DNAJB3 | DNAJB3 | Hsj3; Msj1; MSJ-1 | 414061 | 15504 | 1 D (Mm) | 242 | 26.7 | |
| DNAJB4 | DNAJB4 | Hsc40 | 11080 | 67035 | 1p31.1 | 337 | 37.8 | |
| DNAJB5 | DNAJB5 | Hsc40; HSP40-3 | 25822 | 56323 | 9p13.2 | 348/241 | 39,1/26,9 | |
| DNAJB6 | DNAJB6 | Mrj; mDj4 | 10049 | 23950 | 7q36.3 | 326/241 | 36.1 | |
| DNAJB7 | DNAJB7 | Dj5; mDj5 | 150353 | 57755 | 22q13.2 | 309 | 35.4 | |
| DNAJB8 | DNAJB8 | mDj6 | 165721 | 56691 | 3q21.3 | 232 | 25.7 | |
| DNAJB9 | DNAJB9 | Mdg1; mDj7; ERdj4 | 4189 | 27362 | 7q31 | 223 | 25.5 | |
| DNAJB11 | DNAJB11 | Dj9; ABBP-2; ERdj3 | 51726 | 67838 | 3q28 | 358 | 40.5 | |
| DNAJB12 | DNAJB12 | Dj10; mDj10 | 54788 | 56709 | 10q22.2 | 375 | 41.9 | |
| DNAJB13 | DNAJB13 | Tsarg | 374407 | 69387 | 11q13.4 | 316 | 36.1 | |
| DNAJB14 | DNAJB14 | EGNR9427; FLJ14281 | 79982 | 70604 | 4q23 | 379/294 | 42,5/33,5 | |
| DNAJC | DNAJC1 | DNAJC1 | MTJ1; ERdj1; ERj1p; DNAJl1 | 64215 | 13418 | 0p12.33–p12.32 | 554 | 63.9 |
| DNAJC2 | DNAJC2 | Zrf1; Zrf2; MIDA1 | 27000 | 22791 | 7q22 | 621 | 72.0 | |
| DNAJC3 | DNAJC3 | p58; mp58; Prkri; DNAJc3; p58IPK; DNAJc3b | 5611 | 100037258 | 13q32 | 504 | 57.6 | |
| DNAJC4 | DNAJC4 | HSPf2; Mcg18 | 3338 | 57431 | 11q13 | 135 | 15.2 | |
| DNAJC5 | DNAJC5 | Csp | 80331 | 13002 | 20q13.33 | 198 | 22.1 | |
| DNAJC5B | DNAJC5B | CSP-beta | 85479 | 66326 | 8q12.3 | 199 | 22.5 | |
| DNAJC5G | DNAJC5G | MGC107182 | 285126 | 231098 | 2p23.3 | 189 | 21.4 | |
| DNAJC6 | DNAJC6 | mKIAA0473 | 9829 | 72685 | 1pter-q31.3 | 913 | 100.0 | |
| DNAJC7 | DNAJC7 | Ttc2; mDj11; mTpr2 | 7266 | 56354 | 17q11.2 | 484 | 55.5 | |
| DNAJC8 | DNAJC8 | AL024084; AU019262 | 22826 | 68598 | 1p35.2 | 264 | 31.0 | |
| DNAJC9 | DNAJC9 | AU020082 | 23234 | 108671 | 10q22.3 | 260 | 29.9 | |
| DNAJC10 | DNAJC10 | JPDI; ERdj5 | 54431 | 66861 | 2q32.1 | 793 | 91.1 | |
| DNAJC11 | DNAJC11 | FLJ10737 | 55735 | 230935 | 1p36.23 | 559 | 63.3 | |
| DNAJC12 | DNAJC12 | Jdp1; mJDP1 | 56521 | 30045 | 10q22.1 | 198/107 | 23,4/12,5 | |
| DNAJC13 | DNAJC13 | Rme8; RME-8; Gm1124 | 23317 | 235567 | 3q22.1 | 2243 | 254.4 | |
| DNAJC14 | DNAJC14 | HDJ3; LIP6; DRIP78 | 85406 | 74330 | 12q13.13 | 702 | 78.6 | |
| DNAJC15 | DNAJC15 | DNAJd1 | 29103 | 66148 | 13q14.1 | 150 | 16.4 | |
| DNAJC16 | DNAJC16 | mKIAA0962 | 23341 | 214063 | 1p36.1 | 782 | 90.6 | |
| DNAJC17 | DNAJC17 | C87112 | 55192 | 69408 | 15q15.1 | 304 | 34.7 | |
| DNAJC18 | DNAJC18 | MGC29463 | 202052 | 76594 | 5q31.2 | 358 | 41.5 | |
| DNAJC19 | DNAJC19 | TIM14; TIMM14 | 131118 | 67713 | 3q26.33 | 116 | 12.5 | |
| DNAJC20 | DNAJC20 | JAC1; HSC20 | 150274 | 100900 | 22q12.1 | 235 | 27.4 | |
| DNAJC21 | DNAJC21 | GS3; JJJ1; DNAJA5; | 134218 | 78244 | 5p13.2 | 576/531 | 67,1/62,0 | |
| DNAJC22 | DNAJC22 | FLJ13236; Wurst | 79962 | 72778 | 12q13.12 | 341 | 38.1 | |
| DNAJC23 | DNAJC23 | Sec63; AI649014 | 11231 | 140740 | 6q21 | 760 | 88.0 | |
| DNAJC24 | DNAJC24 | DPH4; zinc finger, CSL-type containing 3 | 120526 | 99349 | 11p13 | 149 | 17.1 | |
| DNAJC25 | DNAJC25 | bA16L21.2.1; DnaJ-like protein | 548645 | X | 9q31.3 | 360 | 42.4 | |
| DNAJC26 | DNAJC26 | GAK; cyclin G associated kinase | 2580 | 231580 | 4p16 | 1311 | 143.2 | |
| DNAJC27 | DNAJC27 | RBJ; Rabj | 51277 | 217378 | 2p23.3 | 273 | 30.9 | |
| DNAJC28 | DNAJC28 | Orf28 open reading frame 28; C21orf55 | 54943 | 246738 | 1q25 | 454 | 51.1 | |
| DNAJC29 | DNAJC29 | SACS; Sacsin | 26278 | 50720 | 13q12 | 4432 | 504.6 | |
| DNAJC30 | DNAJC30 | WBSCR18 | 84277 | 66114 | 7q11.23 | 226 | 26.0 |
HSPA6 and HSPA7 were found only in humans while and although HSPA7 contains an internal frame shift and might be a pseudogene; bypassing the frame shift will result in a protein with a full-length homology to HSPA1A. Thus, the HSPA7 protein with a full-length homology to HSPA1A might be produced, and this has recently been explained elsewhere (Brocchieri et al. 2008).
Patterns of tissue specific HSPH, HSPA, and DNAJ expression
Cellular specialization may require specialized chaperone proteins and therefore may be responsible for part of the chaperone gene expansion. The expression pattern of most chaperone genes is currently unknown. An estimation of the expression pattern can be made by assessing the relative number of EST per tissue using the Unigene database (Schuler 1997). However, some caution should be taken as the Unigene assignments of ESTs to individual genes is not necessarily accurate (i.e., poor sequence quality and related sequences lead to incorrect ‘binning’ of some ESTs).
The expression estimates are displayed in Table 2 (HSPH/HSPA), Table 3 (DNAJA/B), and Table 4 (DNAJC). The peak expression for each tissue is indicated in bold. As expected, HSPA8 shows a high expression in most tissues (Table 2). In contrast, HSPH3, HSPA1L, HSPA6, HSPA7, HSPA12A, and HSPA12B show very low levels in most tissues. HSPH3 and HSPA1L show the highest expression in the testis, which is in agreement with literature (Ito et al. 1998; Held et al. 2006). HSPA6 is absent under non-stress conditions and only expressed upon severe heat shock conditions (Noonan et al. 2007). The expression of HSPA1A is extremely variable, ranging from being absent in lymph (node), parathyroid, and umbilical cord to being expressed at very high levels in the spleen. As for the HSPH/HSPA family, the DNAJ family shows a highly variable expression profile (Tables 3 and 4). The highest expressed members throughout tissues are DNAJA1, DNAJB1, and DNAJB6 indicative of being “housekeeping” DNAJ chaperones although they all are lacking in a few tissues. As for the HSPA/HSPH families, the DNAJ family shows testis-specific members (DNAJB7, DNAJB8, DNAJC5B, and DNAJC5G). In general, with the exception of the testis, most HSP genes do not show an expression restricted to only a single tissue. In addition, peak levels per tissue are highly variable from gene to gene, providing no direct correlative clue for any specific partnerships between the diverse family members.
Table 2.
