Abstract
Image reconstruction for magnetic resonance spectroscopic imaging (MRSI) requires specialized spatial and spectral data processing methods and benefits from the use of several sources of prior information that are not commonly available, including MRI-derived tissue segmentation, morphological analysis and spectral characteristics of the observed metabolites. In addition, incorporating information obtained from MRI data can enhance the display of low-resolution metabolite images and multiparametric and regional statistical analysis methods can improve detection of altered metabolite distributions. As a result, full MRSI processing and analysis can involve multiple processing steps and several different data types. In this paper, a processing environment is described that integrates and automates these data processing and analysis functions for imaging of proton metabolite distributions in the normal human brain. The capabilities include normalization of metabolite signal intensities and transformation into a common spatial reference frame, thereby allowing the formation of a database of MR-measured human metabolite values as a function of acquisition, spatial and subject parameters. This development is carried out under the MIDAS project (Metabolite Imaging and Data Analysis System), which provides an integrated set of MRI and MRSI processing functions. It is anticipated that further development and distribution of these capabilities will facilitate more widespread use of MRSI for diagnostic imaging, encourage the development of standardized MRSI acquisition, processing and analysis methods and enable improved mapping of metabolite distributions in the human brain.
Keywords: MR spectroscopic imaging, data processing, brain
INTRODUCTION
Magnetic resonance spectroscopic imaging (MRSI) permits the spatial distributions of MR-observable compounds to be obtained in a non-invasive manner (1). When applied to in vivo studies, these techniques can be used to map the distributions of several tissue metabolites, potentially indicating areas of altered metabolism even in the absence of structural changes indicated by other imaging modalities. Although several clinical applications of MRSI have been described, the use of these techniques largely remains within the research community and the transfer to more routine clinical applications has been limited. In part, this has been due to the relatively complex and specialized requirements for MRSI data processing and analysis. This complexity not only reflects the combined processing requirements of imaging and NMR spectroscopy, but that additional MRSI-specific processing techniques are available, which can enhance either the data reconstruction, analysis or display.
The reconstruction and analysis of both the spatial and spectral MRSI data dimensions can benefit from the incorporation of prior information, including, for example, knowledge of tissue distributions obtained from higher-resolution MRI (1), information on metabolite spectral patterns and knowledge of normal metabolite concentrations. However, the ability to make use of this information is frequently limited by the lack of suitable software tools and also the lack of information on normal metabolite concentrations for comparison with the individual subject data. This latter point not only reflects the natural variability of tissue metabolite concentrations, which are known to vary by location and subject age (2), but also the considerable variability of MRSI acquisition parameters and processing methods. For most clinical investigations, this has meant that additional data must be acquired from a group of age-matched normal subjects to provide comparative data. To address these limitations, the MIDAS (Metabolite Imaging and Data Analysis System) project is developing a suite of MRSI and MRI data processing, analysis and visualization tools that are suited to repetitive use of 1H MRSI for clinical research studies. In addition to supporting the complex MRI and MRSI processing requirements, this project includes the development of a database of brain metabolite distributions in normal subjects. A secondary aim of this project is to encourage the development of standardized MRSI acquisition and processing protocols by making the MIDAS package available to clinical researchers.
METHODS AND RESULTS
General processing protocols
An MRSI acquisition protocol is typically accompanied by a set of diagnostic MRI studies, but may also include additional MRI measurements to obtain calibration data, e.g. for tissue segmentation or intensity normalization functions. In addition, MRSI protocols vary considerably and require different data processing methods. For example, the choice of TE value, volume selection or lipid nulling will impact whether lipid extrapolation methods (3) are required and the parameters used for spectral fitting; the method used to measure field inhomogeneity or eddy-current correction functions will impact how that information is processed and applied; and the use of a high-speed imaging variants (4–6) may require that an acquisition-specific data resampling be applied prior to MRSI reconstruction. Data processing protocols may therefore vary considerably and must be able to be adapted to different acquisition protocols. In this paper, we review a comprehensive MRI and MRSI data processing protocol. Although the discussion is limited to that required for a specific acquisition protocol and results presented for data obtained at 1.5 T, the proposed methods are sufficiently general that they may be applied to many different protocols. The specific MRSI acquisition method considered in this paper was selected to enable data to be obtained over a large portion of the brain for the purpose of mapping metabolite distributions throughout the brain. While the volumetric format of this acquisition differs from the more commonly used 2D MRSI acquisition methods, it provides a good example for demonstrating the advantages to be gained by integrating the MRSI processing with MRI-derived information. Although not currently provided as a standard sequence on commercial instruments, the technical implementation of this volumetric MRSI acquisition is well within the capabilities of all modern commercial MR instruments.
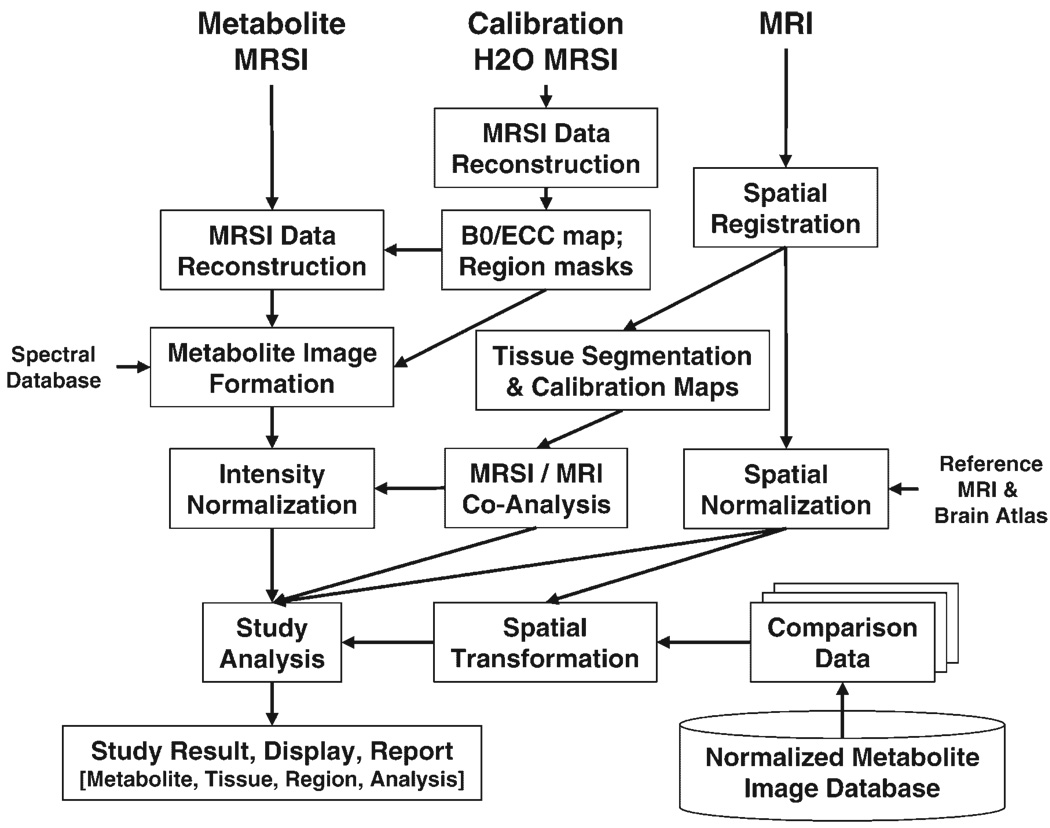
In Fig. 1 is shown a broad overview of the MRI and MRSI processing pathways and the interconnections between these data types. Here it is assumed that the acquisition includes a second MRSI dataset, obtained from the water signal and with parameters identical with those for the metabolite MRSI signal. Termed the ‘water reference’ MRSI, these data are used to provide information that is incorporated into the MRSI processing, including the B0 field inhomogeneity map and masks indicating the subcutaneous lipid regions (lipid mask) and the brain (brain mask). However, this information could equally well be obtained from an MRI acquisition with a different processing path. Specific details of the processing steps are provided in the following sections, but in brief, the reconstructed metabolite images are generated using Fourier transform reconstruction and automated spectral fitting; the data are then calibrated, based on an MRI-derived tissue water image; and the metabolite images then analyzed, which may include correction for cerebrospinal fluid (CSF) partial-volume signal loss, correlation with tissue type and comparison with normal tissue concentrations. This last step requires that comparison data be available for each metabolite, as the mean and standard deviation for each spatial location, which are obtained from a database of normal metabolite values.
Figure 1.
Illustration of the steps and interconnections between MRSI and MRI data types required for full processing and analysis of a single-subject MRSI data set
The result of a complete MRSI/MRI processing procedure will be a multiparametric data set, containing at each MRSI voxel the intensity-normalized spectral parameters and the tissue contributions to that voxel. In addition, spatial transformation parameters will be available that permit image registration with a brain atlas, which may, for example, allow identification of all voxels belonging to a given brain region. An additional piece of information is an estimate of the ‘quality’ of the data at that location, which can be used to exclude spectra of inadequate quality from further analysis.
For routine use it is essential to provide fully automated processing. As indicated in Fig. 1, the complete processing protocol involves multiple steps, the order of which is frequently important, and several of these processing steps can take a significant amount of time. In addition, there is a need to maintain flexibility in the system so that processing pathways can be reconfigured to accommodate different acquisition protocols and processing aims. To satisfy these requirements, the MIDAS system is composed of several independent processing modules, which can be run without any operator intervention in a predefined sequence using a batch processing management module. The individual processing modules only communicate indirectly via the common data management system. Although fully automated processing is possible, it is still recommended that visual checks be included, for example to confirm that the spectral data were not corrupted by subject motion or that the automated spatial registration modules performed correctly.
Data acquisition
MRI and MRSI data of the brain were acquired from 14 normal subjects, aged 27–48 years (average 36 years), at 1.5 T (Siemens Sonata) using the standard head coil. The MRSI acquisition used a volumetric measurement with echo-planar readout (7), a TE of 70 ms and a field of view of 32 × 32 cm in-plane and 18 cm in the inferior–superior direction. To permit imaging of metabolite distributions over a wide region of the brain, the acquisition used an axial 14 cm slab excitation that covered the whole brain. Signals from subcutaneous lipids were reduced using an inversion–recovery preparation, with TI = 210 ms, and a saturation band was placed across the lipid region behind the eyes to suppress movement-associated aliasing from this region. The sequence included acquisition of a water MRSI data set, which was obtained using an interleaved acquisition using parameters identical with those for the metabolite MRSI, but with low flip-angle excitation and no water suppression. Additional details of the MRSI sequence have been provided elsewhere (7).
MRI acquisitions include a T1-weighted MPRAGE sequence, with 0.5 × 0.5 × 1.5 mm resolution and a double spin-echo sequence with TE = 20 and 80 ms, TR = 4600 ms and spatial resolution 0.5 × 0.5 × 3 mm. The MRI and MRSI data were acquired with the same angulation, aligned along the anterior commissure–posterior commissure (AC–PC) line.
Data management
In addition to acquiring the raw MRSI data, a typical study protocol will include multiple MRI data sets and possibly single-voxel MRS measurements. Multiple additional data sets are then generated during processing, as illustrated in Fig. 1. Management of this diverse collection of data becomes of critical importance in addition to the organization of multiple studies under different projects. To address this issue, an Extensible Markup Language (XML) (8) based solution has been implemented that maintains a complete record of subject and study information, all processing steps and data file locations. Additional files are used to maintain a record of all subjects grouped within a single project, and also a list of all projects associated with each investigator. This approach provides a flexible and relatively simple data management system without incurring the challenges of maintaining a sophisticated database system and can be readily adapted to support multiple data types located anywhere across a networked multi-platform environment. All access to the parameter information from the individual processing modules is provided through a set of library functions written in Java, which simplifies support for modules written in several programming languages.
All data, regardless of the source, are first ‘imported’ into the data management system. This process copies all necessary metadata, which are maintained in a hierarchical manner based on the DICOM (9) data model as Subject, Study, Series, Dataset and Frame. Additional subject or study information may also be incorporated at this stage. Typically, this import procedure will also transfer the data from the scanner to the final data storage location. Most data can be imported without any change of the native format; however, subsequent processing operations may save new data files using a simple binary format. Regardless of the file format, a record of all data parameters is maintained in the associated XML file. The results of a processing or data analysis step are also maintained in the XML file, using the same hierarchical organization depending on the source of the input data. Unique data identifiers (UIDs) created at the time of data acquisition are maintained and supplemented with UIDs and time-stamps created for each processing step.
Studies from multiple subjects are grouped together under a ‘Project’ and a subject may be referenced in multiple projects. In this way, any processing done on the subject for one project will be immediately available to other projects that include that same subject. It is also possible to reprocess a dataset using a different processing path without overwriting any existing results. In this case a copy of the subject XML file is created along with the new data files, while maintaining any links to the existing data files.
MRSI data processing
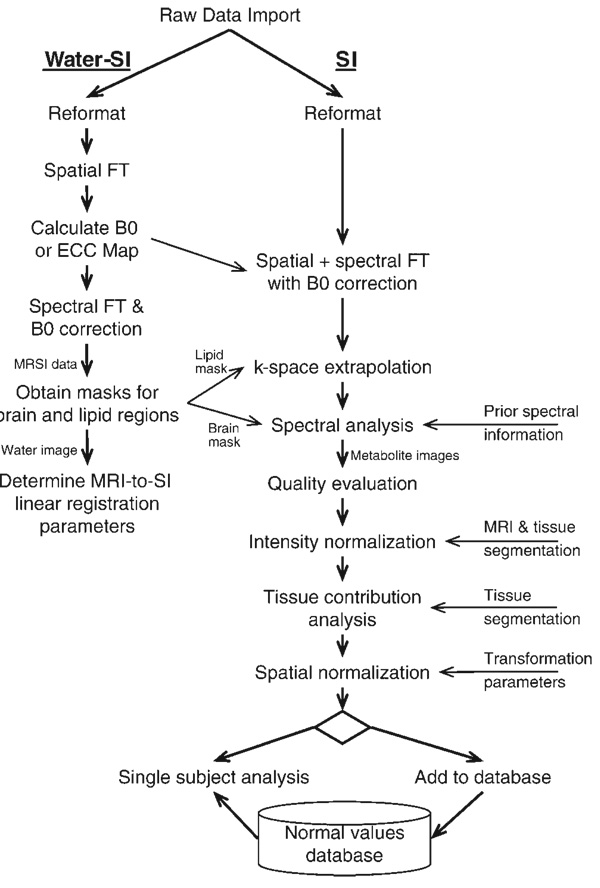
Figure 2 shows a flow diagram that gives a more detailed description of the MRSI portion of Fig. 1 and which describes the specific data processing pathway used for the MRSI data presented in this paper. This includes the processing of the water-reference MRSI dataset from which information is obtained for use in the processing of the metabolite image data.