Expression levels of HspH and HspA genes in various human tissues
| HSPH1 | HSPH2 | HSPH3 | HSPH4 | HSPA1A | HSPA1B | HSPA1L | HSPA2 | HSPA5 | HSPA6 | HSPA7 | HSPA8 | HSPA9 | HSPA12A | HSPA12B | HSPA13 | HSPA14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adipose tissue | 152 | 76 | 0 | 76 | 684 | 608 | 0 | 0 | 608 | 76 | 0 | 989 | 76 | 76 | 0 | 0 | 76 |
| Adrenal gland | 180 | 120 | 0 | 210 | 1740 | 390 | 0 | 60 | 360 | 90 | 0 | 3780 | 510 | 90 | 0 | 90 | 30 |
| Ascites | 99 | 199 | 24 | 124 | 49 | 49 | 0 | 0 | 648 | 0 | 0 | 2720 | 923 | 0 | 0 | 24 | 24 |
| Bladder | 199 | 0 | 66 | 33 | 663 | 1161 | 0 | 0 | 199 | 99 | 33 | 1990 | 431 | 0 | 0 | 232 | 99 |
| Blood | 120 | 24 | 8 | 88 | 555 | 80 | 0 | 8 | 161 | 112 | 8 | 3069 | 386 | 0 | 0 | 40 | 40 |
| Bone | 125 | 153 | 0 | 97 | 153 | 0 | 0 | 0 | 501 | 13 | 27 | 1267 | 320 | 0 | 13 | 55 | 55 |
| Bone marrow | 142 | 61 | 0 | 447 | 40 | 81 | 0 | 40 | 610 | 20 | 0 | 1688 | 447 | 0 | 0 | 40 | 20 |
| Brain | 278 | 50 | 34 | 231 | 850 | 223 | 5 | 634 | 120 | 9 | 3 | 3672 | 338 | 87 | 8 | 130 | 67 |
| Cervix | 82 | 123 | 20 | 103 | 144 | 103 | 0 | 20 | 247 | 0 | 0 | 1257 | 453 | 0 | 20 | 103 | 82 |
| Connective tissue | 80 | 60 | 6 | 120 | 347 | 40 | 13 | 26 | 267 | 13 | 6 | 2145 | 227 | 26 | 0 | 60 | 26 |
| Ear | 183 | 61 | 0 | 0 | 734 | 0 | 0 | 122 | 122 | 0 | 0 | 367 | 61 | 61 | 0 | 0 | 0 |
| Embryonic tissue | 115 | 180 | 0 | 189 | 92 | 37 | 0 | 13 | 671 | 0 | 0 | 1315 | 278 | 13 | 0 | 83 | 92 |
| Esophagus | 246 | 98 | 49 | 689 | 2710 | 1724 | 0 | 246 | 1182 | 98 | 49 | 1921 | 542 | 0 | 0 | 49 | 197 |
| Eye | 61 | 109 | 23 | 85 | 180 | 56 | 14 | 42 | 185 | 14 | 14 | 1043 | 137 | 42 | 0 | 94 | 42 |
| heart | 55 | 33 | 11 | 55 | 1638 | 365 | 11 | 66 | 188 | 33 | 11 | 2170 | 232 | 22 | 66 | 33 | 22 |
| Intestine | 127 | 131 | 4 | 233 | 1028 | 165 | 0 | 72 | 416 | 16 | 8 | 3552 | 271 | 4 | 4 | 33 | 29 |
| Kidney | 159 | 65 | 37 | 301 | 889 | 221 | 9 | 192 | 122 | 9 | 4 | 4628 | 348 | 56 | 42 | 94 | 18 |
| Larynx | 81 | 0 | 0 | 81 | 245 | 81 | 0 | 0 | 613 | 0 | 0 | 695 | 81 | 0 | 0 | 0 | 81 |
| Liver | 62 | 48 | 9 | 302 | 316 | 72 | 14 | 4 | 326 | 0 | 0 | 1238 | 273 | 0 | 0 | 33 | 33 |
| Lung | 124 | 97 | 23 | 198 | 1475 | 162 | 8 | 17 | 210 | 76 | 2 | 1174 | 239 | 0 | 17 | 50 | 20 |
| Lymph | 112 | 90 | 0 | 112 | 0 | 0 | 0 | 0 | 45 | 0 | 0 | 1373 | 315 | 0 | 0 | 0 | 0 |
| Lymph node | 185 | 108 | 0 | 10 | 0 | 10 | 0 | 0 | 174 | 0 | 0 | 566 | 152 | 0 | 0 | 87 | 217 |
| Mammary gland | 71 | 155 | 6 | 1237 | 589 | 97 | 25 | 12 | 719 | 12 | 6 | 2280 | 563 | 6 | 0 | 71 | 58 |
| Mouth | 295 | 191 | 29 | 280 | 1372 | 132 | 0 | 162 | 132 | 0 | 14 | 1918 | 575 | 0 | 0 | 29 | 103 |
| Muscle | 147 | 83 | 27 | 166 | 258 | 55 | 0 | 27 | 110 | 9 | 0 | 2949 | 342 | 27 | 0 | 27 | 27 |
| Nerve | 316 | 63 | 0 | 253 | 2658 | 253 | 0 | 0 | 379 | 0 | 63 | 1202 | 189 | 379 | 0 | 0 | 63 |
| Ovary | 38 | 87 | 0 | 136 | 58 | 87 | 0 | 0 | 428 | 19 | 0 | 1558 | 87 | 29 | 19 | 48 | 38 |
| Pancreas | 69 | 78 | 0 | 130 | 380 | 157 | 0 | 4 | 390 | 18 | 0 | 808 | 125 | 0 | 0 | 27 | 27 |
| Parathyroid | 48 | 0 | 48 | 0 | 0 | 0 | 0 | 0 | 48 | 0 | 0 | 484 | 436 | 48 | 0 | 0 | 339 |
| Pharynx | 0 | 48 | 0 | 48 | 24 | 0 | 0 | 24 | 96 | 0 | 0 | 2144 | 144 | 0 | 0 | 24 | 24 |
| Pituitary gland | 597 | 59 | 0 | 59 | 119 | 0 | 59 | 59 | 537 | 59 | 0 | 1732 | 179 | 119 | 0 | 0 | 59 |
| Placenta | 88 | 84 | 10 | 119 | 165 | 17 | 0 | 225 | 306 | 0 | 56 | 908 | 221 | 14 | 56 | 77 | 70 |
| Prostate | 41 | 115 | 15 | 120 | 1131 | 193 | 5 | 20 | 366 | 10 | 5 | 1560 | 146 | 20 | 0 | 36 | 47 |
| Salivary gland | 0 | 147 | 0 | 49 | 0 | 0 | 0 | 147 | 49 | 0 | 0 | 394 | 98 | 0 | 0 | 49 | 49 |
| Skin | 127 | 189 | 0 | 146 | 222 | 56 | 0 | 146 | 174 | 18 | 4 | 2622 | 411 | 18 | 9 | 14 | 47 |
| Spleen | 36 | 18 | 0 | 55 | 6992 | 1405 | 18 | 92 | 110 | 73 | 0 | 5512 | 295 | 18 | 110 | 18 | 18 |
| Stomach | 205 | 432 | 0 | 72 | 586 | 154 | 0 | 41 | 483 | 30 | 10 | 2038 | 123 | 0 | 10 | 0 | 20 |
| Testis | 365 | 93 | 223 | 374 | 193 | 42 | 105 | 543 | 262 | 0 | 3 | 2079 | 322 | 21 | 9 | 172 | 78 |
| Thymus | 135 | 36 | 12 | 110 | 1821 | 406 | 0 | 12 | 36 | 159 | 0 | 4270 | 258 | 0 | 12 | 49 | 61 |
| Thyroid | 166 | 41 | 20 | 229 | 500 | 312 | 20 | 0 | 1835 | 0 | 0 | 1335 | 584 | 104 | 20 | 0 | 62 |
| Tonsil | 58 | 117 | 0 | 58 | 58 | 0 | 0 | 58 | 0 | 0 | 0 | 469 | 0 | 0 | 0 | 0 | 234 |
| Trachea | 381 | 19 | 152 | 991 | 2345 | 801 | 0 | 57 | 38 | 190 | 57 | 3318 | 209 | 0 | 19 | 247 | 38 |
| Umbilical cord | 0 | 0 | 0 | 798 | 0 | 0 | 0 | 726 | 145 | 0 | 0 | 1597 | 145 | 0 | 72 | 0 | 0 |
| Uterus | 89 | 115 | 17 | 200 | 722 | 273 | 8 | 111 | 585 | 8 | 4 | 2381 | 290 | 21 | 8 | 81 | 34 |
| Vascular | 173 | 0 | 19 | 924 | 962 | 57 | 0 | 19 | 173 | 0 | 0 | 8356 | 423 | 38 | 0 | 115 | 0 |
Number of transcripts per million are indicated.
Table 3.
Expression levels of DnaJA and DnaJB genes in various human tissues
| DNAJA1 | DNAJA2 | DNAJA3 | DNAJA4 | DNAJB1 | DNAJB2 | DNAJB4 | DNAJB5 | DNAJB6 | DNAJB7 | DNAJB8 | DNAJB9 | DNAJB11 | DNAJB12 | DNAJB13 | DNAJB14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adipose tissue | 532 | 76 | 76 | 0 | 1065 | 76 | 152 | 0 | 152 | 0 | 0 | 76 | 0 | 152 | 0 | 76 |
| Adrenal gland | 180 | 60 | 30 | 0 | 210 | 0 | 30 | 60 | 180 | 0 | 0 | 0 | 0 | 120 | 0 | 30 |
| Ascites | 224 | 124 | 149 | 0 | 374 | 74 | 0 | 0 | 324 | 0 | 0 | 0 | 49 | 74 | 0 | 49 |
| Bladder | 165 | 66 | 66 | 99 | 199 | 66 | 99 | 0 | 132 | 0 | 0 | 33 | 0 | 33 | 0 | 0 |
| Blood | 169 | 96 | 88 | 32 | 128 | 24 | 32 | 8 | 161 | 0 | 0 | 32 | 56 | 32 | 0 | 32 |
| Bone | 125 | 194 | 69 | 0 | 125 | 83 | 27 | 27 | 125 | 0 | 0 | 27 | 41 | 27 | 0 | 0 |
| Bone marrow | 183 | 20 | 61 | 20 | 244 | 20 | 122 | 0 | 122 | 0 | 0 | 0 | 40 | 40 | 0 | 0 |
| Brain | 264 | 62 | 115 | 54 | 191 | 84 | 72 | 45 | 221 | 0 | 3 | 66 | 31 | 58 | 0 | 26 |
| Cervix | 268 | 20 | 82 | 82 | 164 | 20 | 20 | 0 | 82 | 0 | 0 | 41 | 82 | 20 | 0 | 0 |
| Connective tissue | 93 | 26 | 53 | 20 | 93 | 26 | 53 | 6 | 200 | 0 | 0 | 60 | 26 | 20 | 6 | 20 |
| Ear | 122 | 0 | 0 | 0 | 61 | 0 | 0 | 0 | 61 | 0 | 0 | 0 | 122 | 0 | 0 | 0 |
| Embryonic tissue | 305 | 46 | 55 | 0 | 120 | 41 | 32 | 37 | 236 | 0 | 0 | 18 | 74 | 27 | 0 | 50 |
| Esophagus | 98 | 98 | 246 | 49 | 246 | 49 | 344 | 0 | 344 | 0 | 0 | 0 | 0 | 147 | 0 | 0 |
| Eye | 208 | 52 | 23 | 42 | 227 | 94 | 23 | 42 | 170 | 4 | 0 | 33 | 66 | 33 | 0 | 37 |
| Heart | 143 | 44 | 33 | 88 | 265 | 22 | 166 | 55 | 199 | 0 | 0 | 99 | 121 | 66 | 0 | 0 |
| Intestine | 165 | 123 | 80 | 29 | 237 | 33 | 12 | 12 | 191 | 0 | 0 | 29 | 42 | 63 | 0 | 25 |
| Kidney | 202 | 23 | 47 | 14 | 197 | 42 | 117 | 32 | 225 | 0 | 0 | 117 | 18 | 84 | 0 | 18 |
| Larynx | 0 | 163 | 0 | 40 | 0 | 40 | 0 | 163 | 204 | 0 | 0 | 0 | 40 | 0 | 0 | 0 |
| Liver | 196 | 96 | 38 | 33 | 177 | 48 | 144 | 9 | 172 | 0 | 0 | 38 | 38 | 24 | 4 | 14 |
| Lung | 171 | 47 | 50 | 70 | 275 | 79 | 23 | 17 | 162 | 0 | 0 | 47 | 62 | 59 | 2 | 41 |
| Lymph | 90 | 22 | 90 | 0 | 67 | 90 | 22 | 0 | 157 | 0 | 0 | 0 | 112 | 22 | 0 | 0 |
| Lymph node | 76 | 43 | 32 | 54 | 76 | 32 | 0 | 10 | 76 | 10 | 0 | 10 | 32 | 65 | 0 | 152 |
| Mammary gland | 129 | 90 | 90 | 71 | 362 | 25 | 25 | 12 | 317 | 0 | 0 | 45 | 38 | 58 | 0 | 51 |
| Mouth | 29 | 88 | 118 | 88 | 88 | 44 | 14 | 0 | 354 | 0 | 0 | 29 | 14 | 29 | 0 | 14 |
| Muscle | 203 | 120 | 101 | 27 | 166 | 55 | 92 | 64 | 166 | 0 | 0 | 27 | 18 | 83 | 0 | 55 |
| Nerve | 189 | 0 | 126 | 189 | 696 | 189 | 189 | 0 | 63 | 0 | 0 | 63 | 63 | 126 | 0 | 0 |
| Ovary | 136 | 87 | 107 | 9 | 185 | 48 | 9 | 38 | 116 | 0 | 0 | 9 | 19 | 19 | 9 | 0 |
| Pancreas | 116 | 27 | 51 | 41 | 199 | 65 | 13 | 4 | 176 | 0 | 0 | 51 | 65 | 55 | 0 | 55 |
| Parathyroid | 0 | 0 | 0 | 48 | 145 | 0 | 0 | 48 | 48 | 0 | 0 | 0 | 48 | 96 | 0 | 0 |
| Pharynx | 578 | 48 | 48 | 24 | 120 | 0 | 0 | 0 | 506 | 0 | 0 | 24 | 24 | 72 | 0 | 96 |
| Pituitary gland | 478 | 59 | 0 | 119 | 298 | 119 | 59 | 59 | 179 | 0 | 0 | 59 | 0 | 0 | 0 | 59 |
| Placenta | 133 | 91 | 49 | 10 | 186 | 66 | 31 | 3 | 154 | 0 | 0 | 169 | 116 | 56 | 0 | 63 |
| Prostate | 99 | 62 | 52 | 26 | 235 | 52 | 10 | 57 | 115 | 0 | 0 | 47 | 104 | 68 | 10 | 20 |
| Salivary gland | 98 | 49 | 197 | 0 | 98 | 0 | 147 | 0 | 98 | 0 | 0 | 98 | 0 | 197 | 0 | 0 |
| Skin | 170 | 61 | 70 | 75 | 226 | 61 | 42 | 51 | 396 | 0 | 0 | 9 | 42 | 61 | 0 | 23 |
| Spleen | 129 | 92 | 55 | 18 | 591 | 18 | 0 | 18 | 295 | 0 | 0 | 55 | 55 | 55 | 0 | 36 |
| Stomach | 205 | 20 | 61 | 41 | 102 | 41 | 41 | 0 | 236 | 0 | 0 | 51 | 102 | 10 | 0 | 10 |
| Testis | 277 | 114 | 63 | 51 | 546 | 30 | 24 | 18 | 253 | 24 | 39 | 87 | 27 | 48 | 15 | 15 |
| Thymus | 12 | 49 | 49 | 24 | 184 | 36 | 73 | 0 | 196 | 0 | 0 | 73 | 24 | 24 | 0 | 12 |
| Thyroid | 208 | 146 | 62 | 20 | 104 | 125 | 41 | 20 | 104 | 20 | 0 | 20 | 146 | 20 | 0 | 0 |
| Tonsil | 880 | 0 | 58 | 0 | 58 | 117 | 0 | 0 | 0 | 0 | 0 | 0 | 58 | 176 | 0 | 0 |
| Trachea | 190 | 57 | 19 | 247 | 228 | 19 | 19 | 0 | 152 | 0 | 0 | 114 | 0 | 19 | 38 | 19 |
| Umbilical cord | 72 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 145 | 0 | 0 |
| Uterus | 286 | 111 | 25 | 42 | 324 | 38 | 47 | 34 | 252 | 0 | 0 | 21 | 34 | 55 | 12 | 25 |
| Vascular | 481 | 38 | 38 | 0 | 211 | 0 | 423 | 0 | 288 | 0 | 0 | 77 | 0 | 19 | 0 | 0 |
Number of transcripts per million are indicated
Table 4.
Expression levels of DnaJC genes in various human tissues
| DNAJC1 | DNAJC2 | DNAJC3 | DNAJC4 | DNAJC5 | DNAJC5B | DNAJC5G | DNAJC6 | DNAJC7 | DNAJC8 | DNAJC9 | DNAJC10 | DNAJC11 | DNAJC12 | DNAJC13 | DNAJC14 | DNAJC15 | DNAJC16 | DNAJC17 | DNAJC18 | DNAJC19 | DNAJC20 | DNAJC21 | DNAJC22 | DNAJC23 | DNAJC24 | DNAJC25 | DNAJC26 | DNAJC27 | DNAJC28 | DNAJC29 | DNAJC30 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adipose tissue | 0 | 0 | 0 | 76 | 0 | 0 | 0 | 0 | 152 | 152 | 0 | 0 | 228 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 304 | 0 | 0 | 152 | 0 | 0 | 76 | 0 |
| Adrenal gland | 60 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | 210 | 210 | 30 | 30 | 90 | 0 | 30 | 60 | 30 | 0 | 0 | 0 | 60 | 0 | 30 | 0 | 30 | 30 | 60 | 60 | 30 | 0 | 0 | 0 |
| Ascites | 24 | 74 | 0 | 24 | 49 | 0 | 0 | 0 | 249 | 199 | 24 | 49 | 124 | 74 | 24 | 349 | 24 | 49 | 0 | 24 | 0 | 24 | 24 | 24 | 49 | 0 | 24 | 99 | 0 | 0 | 0 | 0 |
| Bladder | 0 | 33 | 33 | 0 | 33 | 0 | 0 | 0 | 66 | 132 | 66 | 0 | 0 | 0 | 99 | 99 | 0 | 0 | 33 | 0 | 33 | 0 | 0 | 0 | 0 | 33 | 166 | 66 | 0 | 0 | 0 | 0 |
| Blood | 24 | 40 | 56 | 0 | 80 | 0 | 0 | 0 | 64 | 499 | 16 | 8 | 32 | 0 | 8 | 161 | 0 | 16 | 0 | 8 | 0 | 8 | 40 | 0 | 32 | 24 | 64 | 96 | 0 | 8 | 16 | 40 |
| Bone | 111 | 27 | 0 | 27 | 69 | 0 | 0 | 0 | 69 | 181 | 27 | 97 | 55 | 41 | 55 | 222 | 41 | 0 | 0 | 0 | 0 | 13 | 13 | 0 | 83 | 13 | 69 | 55 | 0 | 13 | 13 | 13 |
| Bone marrow | 0 | 20 | 40 | 40 | 0 | 0 | 0 | 20 | 81 | 183 | 20 | 61 | 20 | 122 | 20 | 40 | 0 | 61 | 0 | 0 | 101 | 0 | 0 | 0 | 162 | 61 | 101 | 0 | 0 | 0 | 20 | 0 |
| Brain | 16 | 15 | 18 | 21 | 64 | 4 | 1 | 231 | 84 | 54 | 17 | 73 | 114 | 19 | 31 | 58 | 40 | 15 | 20 | 119 | 70 | 3 | 17 | 6 | 39 | 9 | 53 | 73 | 22 | 2 | 65 | 24 |
| Cervix | 0 | 82 | 0 | 0 | 20 | 0 | 0 | 0 | 61 | 164 | 20 | 0 | 123 | 82 | 61 | 82 | 20 | 0 | 20 | 0 | 0 | 0 | 20 | 0 | 103 | 20 | 41 | 41 | 0 | 0 | 0 | 41 |
| Connective tissue | 66 | 0 | 6 | 46 | 53 | 6 | 0 | 0 | 33 | 233 | 33 | 100 | 33 | 106 | 113 | 200 | 167 | 13 | 6 | 20 | 6 | 6 | 20 | 0 | 60 | 0 | 80 | 33 | 6 | 0 | 26 | 13 |