Figure 2.
Detailed flow diagram of the processing steps used for the metabolite and the water-reference 1H MRSI data sets
In comparison with the more familiar data processing procedures used for single-voxel MRS studies, MRSI may require several unique processing steps, which are indicated in Fig. 2. For example, corrections for B0 shifts, gradient eddy currents and signal intensity must be applied in a spatially variant manner and k-space extrapolation methods may be used to reduce lipid contamination. In addition, standard spectral processing steps need to be specifically adapted to the MRSI data. Most notably, the spectral analysis must be able to deal with the variable quality of the data, which may include, for example, good-quality spectra with very low signal intensity in regions subject to significant CSF partial volume contribution, regions with unacceptably broadened lineshapes and large residual water and lipid signals, particularly for the whole-brain MRSI method used for this project. Spectra are also acquired from regions of no interest, such as outside of the brain, and should therefore be excluded from the time-consuming spectral analysis. The signal-to-noise ratios (SNRs) are typically lower than those commonly obtained with single voxel methods, requiring that spectral analysis methods be more robust, which may in turn require that a simpler spectral model be used.
The water reference MRSI data are used to address some of the issues associated with MRSI processing. These include the calculation of a B0 map from the frequency of the water resonance, and also masks generated by integrating over the water and lipid spectral ranges. The first of these defines the brain region and is used to limit the voxels selected for spectral analysis (10), while the mask generated from the lipid signal identifies the subcutaneous signal region and is used for lipid k-space extrapolation (3). This latter procedure diminishes ringing and the associated signal contamination from the subcutaneous lipid signal, which may occur as a result of the limited k-space sampling, by incorporating information on the lipid distribution. It is also possible to calculate a space–time data set that contains the time-dependent phase correction function to be used for eddy current correction (ECC) (11) at each voxel location. Finally, the integrated signal from the water-reference MRSI provides a high SNR image that shows significant structural information and which exactly matches the spatial parameters of the metabolite MRSI data and can be used to determine the spatial transformation parameters to register the MRI and the MRSI data (12).
The initial processing step indicated by the ‘reformat’ box in Fig. 2 may include two functions. Currently, raw data exported from a MR instrument in the industry-standard DICOM format is stored with a separate datafile for each encoding step used in a third spatial dimension; therefore, this reformatting step concatenates these data into a single binary file and recalculates the spatial parameter information for these volumetric data. A second processing step may include resampling for non-Cartesian k-space sampling schemes, as was required for the echo-planar acquisition (7) used for this paper. The automated parametric spectral analysis procedure, described in detail elsewhere (10), performs spectral fitting of the complex-value spectral data using a Lorentzian–Gaussian lineshape model and incorporates prior spectral information obtained by spectral simulation using the GAVA program (13). This analysis procedure also takes advantage of the spatial information in the MRSI data by modifying starting values based on local neighborhood information.
A common limitation of automated spectral fitting approaches is that they may obtain a good fit, as defined by a small residual, to bad data. Therefore, the spectral analysis is followed by a simple quality evaluation procedure that creates a ‘Quality Image’ that reflects areas of the image having resultant linewidths and signal intensities considered to be within limits for the acquired data type. This image can be displayed alongside the reconstructed metabolite images, used to mask any displayed metabolite images or incorporated within any subsequent statistical data analysis steps, to provide an indicator of which image regions should be excluded from further consideration or at least viewed with caution. Alternatively, the measures required for evaluating the spectral quality, such as Cramer–Rao lower bounds, linewidth and B0 shifts, can be incorporated into the subsequent data analysis procedures, which is the preferred method when the image registration procedures described in a following section are used.
Metabolite signal intensity normalization
The metabolite images created following image reconstruction and spectral fitting are uncalibrated; therefore, before individual metabolite results (as opposed to ratios) can be compared across studies, it is necessary to apply an intensity normalization procedure. This requires a scaling based on a reference signal (14), for which the tissue water provides a convenient internal reference. This approach has been recommended for single-volume MRS (15) as being readily implemented and having acceptable precision, and has previously been used for MRSI (16). Although signal normalization can be extended to provide absolute metabolite concentration values, this requires measurement of relaxation rates, which is impractical for routine studies; therefore, a normalization to ‘institutional units’ has been implemented, which is valid for comparisons across data acquired at the same field strength and with the same sequence timings (TR, TE, TI), under the assumption of no changes in relaxation rates.
The intensity scaling must take into account spatially-dependent variations in signal reception sensitivity, for which a correction function based on the proton density MRI is used. This image data are modified to correspond to the signal arising from 100% water by taking advantage of the previously-determined tissue distributions as
| (1) |
where MRIPD and MRIH2O are the proton density and resultant 100% water images; ftissue is the fraction of each tissue type, for gray matter (GM), white matter (WM) and CSF determined from tissue segmentation; and ρtissue is the relative water content of each tissue. The term R accounts for residual T1 relaxation effects in the PD image and therefore is a function of the tissue type at each voxel, the T1 of that tissue, using literature values for water relaxation rates (17), and the TR time. This correction term could also include T2 relaxation; however, this was found to be negligible and was therefore not included. The water content of tissue can vary between subjects (18–21), hence the procedure starts with approximate values for ρtissue of CSF = 100%, GM = 75%, WM = 65%, but then proceeds to optimize the GM and WM water content values to minimize entropy over a central slice in MRIH2O, i.e. minimizing the intensity variations over the image relative to CSF. Outlying values are then removed (22) to diminish local intensity changes due to possible tissue misclassification, to produce an image corresponding to 100% water (or 111 m proton concentration). This is then convolved with the spatial response function of the MRSI data and corrected for any change in receiver gain between the MRI and MRSI studies. The resultant image maps the reception sensitivity function at the SI resolution, while also providing a reference signal. The normalized metabolite signal, MetNorm, was then calculated from the spectral fitted image results, Met, as
| (2) |
where the MRI data are convolved and resampled to correspond to the MRSI spatial response function, srfMRSI, and K is a calibration constant that also accounts for the difference in signal intensity between the MRI and MRSI acquisitions. This K-factor is obtained from a measurement in a phantom containing a metabolite of known concentration, using the identical MRI and MRSI imaging protocol except for long TR and short TE to minimize relaxation effects:
| (3) |
where the two I(r̅) terms represent the mean signal intensities from the phantom measurements over some central, homogeneous image region of the MRI and the fitted MRSI results, respectively, and CMet is the reference metabolite concentration.
For the data acquired in this study, the use of a digital receiver system meant that the gain values for the MRI and MRSI were unchanged, for both the calibration and study measurement; therefore, variations of this term did not need to be taken into account.