| Ear | 61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 61 | 122 | 183 | 0 | 0 | 0 | 0 | 61 | 0 | 0 | 0 | 61 | 0 | 0 | 0 | 489 | 0 | 61 | 0 | 0 | 0 | 0 | 61 |
| Embryonic tissue | 37 | 41 | 9 | 23 | 50 | 0 | 4 | 4 | 88 | 189 | 32 | 157 | 78 | 0 | 27 | 115 | 9 | 13 | 13 | 23 | 37 | 4 | 41 | 0 | 74 | 27 | 32 | 64 | 9 | 4 | 74 | 4 |
| Esophagus | 0 | 0 | 197 | 0 | 0 | 0 | 0 | 0 | 147 | 49 | 49 | 197 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 49 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Eye | 28 | 14 | 4 | 75 | 137 | 0 | 0 | 23 | 109 | 128 | 23 | 33 | 47 | 9 | 28 | 132 | 52 | 47 | 9 | 28 | 28 | 9 | 14 | 0 | 66 | 0 | 37 | 42 | 71 | 0 | 28 | 23 |
| Heart | 22 | 22 | 0 | 44 | 33 | 0 | 0 | 11 | 44 | 177 | 33 | 33 | 55 | 0 | 66 | 55 | 121 | 11 | 0 | 0 | 55 | 22 | 33 | 0 | 55 | 0 | 11 | 22 | 33 | 0 | 0 | 66 |
| Intestine | 84 | 29 | 16 | 93 | 123 | 0 | 0 | 12 | 67 | 131 | 72 | 38 | 59 | 16 | 25 | 67 | 72 | 50 | 0 | 0 | 127 | 12 | 21 | 33 | 50 | 12 | 50 | 135 | 8 | 0 | 25 | 25 |
| Kidney | 61 | 37 | 28 | 112 | 18 | 0 | 0 | 18 | 61 | 94 | 37 | 61 | 122 | 18 | 28 | 65 | 94 | 32 | 4 | 23 | 75 | 32 | 14 | 32 | 70 | 14 | 127 | 61 | 37 | 0 | 32 | 32 |
| Larynx | 122 | 40 | 0 | 122 | 81 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 163 | 0 | 0 | 40 | 0 | 0 | 0 | 81 | 0 | 900 | 0 | 0 | 0 | 0 | 0 | 0 | 81 |
| Liver | 72 | 95 | 14 | 0 | 38 | 0 | 0 | 4 | 62 | 134 | 19 | 148 | 24 | 24 | 24 | 100 | 28 | 9 | 14 | 0 | 19 | 4 | 38 | 0 | 81 | 9 | 67 | 19 | 0 | 0 | 23 | 28 |
| Lung | 65 | 32 | 17 | 133 | 109 | 2 | 0 | 14 | 59 | 174 | 50 | 65 | 65 | 20 | 17 | 168 | 29 | 35 | 14 | 5 | 32 | 26 | 23 | 2 | 106 | 5 | 85 | 73 | 32 | 2 | 29 | 35 |
| Lymph | 22 | 22 | 45 | 22 | 90 | 0 | 0 | 45 | 337 | 157 | 0 | 22 | 878 | 45 | 22 | 22 | 22 | 0 | 0 | 0 | 0 | 45 | 0 | 0 | 0 | 0 | 22 | 22 | 0 | 0 | 22 | 0 |
| Lymph node | 0 | 32 | 21 | 130 | 119 | 0 | 0 | 0 | 21 | 76 | 43 | 272 | 32 | 0 | 87 | 43 | 87 | 152 | 21 | 0 | 43 | 0 | 10 | 0 | 43 | 0 | 32 | 141 | 0 | 0 | 32 | 32 |
| Mammary gland | 25 | 58 | 19 | 45 | 116 | 12 | 0 | 6 | 97 | 64 | 19 | 90 | 90 | 19 | 32 | 155 | 38 | 0 | 25 | 6 | 51 | 0 | 38 | 19 | 136 | 19 | 64 | 64 | 6 | 0 | 12 | 25 |
| Mouth | 14 | 0 | 14 | 59 | 0 | 0 | 0 | 0 | 29 | 44 | 0 | 88 | 162 | 0 | 147 | 147 | 29 | 14 | 44 | 0 | 14 | 0 | 0 | 0 | 74 | 0 | 74 | 59 | 0 | 0 | 0 | 0 |
| Muscle | 0 | 0 | 36 | 18 | 110 | 0 | 0 | 18 | 18 | 83 | 0 | 46 | 147 | 18 | 27 | 92 | 36 | 36 | 18 | 46 | 92 | 0 | 27 | 0 | 55 | 18 | 18 | 36 | 27 | 0 | 46 | 0 |
| Nerve | 0 | 0 | 0 | 0 | 63 | 0 | 0 | 253 | 0 | 0 | 63 | 63 | 0 | 0 | 0 | 63 | 0 | 63 | 0 | 0 | 0 | 0 | 0 | 0 | 63 | 0 | 63 | 126 | 0 | 0 | 0 | 0 |
| Ovary | 29 | 38 | 9 | 87 | 77 | 0 | 0 | 0 | 38 | 224 | 19 | 29 | 48 | 0 | 38 | 107 | 0 | 29 | 0 | 9 | 9 | 0 | 38 | 19 | 38 | 9 | 9 | 136 | 29 | 0 | 0 | 38 |
| Pancreas | 37 | 4 | 4 | 55 | 74 | 0 | 0 | 9 | 46 | 65 | 4 | 102 | 32 | 69 | 18 | 69 | 46 | 4 | 0 | 0 | 27 | 4 | 27 | 13 | 51 | 51 | 204 | 46 | 51 | 0 | 4 | 27 |
| Parathyroid | 193 | 0 | 0 | 290 | 0 | 0 | 0 | 0 | 48 | 581 | 0 | 48 | 0 | 387 | 145 | 0 | 96 | 0 | 0 | 0 | 48 | 0 | 0 | 0 | 48 | 0 | 48 | 242 | 0 | 0 | 0 | 96 |
| Pharynx | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 24 | 963 | 0 | 24 | 24 | 0 | 48 | 192 | 48 | 48 | 0 | 0 | 0 | 0 | 24 | 0 | 0 | 24 | 72 | 0 | 0 | 24 | 24 | 0 |
| Pituitary gland | 0 | 0 | 0 | 59 | 59 | 0 | 0 | 59 | 0 | 0 | 0 | 59 | 0 | 0 | 119 | 0 | 119 | 0 | 0 | 59 | 179 | 0 | 0 | 0 | 0 | 0 | 59 | 179 | 0 | 0 | 0 | 0 |
| Placenta | 49 | 17 | 31 | 21 | 17 | 10 | 0 | 42 | 66 | 151 | 7 | 38 | 59 | 0 | 42 | 98 | 42 | 31 | 24 | 7 | 35 | 7 | 17 | 3 | 56 | 3 | 77 | 80 | 7 | 0 | 3 | 3 |
| Prostate | 41 | 26 | 10 | 94 | 94 | 0 | 0 | 0 | 115 | 89 | 15 | 52 | 78 | 10 | 41 | 89 | 73 | 15 | 5 | 5 | 335 | 10 | 41 | 0 | 146 | 20 | 83 | 52 | 26 | 0 | 0 | 26 |
| Salivary gland | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 147 | 0 | 0 | 0 | 0 | 197 | 98 | 49 | 0 | 0 | 49 | 0 | 147 | 0 | 0 | 0 | 0 | 49 | 147 | 295 | 0 | 0 | 0 | 0 |
| Skin | 66 | 47 | 14 | 28 | 85 | 0 | 0 | 14 | 56 | 160 | 42 | 70 | 66 | 9 | 103 | 151 | 33 | 4 | 4 | 4 | 28 | 14 | 23 | 0 | 28 | 0 | 75 | 42 | 0 | 0 | 14 | 18 |
| Spleen | 36 | 73 | 18 | 36 | 55 | 0 | 0 | 0 | 92 | 73 | 0 | 110 | 203 | 0 | 0 | 18 | 55 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 36 | 0 | 18 | 36 | 0 | 0 | 18 | 36 |
| Stomach | 41 | 30 | 10 | 30 | 82 | 0 | 0 | 10 | 113 | 123 | 10 | 82 | 51 | 0 | 41 | 41 | 30 | 0 | 51 | 10 | 0 | 0 | 92 | 10 | 41 | 0 | 72 | 41 | 20 | 10 | 0 | 41 |
| Testis | 18 | 72 | 21 | 66 | 30 | 45 | 39 | 9 | 57 | 81 | 6 | 338 | 141 | 12 | 60 | 162 | 39 | 33 | 6 | 147 | 33 | 3 | 15 | 3 | 102 | 21 | 48 | 87 | 72 | 18 | 9 | 18 |
| Thymus | 73 | 36 | 0 | 12 | 0 | 0 | 0 | 0 | 36 | 12 | 12 | 123 | 110 | 0 | 24 | 159 | 12 | 12 | 0 | 24 | 12 | 0 | 36 | 0 | 36 | 0 | 0 | 123 | 12 | 0 | 0 | 0 |
| Thyroid | 62 | 20 | 0 | 41 | 20 | 0 | 0 | 0 | 125 | 20 | 0 | 83 | 62 | 0 | 41 | 83 | 0 | 41 | 104 | 0 | 41 | 62 | 41 | 0 | 125 | 0 | 41 | 62 | 20 | 0 | 0 | 0 |
| Tonsil | 58 | 0 | 0 | 58 | 0 | 0 | 0 | 0 | 58 | 117 | 0 | 58 | 293 | 0 | 0 | 293 | 58 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Trachea | 38 | 76 | 114 | 0 | 19 | 0 | 0 | 0 | 57 | 38 | 0 | 648 | 133 | 0 | 57 | 38 | 19 | 38 | 38 | 0 | 19 | 0 | 0 | 0 | 209 | 0 | 0 | 114 | 0 | 19 | 38 | 19 |
| Umbilical cord | 290 | 0 | 72 | 0 | 0 | 0 | 0 | 0 | 145 | 0 | 0 | 217 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 145 | 0 | 0 | 0 | 0 | 0 | 72 | 0 |
| Uterus | 34 | 55 | 8 | 4 | 55 | 0 | 0 | 0 | 111 | 149 | 12 | 205 | 85 | 4 | 42 | 175 | 29 | 8 | 4 | 29 | 21 | 4 | 12 | 8 | 76 | 12 | 89 | 34 | 4 | 0 | 76 | 17 |
| Vascular | 19 | 19 | 57 | 0 | 19 | 0 | 0 | 19 | 115 | 57 | 0 | 269 | 19 | 0 | 115 | 38 | 77 | 0 | 0 | 0 | 96 | 0 | 19 | 19 | 19 | 0 | 57 | 0 | 0 | 0 | 77 | 57 |
Number of transcripts per million are indicated
Patterns of developmental expression
We also used the Unigene database to look for developmental specific expression patterns (Table 5). The results show that HSPs are differentially expressed at different developmental stages. HSPA1A shows a large variation throughout different developmental stages. Interestingly, many DNAJC members are expressed during embryogenesis but are repressed in the neonate or infant (DNAC4-6 and DNAJC16-20). Only a minority of the genes were expressed the highest at adult stages.