Tissue segmentation
Information obtained at high spatial resolution from MRI data can be of considerable value for enhancing the analysis of MRSI data. For example, results of MRI tissue segmentation permit the analysis of metabolic changes as a function of tissue type or can be used to account for the relative tissue volume contribution to a specific voxel to account for the differences in normal metabolite concentrations between tissue types (1). In addition, information on the distribution of CSF, which typically contributes no metabolite signals, can be used to correct for signal losses due to partial volume of the tissue within the SI voxel.
In this study, a knowledge-guided tissue segmentation method was implemented (23) that uses the T1-, T2- and proton density-weighted MRIs. Initially, the MRI data were resampled and co-aligned using affine registration, to account for differences in dimensions and for possible subject motion between these data acquisitions. The tissue segmentation procedure then applied a fuzzy c-means clustering algorithm (24) to classify voxels into a number of groups that is larger than the number of tissues being segmented, which were then combined using a set of rules describing the expected relationships and distributions of the different tissue types.
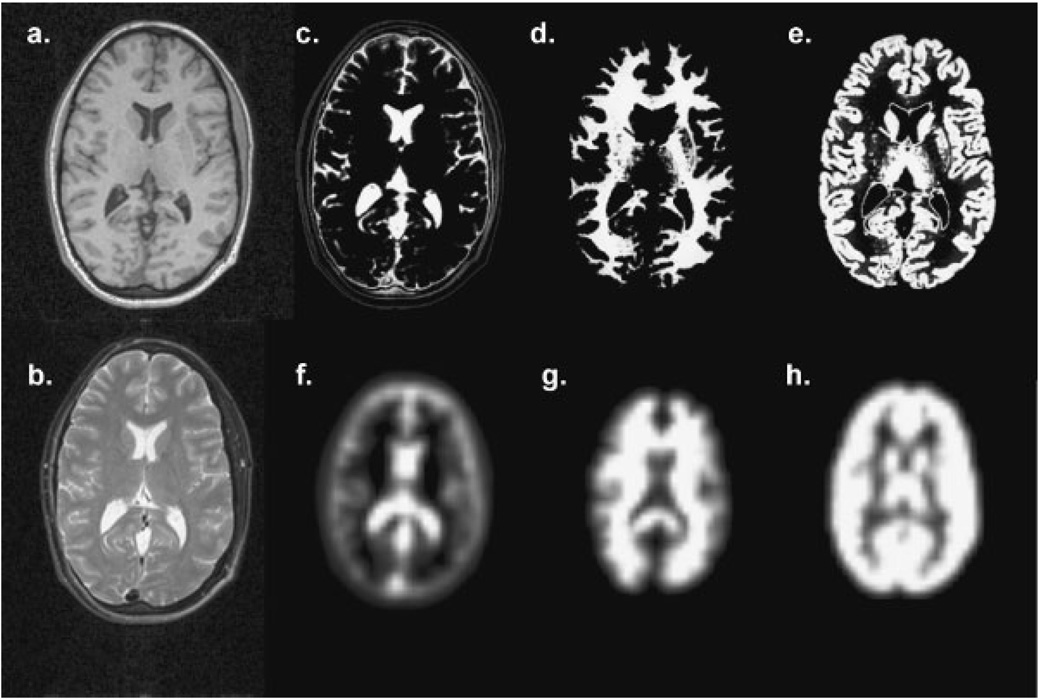
The relative tissue volume contributions corresponding to each voxel in an MRSI dataset can be obtained by convolution of the high spatial resolution segmentation results with the spatial response function of the MRSI data. This operation was implemented using a method similar to that described by Pfefferbaum et al.(25) by inverse Fourier transformation of each MRI-resolution tissue segmentation volume, resampling and zero-filling to correspond to the k-space distribution used for the MRSI reconstruction and forward Fourier transformation. In addition, any differences in field-of-view and spatial position are accounted for. This procedure is applied to each of the GM, WM and CSF image volumes, to obtain the corresponding volume contributions at each MRSI voxel, with exact correspondence between the spatial location and extent as the MRSI voxel. An example of the results of this operation is shown in Fig. 3. In this study, this procedure was applied in a fully volumetric manner; however, for single- or multi-plane MRSI acquisition, the convolution must be performed in-plane only and the segmentation data then averaged over all MRI slices corresponding to the MRSI slice thickness, possibly including the slice-selection profile.
Figure 3.
Illustration of the tissue contribution image processing. In (a) and (b) are shown T1- and T2-weighted MRIs at a single slice and the corresponding tissue segmentation images for CSF, WM and GM in (c), (d) and (e). These tissue maps are convolved with the 3D MRSI spatial response function to form the corresponding images shown in (f), (g) and (h)
Image registration
To permit voxel-based spatial analyses across groups of subjects, it is necessary to co-register all image data. This can be done by applying a spatial transformation to all of the MRSI-resolution images, to co-register the data in a common spatial frame of reference. To compute this transformation both within subject and between subject, spatial transformations must be estimated and applied. Within a single subject study, differences in positioning between series (both structural MRI scans and metabolite data) must be accounted for, which may occur owing to movement between each acquisition. For this study, the high-resolution T1-weighted MRI data were used as the reference image. Rigid transformations between this reference image volume and all of the other target image volumes were then estimated using a fully automated multi-resolution maximization of normalized mutual information (26). To address possible MRSI-to-MRI positioning differences the integrated water reference image was used as the target (indicated in Fig. 2), since this has meaningful structural features in MRSI data coordinates that are suitable for alignment with the MRI data (12). To bring each metabolite map into a common anatomical coordinate system, the MRSI data were spatially transformed to a reference or target brain, for which the MIDAS system uses the MNI BrainWeb simulated MRI data (27). A multi-resolution non-rigid alignment (28) was implemented, with a B-spline-based parameterization of the deformation field between the reference atlas and the T1-weighted high-resolution MRI from the subject, again driven by maximization of normalized mutual information. The transformations from the reference to the subject T1-image coordinate system and between the T1-image and the other subject datasets are then compounded and applied to map each metabolite and tissue density map into the common coordinate system. Finally, to permit regional analysis of brain metabolite levels, a brain atlas was created by labeling the BrainWeb MRI data with lobar-scale anatomical regions. By transforming the labeled atlas into the subject frame of reference, it is then possible to analyze the normalized metabolite results over individual brain regions.