Table 5.
Expression levels of Hsp genes at various developmental stages
| Embryoid body | Blastocyst | Fetus | Neonate | Infant | Juvenile | Adult | |
|---|---|---|---|---|---|---|---|
| HSPH1 | 254 | 32 | 165 | 160 | 168 | 394 | 127 |
| HSPH2 | 98 | 208 | 61 | 0 | 42 | 89 | 132 |
| HSPH3 | 0 | 0 | 15 | 0 | 42 | 0 | 13 |
| HSPH4 | 254 | 224 | 181 | 224 | 0 | 538 | 185 |
| HSPA1A | 84 | 80 | 713 | 448 | 168 | 896 | 274 |
| HSPA1B | 70 | 32 | 155 | 128 | 42 | 215 | 105 |
| HSPA1L | 0 | 0 | 14 | 0 | 0 | 0 | 1 |
| HSPA2 | 0 | 32 | 24 | 320 | 0 | 35 | 67 |
| HSPA5 | 819 | 529 | 174 | 64 | 126 | 269 | 438 |
| HSPA6 | 0 | 0 | 7 | 64 | 0 | 0 | 17 |
| HSPA7 | 0 | 0 | 1 | 0 | 0 | 0 | 12 |
| HSPA8 | 1102 | 1283 | 2208 | 6509 | 8999 | 7371 | 2014 |
| HSPA9 | 339 | 288 | 211 | 577 | 337 | 789 | 265 |
| HSPA12A | 14 | 32 | 22 | 32 | 253 | 107 | 28 |
| HSPA12B | 0 | 0 | 8 | 64 | 0 | 0 | 13 |
| HSPA13 | 84 | 96 | 77 | 0 | 84 | 0 | 54 |
| HSPA14 | 56 | 64 | 52 | 0 | 253 | 35 | 52 |
| DNAJA1 | 197 | 304 | 140 | 64 | 844 | 358 | 158 |
| DNAJA2 | 56 | 80 | 51 | 64 | 84 | 89 | 86 |
| DNAJA3 | 56 | 80 | 40 | 0 | 42 | 125 | 52 |
| DNAJA4 | 0 | 0 | 75 | 0 | 0 | 53 | 45 |
| DNAJB1 | 127 | 96 | 155 | 320 | 211 | 430 | 236 |
| DNAJB2 | 42 | 48 | 47 | 0 | 0 | 53 | 77 |
| DNAJB4 | 56 | 16 | 38 | 256 | 42 | 107 | 36 |
| DNAJB5 | 28 | 80 | 82 | 0 | 168 | 0 | 24 |
| DNAJB6 | 127 | 336 | 147 | 416 | 253 | 251 | 207 |
| DNAJB7 | 0 | 0 | 3 | 0 | 0 | 17 | 0 |
| DNAJB8 | 0 | 0 | 5 | 0 | 0 | 0 | 2 |
| DNAJB9 | 28 | 0 | 22 | 0 | 126 | 89 | 57 |
| DNAJB11 | 42 | 128 | 51 | 32 | 0 | 17 | 66 |
| DNAJB12 | 42 | 16 | 33 | 64 | 42 | 0 | 51 |
| DNAJB13 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| DNAJB14 | 42 | 48 | 26 | 64 | 42 | 17 | 38 |
| DNAJC1 | 70 | 0 | 61 | 128 | 0 | 107 | 44 |
| DNAJC2 | 56 | 64 | 33 | 32 | 42 | 17 | 35 |
| DNAJC3 | 28 | 0 | 22 | 32 | 0 | 17 | 17 |
| DNAJC4 | 28 | 0 | 58 | 0 | 0 | 0 | 46 |
| DNAJC5 | 84 | 80 | 31 | 0 | 0 | 35 | 79 |
| DNAJC5B | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| DNAJC5G | 0 | 16 | 0 | 0 | 0 | 0 | 2 |
| DNAJC6 | 14 | 0 | 58 | 0 | 0 | 71 | 27 |
| DNAJC7 | 70 | 64 | 63 | 128 | 0 | 107 | 90 |
| DNAJC8 | 127 | 192 | 105 | 160 | 84 | 161 | 131 |
| DNAJC9 | 42 | 32 | 36 | 32 | 126 | 17 | 23 |
| DNAJC10 | 240 | 144 | 73 | 160 | 84 | 358 | 72 |
| DNAJC11 | 56 | 128 | 72 | 64 | 42 | 35 | 52 |
| DNAJC12 | 0 | 0 | 40 | 0 | 0 | 0 | 30 |
| DNAJC13 | 28 | 16 | 77 | 0 | 42 | 0 | 47 |
| DNAJC14 | 155 | 160 | 81 | 32 | 0 | 35 | 115 |
| DNAJC15 | 14 | 16 | 59 | 0 | 0 | 0 | 50 |
| DNAJC16 | 0 | 16 | 12 | 0 | 0 | 53 | 26 |
| DNAJC17 | 0 | 32 | 5 | 0 | 0 | 0 | 14 |
| DNAJC18 | 14 | 64 | 118 | 0 | 42 | 0 | 16 |
| DNAJC19 | 14 | 64 | 84 | 0 | 0 | 17 | 68 |
| DNAJC20 | 14 | 0 | 21 | 0 | 0 | 0 | 8 |
| DNAJC21 | 70 | 48 | 36 | 32 | 0 | 0 | 35 |
| DNAJC22 | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| DNAJC23 | 70 | 80 | 81 | 64 | 0 | 143 | 94 |
| DNAJC24 | 28 | 32 | 7 | 0 | 0 | 0 | 6 |
| DNAJC25 | 56 | 16 | 56 | 64 | 0 | 0 | 67 |
| DNAJC26 | 56 | 96 | 47 | 0 | 0 | 35 | 69 |
| DNAJC27 | 0 | 16 | 51 | 0 | 0 | 0 | 13 |
| DNAJC28 | 0 | 16 | 14 | 0 | 0 | 0 | 1 |
| DNAJC29 | 70 | 112 | 75 | 64 | 0 | 17 | 14 |
| DNAJC30 | 14 | 0 | 47 | 0 | 0 | 35 | 22 |
Number of transcripts per million
Heat-induced transcription
Although HSPs were originally identified as heat inducible proteins, most members are identified according to presence of typical domains such as the HSP70 ATPase domain or the HSP40 DNAJ domain. For most of these members, it is currently unknown whether they are induced by heat. To investigate this, we used Affymetrix gene array data (Page et al. 2006) and performed a biased search on the heat inducibility for HSPH, HSPA, and DNAJ members (Table 6). We used an arbitrary threshold of twofold induction to define heat inducibility. Using this threshold, we found that HSPH1, HSPA1A, HSPA1B, HSPA1L, HSPA6, DNAJB1, DNAJB2, DNAJB4, and DNAJB6 are the major heat-inducible genes in Hela cells. Thus, the majority of HSPs are not heat inducible. Of course, it must be noted that these patterns could be different for other cell lines and other heat conditions.
Table 6.
Heat-induced transcription of hsp genes
| Family | Gene Symbol | 0.5 hour | 2 hours | 4 hours | Probe Set ID |
|---|---|---|---|---|---|
| HSPH | HSPH1 | 0.9 | 2.6 | 3.1 | 206976_s_at |
| HSPH2 | 0.9 | 1.5 | 1.3 | 208814_at | |
| HSPH3 | 0.9 | 1.8 | 2.0 | 205543_at | |
| HSPH4 | 1.0 | 1.0 | 1.2 | 200825_s_at | |
| HSPA | HSPA1A | 1.1 | 2.3 | 2.0 | 200799_at |
| HSPA1A /// HSPA1B | 1.2 | 4.4 | 3.3 | 200800_s_at | |
| HSPA1L | 0.9 | 2.6 | 1.1 | 210189_at | |
| HSPA2 | 0.9 | 1.0 | 0.9 | 211538_s_at | |
| HSPA5 | 0.9 | 1.2 | 1.4 | 211936_at | |
| HSPA6 | 1.3 | 64.1 | 5.8 | 117_at | |
| HSPA8 | 0.9 | 1.1 | 0.9 | 208687_x_at | |
| HSPA9B | 0.9 | 0.9 | 1.0 | 200690_at | |
| HSPA12A | 0.9 | 0.8 | 0.7 | 214434_at | |
| HSPA13 | 0.9 | 0.8 | 0.8 | 202557_at | |
| HSPA14 | 1.0 | 0.9 | 0.9 | 219212_at | |
| DNAJA | DNAJA1 | 1.0 | 1.5 | 1.5 | 200880_at |
| DNAJA2 | 0.9 | 0.9 | 0.8 | 209157_at | |
| DNAJA3 | 0.9 | 0.9 | 0.7 | 205963_s_at | |
| DNAJA4 | 1.1 | 1.9 | 1.2 | 220395_at | |
| DNAJB | DNAJB1 | 1.0 | 5.7 | 2.8 | 200664_s_at |
| DNAJB2 | 1.1 | 2.1 | 1.4 | 202500_at | |
| DNAJB4 | 0.8 | 3.0 | 1.0 | 203810_at | |
| DNAJB5 | 0.9 | 1.0 | 1.0 | 212817_at | |
| DNAJB6 | 0.8 | 1.7 | 2.0 | 208810_at | |
| DNAJB9 | 0.6 | 1.2 | 0.8 | 202843_at | |
| DNAJB12 | 1.0 | 0.8 | 0.9 | 202865_at | |
| DNAJB12 | 1.1 | 1.1 | 1.2 | 214338_at | |
| DNAJB14 | 0.6 | 0.7 | 0.4 | 219237_s_at | |
| DNAJC | DNAJC1 | 1.3 | 1.0 | 1.1 | 218409_s_at |
| DNAJC3 | 1.0 | 0.8 | 1.0 | 208499_s_at | |
| DNAJC4 | 1.0 | 1.0 | 1.1 | 206781_at | |
| DNAJC6 | 0.8 | 0.7 | 0.7 | 204720_s_at | |
| DNAJC7 | 1.0 | 1.2 | 1.4 | 202416_at | |
| DNAJC8 | 0.8 | 0.8 | 0.9 | 212490_at | |
| DNAJC9 | 0.9 | 0.9 | 1.0 | 213088_s_at | |
| DNAJC10 | 0.9 | 0.8 | 0.8 | 221782_at | |
| DNAJC11 | 1.0 | 0.9 | 1.0 | 215792_s_at | |
| DNAJC12 | 1.0 | 0.8 | 0.8 | 218976_at | |
| DNAJC13 | 1.0 | 0.9 | 0.8 | 212467_at | |
| DNAJC15 | 1.0 | 1.0 | 1.0 | 218435_at | |
| DNAJC16 | 0.7 | 0.8 | 0.5 | 212908_at | |
| DNAJC17 | 1.1 | 1.0 | 1.2 | 219861_at | |
| DNAJC22 | 1.0 | 1.0 | 1.0 | 216595_at | |
| DNAJC23 | 0.8 | 0.8 | 0.7 | 201914_s_at | |
| DNAJC26 | 1.1 | 1.0 | 1.1 | 202281_at | |
| DNAJC28 | 0.9 | 1.1 | 1.0 | 220372_at | |
| DNAJC29 | 1.0 | 0.6 | 0.7 | 213262_at |
Affymetrix gene array data.