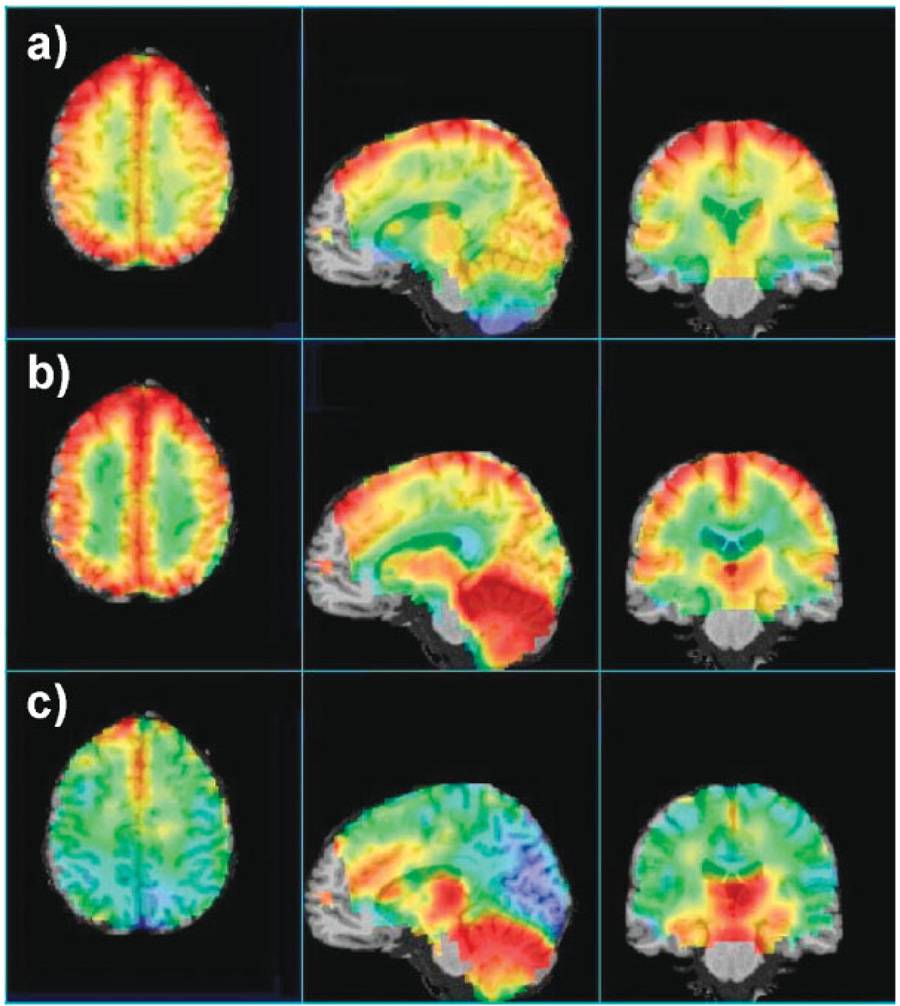
An example of the image registration procedure is shown in Fig. 4. Here, the data from 14 MRSI study results have been combined to create maps of the average N-Acetylaspartate (NAA), creatine and choline over the whole brain. The metabolite images have been intensity-normalized and corrected for CSF volume contribution. The average metabolite value was not calculated at a voxel if any one of the MRSI data sets had a fitted linewidth result of greater than 10 Hz, with the result that data were excluded in the lower frontal brain and temporal lobe. However, with the acquisition of a larger number of data sets, it is likely that additional information for these regions can be obtained with sufficient statistical confidence. These preliminary images show striking regional differences of metabolite concentrations. Some of these variations include higher NAA and creatine in cerebral GM than in cerebral WM, decreased choline in occipital GM and increased creatine and choline in the cerebellum and increased choline in the thalamus, in general agreement with previous studies (29–33), but with much greater regional definition. Some localized intensity changes of choline in frontal brain regions remain owing to incorrect fitting, arising from imperfect water suppression in these regions; however, this will be addressed by improving the quality evaluation procedures and by removing outliers in the mean value determination.
Figure 4.
Maps of the average metabolite levels from 14 subjects selected from the three-dimensional volume, showing the (a) NAA, (b) creatine and (c) choline distributions along three orthogonal directions. The metabolite images were CSF corrected and intensity normalized, and are shown in color superimposed on the reference MRI, with red indicating high data values, yellow intermediate and green–blue low. The color scales were adjusted separately for the best visualization of each set of metabolite images
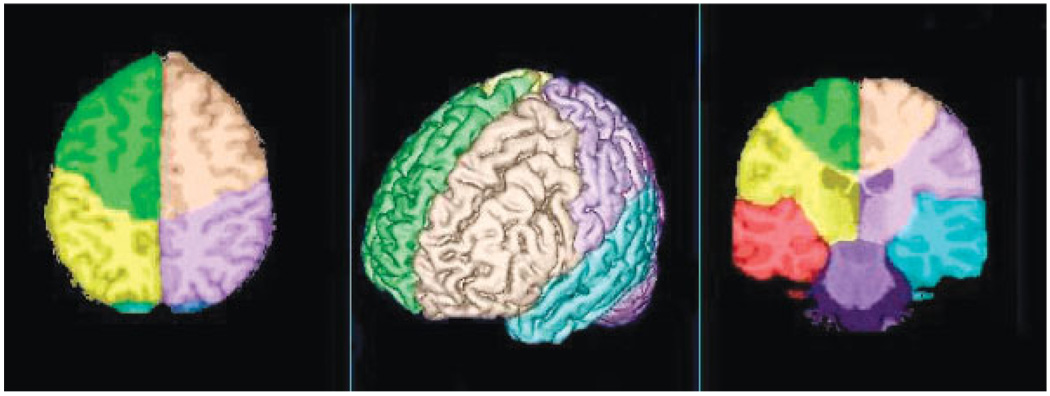
The brain atlas, which is also defined in the same reference MRI spatial coordinates, is illustrated in Fig. 5. Two orthogonal image slices and a volume-rendered brain image are shown with the lobar-scale labeling super-imposed in different colors.
Figure 5.
Illustration of the anatomical atlas with each lobar region indicated in color and overlaid on a rendering of the template brain
Metabolite database formation
For many clinical applications, MRSI-observed metabolic changes may not be visible as a focal change of the detected metabolite signals, but may exist as a diffuse change over a wider tissue region. When combined with the generally low SNRs common for in vivo MRSI and the known variability of metabolites over different brain regions, the analysis of this type of data can benefit from both signal averaging over some predefined brain region and comparisons with normal values for that same region. For example, it may be of value to determine the mean metabolite concentration in WM over a lobar-scale brain region and to compare the result with normal values for the same region. The general implementation of this type of analysis requires the formation of a reference dataset that provides mean and standard deviations of the normal metabolite values over all brain regions of interest and a mechanism to compare individual subject metabolite data with this reference. Ideally, the reference data should be generated from a large, statistically significant, group of normal subjects for the corresponding subject characteristics, such as age and gender. Previous studies indicate that the major variable to be taken into account is age (2,34); however, the creation of the MIDAS database based on a larger sample of the population may eventually provide further insight into these subject-dependent variables.
The formation of the reference metabolite information requires that results from multiple studies be combined in a standardized spatial coordinate system, as described in the previous section, where voxel-based statistics can be applied to the combined data. For this study, the metabolite database was generated by first transforming each normal-subject metabolite image dataset into the MNI reference frame using the following procedure:
Determine non-linear registration parameters between the T1-weighted MRI from the subject and the reference MRI data set.
Determine affine registration parameters between the water-reference MRSI and the subject T1-weighted MRI.
Transform all metabolite images to the standardized spatial coordinate system by applying the combined transforms determined in steps (1) and (2) and saving the spatially normalized metabolite images.
Generate group metabolite image statistics as needed, from the collection of spatially normalized metabolite image data. For example, mean and standard deviations of normal metabolite distributions can be calculated as a function of age group.
The comparison of individual subject data against the group-mean metabolite images can be done in either the subject or the reference spatial coordinate system, by making appropriate use of the spatial transformation parameters determined for that subject.
In addition to creating a database for the normal-subject group data, the MIDAS database may also include results from clinical studies, thereby enabling group statistics to be generated for any study cohort and for comparisons between cohorts.