Data are shown as fold change compared to an unheated control
Subcellular localization
Determination of HSP localization is essential to understand its biochemical function. Unfortunately, high-throughput analysis of HSP localization without the use of possible interfering tags is currently impossible due to the lack of specific antibodies. As subcellular localization signals share common characteristics, computational methods have been developed to predict the subcellular localization of proteins (Sprenger et al. 2006). We selected several publicly available localization prediction methods, which accept large batches of protein sequences and which were able to predict all of the major subcellular localizations. The selected methods were Wolf PSORT (Horton et al. 2007), pTarget (Guda 2006), CELLO (Yu et al. 2006), Multiloc (Hoglund et al. 2006) and Proteome Analyst (Szafron et al. 2004). In addition, we searched the human protein database (Mishra et al. 2006) for experimentally verified HSP localizations. As can be seen from Table 7, there are large variations in the prediction using the various programs. Therefore, we first searched for the prediction method that showed the highest accuracy for biochemical verified HSP members such as HSPA1A/HSP70 (cytosol/nucleus), HSPA1B/HSP72 (cytosol/nucleus), HSPA8/Hsc70 (cytosol/nucleus), HSPA5/Bip (ER), HSPA9/Grp75 (mitochondria), DNAJA3/Tid1 (Mitochondria), DNAJB1/HSP40 (cytosol/nucleus), DNAJB9/ERdJ4 (ER) DNAJB11/Erdj3 (ER), DNAJC1/ERdJ1 (ER), DNAJC10/ERdj5 (ER), and DNAJC19/TIM14 (mitochondria). Out of these 12 known localized proteins, the following number of correct predictions was found: Wolf PSORT: 7; pTarget: 10; CELLO: 8; Multiloc: 7; and Proteome Analyst: 10. Thus, Proteome Analyst and CELLO showed the highest correct prediction. However, it should be noted that at this stage, all prediction are potentially unreliable and should be used carefully. The scoring of the most reliable prediction method does rely on a relatively low number of verified chaperone proteins and the most reliable prediction program could therefore change in the future once more proteins will be experimentally verified.
Table 7.
Prediction of Hsp subcellular localization
| Wolf | Ptarget | Cello | Multiloc | Prot. analyst | Consensus | Experimental | Prenylation | ||
|---|---|---|---|---|---|---|---|---|---|
| HSPH | HSPH1 | c | c | c | c | c | c | – | No |
| HSPH2 | c | c | n | c | c | c | g | No | |
| HSPH3 | c | c | n | n | c | c | c | No | |
| HSPH4 | e | m | c | e | e | e | e | No | |
| HSPA | HSPA1A | c | c | c | p | c | c | c | No |
| HSPA1B | c | c | c | p | c | c | c | No | |
| HSPA1L | c | c | c | p | c | c | c | No | |
| HSPA2 | c | c | c | p | c | c | n | No | |
| HSPA5 | e | c | e | e | e | e | e | No | |
| HSPA6 | c | c | c | n | c | c | n | No | |
| HSPA8 | c | c | c | p | c | c | c | No | |
| HSPA9 | m | m | m | m | m | m | m | No | |
| HSPA12A | m | – | m | m | – | m | – | No | |
| HSPA12B | c | n | m | p | – | – | – | No | |
| HSPA13 | e | e | c | e | e | e | e | No | |
| HSPA14 | x | p | c | c | c | c | – | No | |
| DNAJA | DNAJA1 | c | c | n | c | e | c | c | Yes FT |
| DNAJA2 | c | c | n | n | e | cn | c | No | |
| DNAJA3 | m | m | m | m | m | m | m | No | |
| DNAJA4 | c | c | n | c | e | c | a | Yes FT | |
| DNAJB | DNAJB1 | n | n | c | c | c | c | c | No |
| DNAJB2a | n | n | c | c | – | cn | c | No | |
| DNAJB2b | n | n | o | c | – | n | – | Yes FT GGT1 | |
| DNAJB4 | c | c | c | c | e | c | – | No | |
| DNAJB5 | c | n | c | c | e | c | – | No | |
| DNAJB6a | n | c | x | c | – | c | c | No | |
| DNAJB6b | c | c | c | c | – | c | c | No | |
| DNAJB7 | n | c | o | n | – | n | – | No | |
| DNAJB8 | c | c | c | n | – | c | – | No | |
| DNAJB9 | x | e | c | x | c | xc | e | No | |
| DNAJB11 | x | e | c | e | e | e | e | No | |
| DNAJB12 | m | n | c | n | e | n | – | No | |
| DNAJB13 | c | c | c | c | c | c | – | No | |
| DNAJB14a | c | c | c | n | e | c | – | No | |
| DNAJB14b | c | c | x | n | c | c | – | No | |
| DNAJC | DNAJC1 | a | n | n | e | n | n | e | No |
| DNAJC2 | n | – | n | n | n | n | – | No | |
| DNAJC3 | x | c | c | e | c | c | c | No | |
| DNAJC4 | c | n | n | n | c | n | – | No | |
| DNAJC5 | c | c | x | x | e | cx | v | No | |
| DNAJC5B | c | c | x | a | e | c | – | No | |
| DNAJC5G | c | n | x | c | e | c | – | No | |
| DNAJC6 | n | n | n | n | c | n | n | No | |
| DNAJC7 | n | c | n | c | c | c | c | No | |
| DNAJC8 | n | n | n | n | e | n | l | No | |
| DNAJC9 | c | e | c | c | – | c | – | No | |
| DNAJC10 | a | e | c | g | e | e | e | No | |
| DNAJC11 | c | c | n | n | – | cn | – | No | |
| DNAJC12a | c | c | n | n | – | cn | – | No | |
| DNAJC12b | c | c | n | c | – | c | – | No | |
| DNAJC13 | a | – | c | c | c | c | d | No | |
| DNAJC14 | c | n | m | n | e | n | e | No | |
| DNAJC15 | x | c | m | n | m | m | – | No | |
| DNAJC16 | e | e | m | g | c | e | – | No | |
| DNAJC17 | c | c | n | c | n | c | – | No | |
| DNAJC18 | c | c | n | n | e | cn | – | No | |
| DNAJC19 | x | m | m | m | m | m | m | No | |
| DNAJC20 | m | m | n | m | m | m | m | No | |
| DNAJC21a | n | n | n | n | n | n | – | No | |
| DNAJC21b | n | c | n | n | n | n | – | No | |
| DNAJC22 | a | e | a | y | ce | ae | no | ||
| DNAJC23 | a | n | n | e | e | ne | e | No | |
| DNAJC24 | x | c | n | c | c | c | – | No | |
| DNAJC25 | a | c | a | e | c | ac | a | No | |
| DNAJC26 | a | m | n | n | c | n | c | No | |
| DNAJC27 | c | c | c | c | g | c | – | No | |
| DNAJC28 | m | m | m | m | – | m | – | No | |
| DNAJC29 | c | c | n | n | – | cn | – | No | |
| DNAJC30 | m | e | m | x | c | m | e | No | |
Legend: n nuclear, m mitochondrial, g Golgi, e er, p peroxisomes, x extracellular, a plasma membrane, o outer membrane, v cytoplasmic vesicle, l nucleolus, d endosome, k cytoskeleton, y lysosome, FT farnesyltransferase, GGT1 geranylgeranyltransferase 1
We used the PrePS webserver to predict farnesylation of chaperones. As shown in Table 7, DNAJA1 and DNAJA4 are predicted to be prenylated by farnesyltransferase, which is in agreement with the literature (Terada and Mori 2000). In addition, DNAJB2b was predicted to be prenylated by geranylgeranyltransferase I as shown in the literature (Chapple and Cheetham 2003).
Homology of HSPH, HSPA, and DNAJ paralogs
Next, we computed protein similarity trees based on the alignments of the HSP protein sequences using the Neighboring–joining clustering method (Gascuel and Steel 2006). Figure 1 shows the output of these alignments depicted as phylograms. Three subfamilies can be derived from Fig. 1A. As expected, the first contains all the HSPH/HSP110 members. The second subfamily contains the cytosolic predicted HSPA proteins HSPA1A, HSPA1B, HSPA1L, HSPA2, HSPA6, and HSPA8 and is flanked by the ER-localized HSPA5 and the mitochondrial-localized HSPA9 protein. The third subfamily consists of the distantly related HSPA12A and HSPA12B proteins. Thus, a high number of highly related HSPA proteins are localized in the cytosolic/nuclear compartment. To date, the reason for so many highly related cytosolic HSPA proteins is unknown.
DNAJ proteins can be divided in three subfamilies on the basis of the primary amino acid composition and are classified as type A, B and C proteins (Hennessy et al. 2005). Type A proteins are the closest human orthologs of the Escherichia coli DNAJ and contain, besides an extreme N-terminal J-domain, a glycine/phenylalanine-rich region, a cysteine rich region, and a variable C-terminal domain. Type B proteins contain an N-terminal J-domain, a glycine/phenylalanine-rich region but lack the cysteine rich region. Type C DNAJ proteins contain only the J domain that is not necessarily restricted at the N-terminus but can be positioned at any place within the protein (Hennessy et al. 2005). The DNAJA is a highly related family of proteins and DNAJA3 (the mitochondrial localized member) is the most distantly related member (Fig. 1B). For the DNAJB family, three major subfamilies are found (Fig. 1C). The first consists of the members DNAJB2, DNAJB6, DNAJB7, and DNAJB8, the second of the members DNAJB1, DNAJB4, DNAJB5, DNAJB9, DNAJB11, and DNAJB13 and the third of the members DNAJB12 and DNAJB14. Although different C-termini could be defined based on the primary amino acid level within the DNAJB family, at present, no clear biochemical function can be assigned to one of these subfamilies. The DNAJC family (Fig. 1D) shows the highest divergence of all families. Based on these results we decided to clone the HSPH, HSPA, DNAJA, and DNAJB family. As the DNAJC family is highly diverse, we omitted this family for library construction.
Cloning the HSPH, HSPA, and DNAJ gene families
Selection of an expression system
For cloning a human expression library to perform reverse genetic screens, we used a robust and versatile system with a high degree of flexibility: the Flp-In T-Rex tetracycline inducible expression system. The core promoter of this construct contains the full human cytomegalovirus (CMV) promoter followed by two tetracycline repressor binding sites. Thus, in cell systems engineered to express the tetracycline repressor, tetracycline can be used for regulated expression of the gene of interest, whereas full CMV strength promoter activity will be achieved in cell systems that do not contain the tetracycline repressor (Knopf et al. 2008). In addition to regulated expression, the vector contains an FRT recombination site for the Flp recombinase-mediated stable integration of the vector at a specific site in an engineered FRT site-harboring cell line (Garcia-Otin and Guillou 2006). The eukaryotic selection marker lacks a start codon, which selects for a site-specific integration in the target genome. We selected the Flp-In T-Rex 293 cell line, a modified human embryonic kidney (Hek-293) cell line that expresses the tetracycline repressor and harbors a single copy of the FRT site at an active site in the genome. The Hek-293 cell line has been used extensively as a model for protein-folding diseases and is widely known for its ease of manipulation (Graham et al. 1977). A summary of the Flp-In T-Rex system is depicted in Fig. 2.
Construction of vector fusion tags
Specific antibodies against most of the recently identified human heat shock proteins are not available. To verify the expression of the different proteins, we used a subset of frequently used protein tags. In some cases, protein tags interfere with the native function of the protein (Muller-Taubenberger 2006). Therefore, caution must be taken with the interpretation of the results obtained. In general, experiments using this library can always be confirmed using the non-tagged version. Although the protein expression cannot be confirmed with the untagged version, one can easily compare the biological effects detected in a particular assay.
As a first step toward a vector library for the expression of different heat shock proteins, we selected different protein tags harboring different biological properties. eGFP was selected for subcellular localization studies. As a second (smaller) tag, we used the V5 tag, consisting of only 14 amino acids for which high affinity antibodies are commercially available. In addition, we used a hexa-histidine tag for protein precipitation experiments (Fig. 2).
To reduce cloning efforts, an N-terminal fusion tag was preferred. In this setting, we could maintain the natural stop codon in the gene of interest, which allows for simple shuttling from tagged to non-tagged constructs. However, it should be mentioned that N-terminal fusion tags could interfere with the import in subcellular organelles such as the ER or mitochondria and non-tagged versions are in such cases preferred.
An overview of the fusion tag cloning primers and procedure is shown in Fig. 2. The polymerase chain reaction (PCR) product of the eGFP gene lacking a stop-codon was cloned in pCDNA5/FRT/TO. For V5 and His tags, the corresponding oligos were annealed and cloned directly in the pCDNA5/FRT/TO vector.
Cloning the chaperone library
The focus of our gene library is on the cytosolic and nuclear expressed chaperones. Therefore, we selected the HSP70/HSPA proteins, which are putatively expressed in the cytosol or the nucleus (Table 7). For the HSP40/DNAJ family of proteins, we selected the major part of the DNAJA and DNAJB subfamily, which are the closest orthologs to E. coli DNAJ. As a certain human cell type typically only expresses a subset of its genes, we used pooled RNA from 10 different human cell lines as a source for cDNA synthesis and gene amplification. No amplification products were obtained for DNAJB4, DNAJB5, and DNAJB8. Instead, these genes were amplified from commercially obtained cDNA plasmids (Open Biosystems, Huntsville, AL). In addition, the HSPA6 gene was not amplified from the pooled cDNA. As this gene did not contain any introns, we amplified it directly from human chromosomal DNA. The yeast HSP70 gene SSA1 and the prokaryotic HSP70 gene DNAK were amplified from genomic Saccharomyces cerevisiae and E. coli DNA, respectively. An overview of the cloning procedure can be found in Fig. 2 and the cloning details can be found in Table 8. We used a nested PCR approach for the HSPH gene family as the start and the end of the members in this gene family are similar. The PCR products were purified, digested, and cloned in the pCDNA5/FRT/TO GFP vector. The constructs were sequence verified for the presence of the correct insert. Thereafter, expression was verified by Western blot analysis (data not shown) and the genes were subcloned in the pCDNA5/FRT/TO V5, pCDNA5/FRT/TO HIS, and the pCDNA5/FRT/TO vector.
Validation of the library
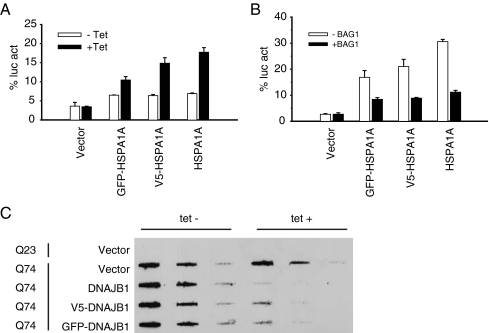
To test the effect of the different protein tags, two different biochemical assays were used. First, the effect of the tag on the ability of HSPA1A to assist the refolding of heat-denatured luciferase was tested (Michels et al. 1997, 1999). Therefore, we used constructs containing the HSPA1A gene downstream and in frame with the GFP tag, the V5 tag, and the pCDNA5/FRT/TO vector lacking a tag and compared the efficacy in the stimulation of luciferase refolding. The GFP tag significantly reduced the activity of the HSPA1A protein (Fig. 3A), whereas the V5 tag showed little to no significant effect on HSPA1A activity. Yet, modulation of HSPA1A related refolding by the co-factor BAG-1 (Nollen et al. 2000) could be achieved with all tagged versions (Fig. 3B). Thus, HSPA1A N-terminally tagged with eGFP may be less active related to non-tagged versions but seems unaffected in its ability to cooperate with its cofactors.
Fig. 3.
The effect of different fusion tags on HSPA1A (A) and (B) or DNAJB1 (C). (A) Luciferase refolding assay using GFP, V5, and untagged HSPA1A versions. Cells were transfected with different tagged versions of HSPA1A together with a plasmid encoding firefly luciferase. HSPA1A expression was induced using tetracycline. The day after transfection, the cells were heated at 37°C or 45°C for 30 min and reincubated for 1 h at 37°C to allow luciferase refolding. Thereafter, cells were lysed and measured for luciferase activity. The percentage of luciferase activity is plotted relative to the activity in unheated control cells (100%). (B) Modulation of tagged HSPA1A versions by BAG-1. Cells were treated as in (A) but also co-transfected with a BAG-1 encoding plasmid as indicated. (C) Filter trap assay showing aggregation of expanded Huntingtin. GFP, V5 and untagged versions of DNAJB1 were used as indicated. Cells were transfected with different tagged versions of DNAJB1 together with a plasmid encoding GFP-tagged Huntingtin containing either 23Q or 74Q. DNAJB1 was induced by tetracycline. Two days after transfection, cells were lysed and the lysates were loaded on to a cellulose acetate membrane. After transblotting, blots were immunostained for GFP to detect aggregated Huntingtin. GFP-tagged DNAJB1 alone did not show any signal on the membrane (not shown)
To test the effect of tagging DNAJ-like proteins, we used a filter trap assay to detect aggregation of polyglutamine proteins such as mutant Huntingtin. Aggregated Huntingtin is SDS-insoluble and retains trapped in a non-protein binding cellulose acetate membrane, and DNAJB1 is known to be able to inhibit this aggregation (Carra et al. 2005; Rujano et al. 2007). We used constructs containing the DNAJB1 gene downstream and in frame with the GFP tag, the V5 tag, and the pCDNA5/FRT/TO vector lacking a tag and compared the efficacy of DNAJB1 in the suppression of mutant Huntingtin aggregation. As shown in Fig. 3C, untagged DNAJB1 strongly suppresses mutant Huntingtin aggregation containing a polyQ tract of 74 residues. Both the V5- and the GFP-tagged showed an equal slight reduction on the aggregation suppression but yet retained substantial activity. Thus, N-terminal tagging sometimes does influence the maximal activity of the chaperones tested. This implies that after performing experiments with our tagged HSPs, confirmation with untagged versions is required.