Statistical analysis
The overall result of the data processing described in the previous sections is spatially-coregistered MRI and MRSI data from a single subject and spatially registered comparison data from a cohort of normal subjects. Several types of statistical analysis can be applied to these data, depending on the aims of the study. In applications with a strong prior anatomical hypothesis of metabolite alterations in a specific location in the brain, the classical approach of two sample comparisons by Student’s t-test may be applied to evaluate the null hypothesis. For non-Gaussian probability distributions, a Wilcoxon rank test can be used; however, in the absence of prior anatomical information regarding metabolite alterations in the brain, the statistical analysis must proceed by simultaneously assessing the MRSI data at each voxel individually. Furthermore, because a metabolite signal may arise from both GM and WM tissue, it is critical to differentiate between metabolite changes of GM, WM and other tissue types. Several statistical models that are special cases of the General Linear Model are provided.
Previous studies have demonstrated that tissue-dependent changes of metabolites can occur with disease (35). For this type of analysis, a regression of the normalized metabolite signal against the tissue content of a group of voxels can be used. A linear regression model estimates the variation of metabolite intensity, Yn, over a range of n MRSI voxels in terms of contributions from GM, WM and other tissue types, i.e.
| (4) |
where βGM and βWM represent the signal contributions from cortical GM and WM, respectively, and are synonymous with concentrations in this context. and are the weights of GM and WM in each voxel, respectively, and Bias is the signal component from spurious contributions of other tissue types, such as WM lesions and subcortical GM. The bias term is, by hypothesis, a function of the weights and , because the lower their sum for a voxel the more likely it is that other than GM and WM tissue are contributing to the signal. A major exception to this is when the rest of the voxel contains CSF or non-brain tissue, as both contribute nothing to the signal. The error term represents simply the error from estimations of the metabolite signal itself and is assumed to approximate a Gaussian. The model can be fitted using standard minimum least squares. To evaluate variations of βGM and βWM between subjects, for example, analysis of variance (ANOVA) methods can be used.
A more robust and rigorous approach to evaluate variations of βGM and βWM between subjects is to use simultaneously the MRSI data from all subjects in a study and explain metabolite variations in terms of both tissue contributions and differences among subjects. A mixed effects linear regression model is provided to conduct this type of analysis:
| (5) |
In addition to the coefficients for the model in eqn. (4), eqn. (5) contains random coefficients, , for each subject, k, and tissue type (i.e. GM or WM) and minor contributions from other brain tissue types, Bias . As an example, Table 1 lists results from a mixed effects analysis of NAA differences in GM and WM in superior brain regions (encompassing frontal, parietal and occipital lobes) in 14 normal subjects. Normalized metabolite concentrations were regressed against GM and WM voxel compositions as fixed effects and against subjects as random effects to account for between-subject variations. Both NAA and creatine had higher concentrations in GM than WM, by 20 and 60%, respectively, and choline in GM was approximately 50% of the concentration in WM. These relative concentrations can also be seen in the image data shown in Fig. 4.
Table 1.
Results of the mixed effects linear regression for metabolite concentrations in gray and white matter in superior brain regions (in institutional units), shown as mean values ± standard errors
| Tissue | Cho | Cr | NAA |
|---|---|---|---|
| GM | 0.54 ± 0.03 | 1.88 ± 0.02 | 2.64 ± 0.03 |
| WM | 1.09 ± 0.02 | 1.17 ± 0.01 | 2.19 ± 0.02 |
The ultimate goal for statistical analysis of MRSI data is assessment of metabolite abnormalities for individual subject diagnosis. The usefulness of MRSI for this purpose still remains to be demonstrated; however, a major obstacle towards achieving this goal has been the lack of uniform MRSI procedures and large MRSI databases that permit comparisons of individual MRSI data to normal values. Part of the statistical analysis provided in the MIDAS package is therefore to determine the norm of regional metabolite distributions based on data from a population of well-characterized healthy subjects. The statistical analysis tools provide a mechanism for querying MRSI data and study information (via SQL queries) using R, an open source statistical computing and graphics package (36). The results can be exported in standard formats, including the widely used Analyze image format (37).
Display methods
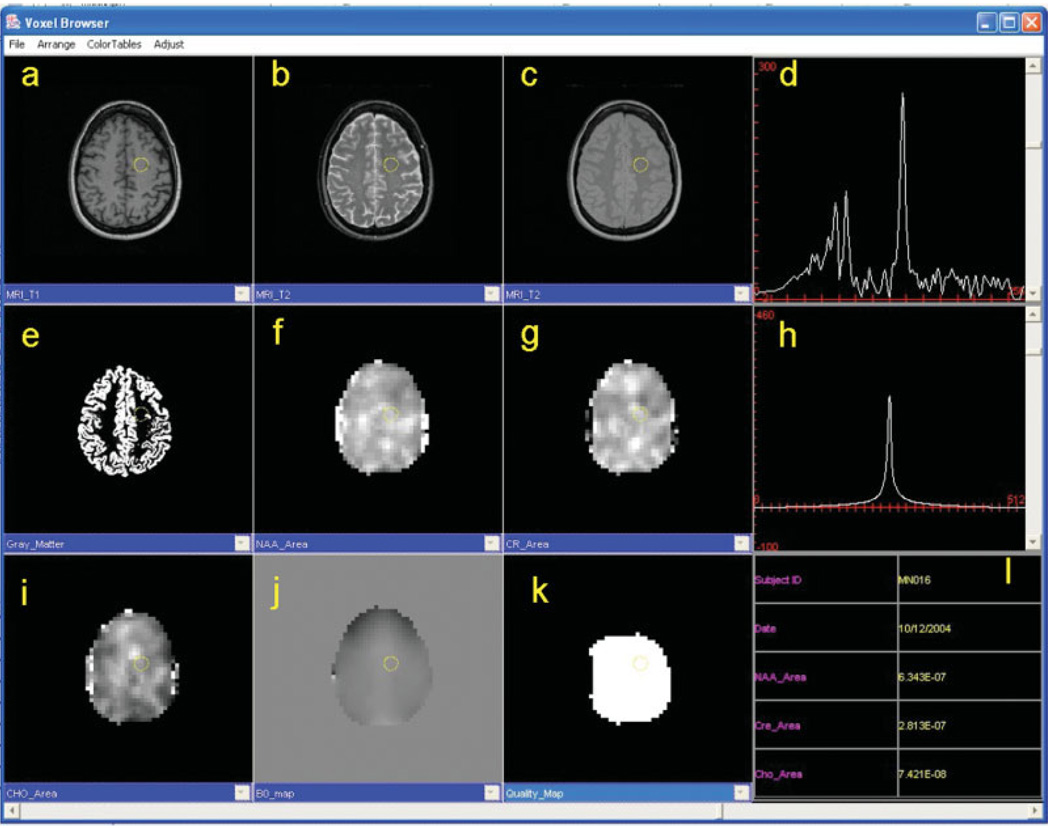
The requirements for visual review of MRSI data continue to evolve, but may include display of MRI and metabolite images as well as of individual spectra. Although the aim of the MRSI processing described in this paper is the generation of metabolite images that can be viewed directly, there remains a need to view spectra from individual voxels for both visual analysis of the relative metabolite intensities and for verifying the quality of the data. While a number of additional display and processing options may be of interest to the more research-oriented user (38), these could be a distraction for the clinical user who may require different features, such as viewing multiple study results taken at different time periods (39). Therefore, to satisfy these different visualization requirements, two display modes are provided with the MIDAS package, one more oriented to the researcher and the other to the clinician. In Fig. 6 is shown an example of this clinical-display function, which illustrates the display of multiple images having different contrast characteristics and different images resulting from processing (e.g. spectroscopic images) in a topographically correlated manner, together with the voxel spectral data. The image data type displayed in each window is fully configurable and a mouse click on any image voxel updates the spectral display. User-defined window and level adjustments for all image windows can be made independently of each other and a slider (bottom) moves all displayed images through the third dimension in concert. Multiple viewers may be opened to view and compare different studies and alternative display methods are available, including orthogonal sections through volume data sets and combined metabolite and MRI image display.
Figure 6.
Example screen shot of the MIDAS Viewer. Any acquired or derived data can be displayed, including conventional MR images with T1-, T2- and proton density-weighting (a–c), tissue content images (e.g. GM) (e), spectroscopic images of NAA, choline and creatine signal area (f, g, i), the B0 image (j) and the Quality map image (k). The metabolite spectrum (d) and the water signal obtained from the reference acquisition (h) can be displayed from any user-selected voxel, in addition to the corresponding study information and numerical results (e.g. area of choline, creatine and NAA signals). Image window and level adjustments can be made independently of each other and a slider (bottom) moves all images through the third dimension
DISCUSSION
MRSI acquisition protocols are now widely implemented on commercial MR systems and routinely carried out as part of many diagnostic MR procedures; however, the data processing methods remain less well developed. The reasons for this include the inherent complexity of a comprehensive MRSI processing protocol, the high degree of variability of the resultant data still seen in MRSI acquisitions and difficulties in obtaining comparative quantitative information on metabolite distributions. The processing methods described in this paper aim to address some of these remaining limitations. Several important features make this possible. First, the fully automated reconstruction and spectral analysis methods are essential to process the large amounts of data acquired in a typical MRSI study. Second, signal normalization is required to make full use of the available information and permit comparisons with normal values. Furthermore, this capability must also be fully integrated within the clinical acquisition protocol, without requiring extra time. Third, morphological operations including tissue segmentation and image registration provide effective approaches to improving the information content on individual MRSI studies and to the formation of reference metabolite information obtained from groups of subjects. Finally, comprehensive display and statistical analysis functions are necessary. All of these functions must also be sufficiently adaptable to allow different imaging protocols to be processed.
In this initial demonstration of the MIDAS software environment, multiple MRSI data have been processed in a fully automated manner and combined in a common spatial frame of reference. The resultant mean metabolite image data demonstrate considerable regional and tissue-specific variations throughout the whole brain, further emphasizing the need for regional comparisons of individual subject data with normal values. Statistical analysis of these spatially transformed data has also been demonstrated, to provide quantitative measurement of tissue-specific normalized metabolite signal intensities. In this study, the analysis was performed for voxels located in a superior brain region, although as the brain atlas is fully integrated with the analysis methods it will be possible to provide automatic analyses over all brain regions defined in the atlas.
The MIDAS software package and the comparative normal subject data are made available to the research community and further information on obtaining this package can be obtained from the corresponding author. It is expected that by increasing the number of users of this system, the brain metabolite database will be expanded to include additional acquisition protocols and field strengths. As improved statistics on distributions of brain metabolites in normal subject are obtained, this in turn will permit improved analysis of individual studies.
Acknowledgements
This work was supported by NIH Biomedical Research Partnership grant R01EB00822. Components of this package were also developed under NIH grants R01EB00730 and R01MH65392 and a Whitaker Foundation Biomedical Engineering Grant, RG-01-0115.
Abbreviations used
- AC–PC
anterior commissure–posterior commissure
- CSF
cerebrospinal fluid
- DICOM
Digital Imaging and Communications in Medicine
- ECC
eddy current correction
- GM
gray matter
- MIDAS
Metabolite Imaging and Data Analysis System
- MNI
Montreal Neurological Institute
- MPRAGE
magnetization prepared rapid acquisition gradient-echo
- MRSI
magnetic resonance spectroscopic imaging
- SI
signal intensity
- SNR
signal-to-noise ratio
- WM
white matter
- XML
Extensible Markup Language
REFERENCES
- 1.Maudsley AA. MR spectroscopic imaging. In: Toga AW, Mazziotta JC, editors. Brain Mapping: the Methods. Amsterdam: Academic Press; 2002. pp. 351–378. [Google Scholar]
- 2.Angelie E, Bonmartin A, Boudraa A, Gonnaud PM, Mallet JJ, Sappey-Marinier D. Regional differences and metabolic changes in normal aging of the human brain: proton MR spectroscopic imaging study. AJNR. 2001;22:119–127. [PMC free article] [PubMed] [Google Scholar]
- 3.Haupt CI, Schuff N, Weiner MW, Maudsley AA. Removal of lipid artifacts in 1H spectroscopic imaging by data extrapolation. Magn. Reson. Med. 1996;35:678–687. doi: 10.1002/mrm.1910350509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Posse S, Tedeschi G, Risinger R, Ogg R, LeBihan D. High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn. Reson. Med. 1995;33:34–40. doi: 10.1002/mrm.1910330106. [DOI] [PubMed] [Google Scholar]
- 5.Ebel A, Soher BJ, Maudsley AA. Assessment of 3D 1H NMR echo-planar spectroscopic imaging using automated spectral analysis. Magn. Reson. Med. 2001;46:1072–1078. doi: 10.1002/mrm.1301. [DOI] [PubMed] [Google Scholar]
- 6.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn. Reson. Med. 1998;39:889–898. doi: 10.1002/mrm.1910390606. [DOI] [PubMed] [Google Scholar]
- 7.Ebel A, Maudsley AA. Improved spectral quality for 3D MR spectroscopic imaging using a high spatial resolution acquisition strategy. Magn. Reson. Imaging. 2003;21:113–120. doi: 10.1016/s0730-725x(02)00645-8. [DOI] [PubMed] [Google Scholar]
- 8.Extensible Markup Language. World Wide Web Consortium. www.w3.org/XML.