Conclusion/Discussion
The HSPH/HSPA and DNAJ families are large gene families with many poorly studied individual members. We used bioinformatics approaches to study the expression, the localization, and the homology of these families. These approaches generated large datasets, which will be useful for the systematic biochemical analysis of these family members. It was found that HSPs are expressed at highly variable levels in different tissues. So far, no clear patterns were seen for paired expression of certain members within, e.g., the HSPA and DNAJ family in most tissues. Although it is valid to search for the highest expressing tissue for a particular transcript, it is difficult to compare the level of different transcripts for a particular tissue. This is thought to be partly because different messengers show a different half-life and partly because different transcripts show large differences in the window of bottom-to-peak expression making such a comparison difficult. Interestingly, we did find some pattern for the testis. A testis-specific HSPA transcript was found (HSPA1L) as well as testis-specific DNAJ members (DNAJB7, DNAJB8, DNAJC5B, and DNAJC5G). This could indicate that HSPA1L cooperates with one of these DNAJ members.
We also studied the expression levels of HSPs during various developmental stages. The results of the peak expression per transcript show that there is a wide variation in HSP expression throughout different developmental stages but many DNAJC members peak at the blastocyst and fetal stages, indicating that there is a need for specialized DNAJ members early in development.
Surprisingly, the heat inducibility of the different HSPs was restricted to only a couple of members within each family (HSPH1, HSPA1A/B, HSPA6, DNAJB1, DNAJB2, DNAJB4, and DNAJB6). This could mean that HSPA1A or HSPA6 cooperate with HSPH1 and one of these DNAJ members following stress conditions. Interestingly, no heat-inducible DNAJC member was found indicating that DNAJC members do not function in the stress response. It should be noted, however, that the array did not contain probes corresponding to all DNAJC members.
Analyzing the cellular distribution and homology of different HSP members showed that a very homologous subfamily of the HSPA family is predicted to be expressed in the cytosol (HSPA1A/B, HSPA1L, HSPA2, HSPA6, and HSPA8). This indicates that only a minority of the gene duplication occurred as a result of the compartmentalization. It is unclear at this stage if this homologous subgroup of HSPA chaperones is regulated by the same subset of co-factors and if they bind the same subset of client proteins. It will be highly interesting to answer these questions by using available biochemical approaches. For this purpose, we cloned a large collection of chaperone-encoded genes in a tetracycline-inducible vector system. Different tags with different properties were used in order to detect expression levels (V5), study subcellular localization in living or fixed cells (GFP/V5), or to enrich the expressed protein from crude cell lysates (His). In addition, non-tagged versions were made to verify obtained biological effects. This expression library will be useful to systematically study the biochemical and cell biological features of these poorly characterized HSPs and might help answer the intriguing question why we have so many HSPA and DNAJ chaperones.
Acknowledgements
We would like to thank Maria A.W.H. van Waarde for expert assistance on biochemical analysis. We thank Eefje Pelster, Alette H. Faber, and Reinier Bron for assistance on gene cloning. Lenja Bystrykh (Department of Cell Biology, Stem Cell Biology Section, University Medical Center Groningen, The Netherlands) is kindly acknowledged for help on the Unigene EST collection and Ron Dirks (section Biochemistry, Radbout University, Nijmegen, The Netherlands) for help on the Affimetrix gene array data. Russell S. Thomas (CIIT Centers for Health Research, NC, USA) is acknowledged for providing the array data sets. This work was supported by Innovatiegerichte Onderzoeksprogramma Genomics Grant IGE03018.
References
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Brocchieri L, Conway de ME, Macario AJ. Chaperonomics, a new tool to study ageing and associated diseases. Mech Ageing Dev. 2007;128:125–136. doi: 10.1016/j.mad.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Brocchieri L, Conway de ME, Macario AJ. HSP70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol Biol. 2008;8:19. doi: 10.1186/1471-2148-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra S, Sivilotti M, Chavez Zobel AT, Lambert H, Landry J. HSPB8, a small heat shock protein mutated in human neuromuscular disorders, has in vivo chaperone activity in cultured cells. Hum Mol Genet. 2005;14:1659–1669. doi: 10.1093/hmg/ddi174. [DOI] [PubMed] [Google Scholar]
- Chapple JP, Cheetham ME. The chaperone environment at the cytoplasmic face of the endoplasmic reticulum can modulate rhodopsin processing and inclusion formation. J Biol Chem. 2003;278:19087–19094. doi: 10.1074/jbc.M212349200. [DOI] [PubMed] [Google Scholar]
- Garcia-Otin AL, Guillou F. Mammalian genome targeting using site-specific recombinases. Front Biosci. 2006;11:1108–1136. doi: 10.2741/1867. [DOI] [PubMed] [Google Scholar]
- Gascuel O, Steel M. Neighbor-joining revealed. Mol Biol Evol. 2006;23:1997–2000. doi: 10.1093/molbev/msl072. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Lumia V, Creti R, Macario EC, Sanangelantoni A, Cammarano P. Discontinuous occurrence of the HSP70 (dnaK) gene among Archaea and sequence features of HSP70 suggest a novel outlook on phylogenies inferred from this protein. J Bacteriol. 1999;181:434–443. doi: 10.1128/jb.181.2.434-443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda C. pTARGET: a web server for predicting protein subcellular localization. Nucleic Acids Res. 2006;34:W210–W213. doi: 10.1093/nar/gkl093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman J, Eggen BJ, Rozema T, Damman K, Kampinga HH, Coppes RP. Radiation and transforming growth factor-beta cooperate in transcriptional activation of the profibrotic plasminogen activator inhibitor-1 gene. Clin Cancer Res. 2005;11:5956–5964. doi: 10.1158/1078-0432.CCR-05-0427. [DOI] [PubMed] [Google Scholar]
- Held T, Paprotta I, Khulan J, et al. HSPA4l-deficient mice display increased incidence of male infertility and hydronephrosis development. Mol Cell Biol. 2006;26:8099–8108. doi: 10.1128/MCB.01332-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of HSP40–HSP70 interactions. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund A, Donnes P, Blum T, Adolph HW, Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;22:1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Ando A, Ando H, Ando J, Saijoh Y, Inoko H, Fujimoto H. Genomic structure of the spermatid-specific HSP70 homolog gene located in the class III region of the major histocompatibility complex of mouse and man. J Biochem. 1998;124:347–353. doi: 10.1093/oxfordjournals.jbchem.a022118. [DOI] [PubMed] [Google Scholar]
- Kampinga HH. Chaperones in preventing protein denaturation in living cells and protecting against cellular stress. Handb Exp Pharmacol. 2006;172:1–42. doi: 10.1007/3-540-29717-0_1. [DOI] [PubMed] [Google Scholar]
- Knopf CW, Zavidij O, Rezuchova I, Rajcani J. Evaluation of the T-REx transcription switch for conditional expression and regulation of HSV-1 vectors. Virus Genes. 2008;36:55–66. doi: 10.1007/s11262-007-0178-9. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Koranda M, Benetka W, Schneider G, Sirota FL, Eisenhaber F. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AA, Nguyen VT, Konings AW, Kampinga HH, Bensaude O. Thermostability of a nuclear-targeted luciferase expressed in mammalian cells. Destabilizing influence of the intranuclear microenvironment. Eur J Biochem. 1995;234:382–389. doi: 10.1111/j.1432-1033.1995.382_b.x. [DOI] [PubMed] [Google Scholar]
- Michels AA, Kanon B, Konings AW, Ohtsuka K, Bensaude O, Kampinga HH. HSP70 and HSP40 chaperone activities in the cytoplasm and the nucleus of mammalian cells. J Biol Chem. 1997;272:33283–33289. doi: 10.1074/jbc.272.52.33283. [DOI] [PubMed] [Google Scholar]
- Michels AA, Kanon B, Bensaude O, Kampinga HH. Heat shock protein (HSP) 40 mutants inhibit HSP70 in mammalian cells. J Biol Chem. 1999;274:36757–36763. doi: 10.1074/jbc.274.51.36757. [DOI] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, et al. Human protein reference database – 2006 update. Nucleic Acids Res. 2006;34:D411–D414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Taubenberger A. Application of fluorescent protein tags as reporters in live-cell imaging studies. Methods Mol Biol. 2006;346:229–246. doi: 10.1385/1-59745-144-4:229. [DOI] [PubMed] [Google Scholar]
- Nollen EA, Brunsting JF, Song J, Kampinga HH, Morimoto RI. Bag1 functions in vivo as a negative regulator of HSP70 chaperone activity. Mol Cell Biol. 2000;20:1083–1088. doi: 10.1128/MCB.20.3.1083-1088.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Giardina C, Hightower LE. HSP70B’ regulation and function. Cell Stress Chaperones. 2007;12:219–229. doi: 10.1379/CSC-278.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Page TJ, Sikder D, Yang L, Pluta L, Wolfinger RD, Kodadek T, Thomas RS. Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol Biosyst. 2006;2:627–639. doi: 10.1039/b606129j. [DOI] [PubMed] [Google Scholar]
- Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DNAJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano MA, Kampinga HH, Salomons FA. Modulation of polyglutamine inclusion formation by the HSP70 chaperone machine. Exp Cell Res. 2007;313:3568–3578. doi: 10.1016/j.yexcr.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci U S A. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD. Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J Mol Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- Sprenger J, Fink JL, Teasdale RD. Evaluation and comparison of mammalian subcellular localization prediction methods. BMC Bioinformatics. 2006;7(Suppl 5):S3. doi: 10.1186/1471-2105-7-S5-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafron D, Lu P, Greiner R, et al. Proteome Analyst: custom predictions with explanations in a web-based tool for high-throughput proteome annotations. Nucleic Acids Res. 2004;32:W365–W371. doi: 10.1093/nar/gkh485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K, Mori M. Human DNAJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J Biol Chem. 2000;275:24728–24734. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]