- 9.DICOM Standards Committee. Digital Imaging and Communications in Medicine (DICOM) National Electrical Manufacturers Association; [Google Scholar]
- 10.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn. Reson. Med. 1998;40:822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- 11.Johnson G, Jung KJ, Wu EX, Hilal SK. Self-correction of proton spectroscopic images for gradient eddy current distortions and static field inhomogeneities. Magn. Reson. Med. 1993;30:255–261. doi: 10.1002/mrm.1910300217. [DOI] [PubMed] [Google Scholar]
- 12.Rousseau F, Maudsley AA, Ebel A, Darkazanli A, Weber P, Sivasankaran K, Yu Y, Studholme C. Evaluation of sub-voxel registration accuracy between MRI and 3D MR spectroscopy of the brain. Proc. SPIE. 2005;5747:1213–1221. [Google Scholar]
- 13.Aygula Z, Soher BJ, Young K, Maudsley AA. GAVA–s graphical pulse sequence simulation, display and storage environment. Proceedings of the International Society for Magnetic Resonance in Medicine; Toronto. 2003. p. 852. [Google Scholar]
- 14.Podo F, Henriksen O, Bovée WM, Leach MO, Leibfritz D, De Certaines JD. Absolute metabolite quantification by in vivo NMR spectroscopy: I. Introduction, objectives and activities of a concerted action in biomedical research. Magn. Reson. Imaging. 1998;16:1085–1092. doi: 10.1016/s0730-725x(98)00117-9. [DOI] [PubMed] [Google Scholar]
- 15.Keevil SF, Barbiroli B, Brooks JC, Cady EB, Canese R, Carlier P, Collins DJ, Gilligan P, Gobbi G, Hennig J, Kügel H, Leach MO, Metzler D, Mlynárik V, Moser E, Newbold MC, Payne GS, Ring P, Roberts JN, Rowland IJ, Thiel T, Tkác I, Topp S, Wittsack HJ, Podo F. Absolute metabolite quantification by in vivo NMR spectroscopy: II. A multicentre trial of protocols for in vivo localised proton studies of human brain. Magn. Reson. Imaging. 1998;16:1093–1106. doi: 10.1016/s0730-725x(98)00118-0. [DOI] [PubMed] [Google Scholar]
- 16.Alger JR, Symko SC, Bizzi A, Posse S, DesPres DJ, Armstrong MR. Absolute quantitation of short TE brain 1H-MR spectra and spectroscopic imaging data. JCAT. 1993;17:191–199. doi: 10.1097/00004728-199303000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Haacke EM, Brown RW, Thompson MR, Ventkatesan R. Magnetic Resonance Imaging: Physical Principles and Sequence Design. New York: Wiley; 1999. [Google Scholar]
- 18.Chang L, Ernst T, Poland RE, Jenden DJ. in vivo proton magnetic resonance spectroscopy of the normal aging human brain. Life Sci. 1996;58:2049–2056. doi: 10.1016/0024-3205(96)00197-x. [DOI] [PubMed] [Google Scholar]
- 19.Ernst T, Kreis R, Ross BD. Absolute quantitation of water and metabolites in the human brain. I. Compartments and water. J. Magn. Reson. B. 1993;102:1–8. [Google Scholar]
- 20.Fatouros PP, Marmarou A. Use of magnetic resonance imaging for in vivo measurements of water content in human brain: method and normal values. J. Neurosurg. 1999;90:109–115. doi: 10.3171/jns.1999.90.1.0109. [DOI] [PubMed] [Google Scholar]
- 21.Kövér F, Schwarcz A, Pál J, Bogner P, Vajna T, Vandon G, Dóczi T. Fast method for longitudinal relaxation time and water content mapping of the human brain on a clinical scanner. Acta Neurochir. 2004;146:1341–1346. doi: 10.1007/s00701-004-0370-9. [DOI] [PubMed] [Google Scholar]
- 22.Cleveland WS, Devlin SJ. Locally weighted regression: an approach to regression analysis by local fitting. J. Am. Stat. Assoc. 1988;83:569–610. [Google Scholar]
- 23.Clark MC, Hall LO, Goldgof DB, Clark L, Velthuizen R, Silbiger ML. MRI segmentation using fuzzy clustering techniques: integrating knowledge. IEEE Eng. Med. Biol. Mag. 1994;13:730–742. [Google Scholar]
- 24.Cheng TW, Goldgof DB, Hall LO. Fast fuzzy clustering. Fuzzy Sets Syst. 1998;93:49–56. [Google Scholar]
- 25.Pfefferbaum A, Adalsteinsson E, Spielman D, Sullivan EV, Lim KO. in vivo spectroscopic quantification of the N-acetyl moiety, creatine and choline from large volumes of brain gray and white matter: Effects of normal aging. Magn. Reson. Med. 1999;41:276–284. doi: 10.1002/(sici)1522-2594(199902)41:2<276::aid-mrm10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 26.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recog. 1999;32:71–86. [Google Scholar]
- 27.Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans. Med. Imaging. 1998;17:463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- 28.Studholme C, Novotny E, Zubal IG, Duncan JS. Estimating tissue deformation between functional images induced by intracranial electrode implantation using anatomical MRI. Neuroimage. 2001;13:561–576. doi: 10.1006/nimg.2000.0692. [DOI] [PubMed] [Google Scholar]
- 29.Schuff N, Ezekiel F, Gamst A, Amend D, Capizzano AA, Maudsley AA, Weiner MW. Region and tissue differences of metabolites in normally aged brain using 1H magnetic resonance spectroscopic imaging. Magn. Reson. Med. 2001;45:899–907. doi: 10.1002/mrm.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs MA, Horska A, van Zijl PC, Barker PB. Quantitative proton MR spectroscopic imaging of normal human cerebellum and brain stem. Magn. Reson. Med. 2001;46:699–705. doi: 10.1002/mrm.1248. [DOI] [PubMed] [Google Scholar]
- 31.Pfefferbaum A, Adalsteinsson E, Spielman D, Sullivan EV, Lim KO. in vivo brain concentrations of N-acetyl compounds, creatine and choline in Alzheimer disease. Arch. Gen. Psychiatry. 1999;56:185–192. doi: 10.1001/archpsyc.56.2.185. [DOI] [PubMed] [Google Scholar]
- 32.Barker PB, Szopinski K, Horska A. Metabolic heterogeneity at the level of the anterior and posterior commissures. Magn. Reson. Med. 2000;43:348–354. doi: 10.1002/(sici)1522-2594(200003)43:3<348::aid-mrm5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 33.Geurts JJ, Barkhof F, Castelijns JA, Uitdehaag BM, Polman CH, Pouwels PJ. Quantitative 1H-MRS of healthy human cortex, hippocampus and thalamus: metabolite concentrations, quantification precision and reproducibility. J. Magn. Reson. Imaging. 2004;20:366–371. doi: 10.1002/jmri.20138. [DOI] [PubMed] [Google Scholar]
- 34.Komoroski RA, Heimberg C, Cardwell D, Karson CN. Effects of gender and region on proton MRS of normal human brain. Magn. Reson. Imaging. 1999;17:427–433. doi: 10.1016/s0730-725x(98)00186-6. [DOI] [PubMed] [Google Scholar]
- 35.Schuff N, Capizzano AA, Du AT, Amend DL, O’Neill J, Norman D, Jagust WJ, Chui HC, Kramer JH, Reed BR, Miller BL, Yaffe K, Weiner MW. Different patterns of N-acetylaspartate loss in subcortical ischemic vascular dementia and AD. Neurology. 2003;61:358–364. doi: 10.1212/01.wnl.0000078942.63360.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The R Foundation for Statistical Computing. www.r-project.org.
- 37.Analyze. http://www.mayo.edu/bir/Software/Analyze/Analyze.html.
- 38.Maudsley AA, Lin E, Weiner MW. Spectroscopic imaging display and analysis. Magn. Reson. Imaging. 1992;10:471–485. doi: 10.1016/0730-725x(92)90520-a. [DOI] [PubMed] [Google Scholar]
- 39.Alger JR, Frew AJ, Cloughesy TF, Del Vecchio W, Villablanca JP, Curran JG. Novel methodology for the archiving and interactive reading of clinical magnetic resonance spectroscopic imaging. Magn. Reson. Med. 2002;48:411–418. doi: 10.1002/mrm.10239. [DOI] [PubMed] [Google Scholar]