Abstract
DNA–protein cross-links (adducts) are formed in cellular DNA under a variety of conditions, particularly following exposure to an α,β-unsaturated aldehyde, acrolein. DNA–protein cross-links are subject to repair or damage-tolerance processes. These adducts serve as substrates for proteolytic degradation, yielding DNA–peptide lesions that have been shown to be actively repaired by the nucleotide excision repair complex. Alternatively, DNA–peptide cross-links can be subjected to replication bypass. We present new evidence about the capabilities of DNA polymerases to synthesize DNA past such cross-links. DNAs were constructed with site-specific cross-links, in which either a tetrapeptide or dodecylpeptide was covalently attached at the N2 position of guanine via an acrolein adduct, and replication bypass assays carried out with members of the DinB family of polymerases, human polymerase (pol)κ and Escherichia coli (E. coli) pol IV, and various E. coli polymerases that do not belong to the DinB family. Pol κ was able to catalyze both the incorporation and extension steps with an efficiency that was qualitatively indistinguishable from control (undamaged) substrates. Fidelity was comparable on all these substrates, suggesting that pol κ would have a role in the low mutation frequency associated with replication of these adducts in mammalian cells. When the E. coli orthologue of pol κ, damage-inducible DNA polymerase, pol IV, was analyzed on the same substrates, pause sites were detected opposite and three nucleotides beyond the site of the lesion, with incorporation opposite the lesion being accurate. In contrast, neither E. coli replicative polymerase, pol III, nor E. coli damage-inducible polymerases, pol II and pol V, could efficiently incorporate a nucleotide opposite the DNA–peptide cross-links. Consistent with a role for pol IV in tolerance of these lesions, the replication efficiency of DNAs containing DNA–peptide cross-links was greatly reduced in pol IV-deficient cells. Collectively, these data indicate an important role for the DinB family of polymerases in tolerance mechanisms of N2-guanine linked DNA–peptide cross-links.
Introduction
DNA–protein cross-links are formed in cells not only as a consequence of routine DNA metabolism, but also from exposure to a variety of chemical toxicants and metals (1). In particular, these lesions have been detected in DNA following treatment of cultured human cells with an α,β-unsaturated aldehyde, acrolein (2, 3). Among aldehydes that can induce DNA–protein cross-links in vitro, acrolein is one of the most potent (4). It is cytotoxic and mutagenic (5–7), and has tumor initiating activity (8). Acrolein is ubiquitous in the environment, but more importantly, it is generated in vivo, mainly as a product of peroxidation of fatty acids (9–11). Thus, acrolein may represent a significant endogenous causative factor for induction of DNA–protein cross-links.
Being a strong bifunctional electrophile, acrolein reacts with DNA to modify nucleobases at various positions and in particular, at exocyclic nitrogen (N2) of deoxyguanosine (dG1) (12–16). The major product of reaction between N2-dG and acrolein is γ-hydroxypropano-deoxyguanosine (γ-HOPdG) (12), a mutagenic adduct (17) that has been detected in cellular DNA isolated from various tissues both following acrolein exposure and as endogenous lesion (10, 18–20). The γ-HOPdG adduct undergoes spontaneous ring-opening and closing (Fig. 1A) with the equilibrium being shifted toward the ring-opened form in duplex DNA (21, 22). The free aldehyde of the ring-opened γ-HOPdG can further react with amines in peptides and proteins to yield Schiff base-mediated linkages (23, 24). Similar to the acrolein adduct, N2-dG adducts of other biologically important α,β-unsaturated aldehydes, crotonaldehyde and trans-4-hydroxynonenal, have also been shown to form Schiff base-mediated DNA–peptide cross-links (23). Therefore, elucidation of a role for acrolein and other related compounds in living organisms cannot be complete without detailed understanding of cellular processing of DNA–protein cross-links.
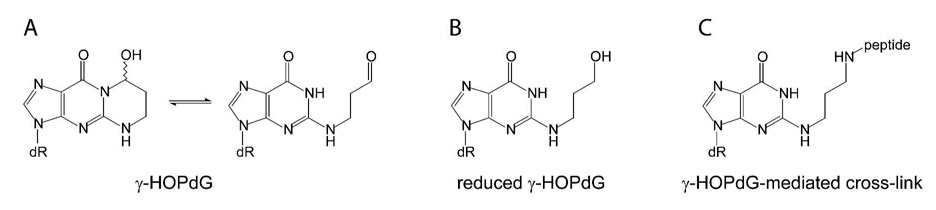
Fig. 1.
Structure of DNA adducts used in the current study. A. γ-hydroxypropano deoxyguanosine shown as the equilibrium between the ring-closed and ring-opened forms; B. reduced γ-hydroxypropano deoxyguanosine; C. reduced γ-hydroxypropano deoxyguanosine-mediated peptide cross-link.
Although a subset of DNA–protein cross-links can undergo spontaneous reversal, others require active removal by DNA repair machinery or are processed through damage avoidance and tolerance pathways. Prokaryotic nucleotide excision repair (NER) has been demonstrated to function on DNA–protein adducts as large as 16,000 Da (25), while comparably sized cross-linked protein adducts were refractory to excision by the human NER complex (26–28). However, these data showing a lack of repair of intact proteins by human NER, do not rule out a role for this pathway in the ultimate processing of these lesions, since there is a literature precedent suggesting an initial proteolytic processing of these lesions prior to excision. In this regard, data obtained from the Zhitkovich’s group (29) demonstrated that repair of formaldehyde-induced DNA–protein cross-links was significantly reduced in cells treated with lactacystin, a proteasome inhibitor. It was hypothesized that the cross-linked proteins were proteolytically degraded into substrates suitable for completion of the repair process. Consistent with these results, DNA–peptide cross-links serve as much better substrates relative to DNA–protein cross-links for both prokaryotic (25, 30) and human NER (26–28). Baker et al. also demonstrated that incubation with the 26S proteasome inhibitor, MG132, resulted in a greater than 50% reduction in the intracellular repair of plasmids carrying site-specific DNA–protein cross-links (28). Collectively, these data indicate that biological processing of covalently linked protein adducts can proceed via a proteolytic degradative process, converting DNA–protein cross-links into DNA–peptide cross-links.
NER processing of DNA–peptide cross-links could be sufficiently delayed to make these lesions available for DNA replication bypass and potentially lead to mutations. Recently, we have assessed the mutagenic properties of Lys-Trp-Lys-Lys cross-links during extrachromosomal replication in mammalian cells. When this tetrapeptide was linked to the N2position of dG via the acrolein-derived γ-HOPdG, lesion bypass was only moderately mutagenic (31). Previous investigations have suggested that the efficient and non-mutagenic replication bypass of bulky N2-dG adducts can be performed by DNA polymerases of the DinB family, polymerase (pol) κ in mammalian cells and its bacterial orthologue, pol IV, in Escherichia coli (E. coli) (32–39). Here, we hypothesized that human pol κ as well as pol IV also function in replication bypass of N2-dG–peptide cross-links. In order to test this hypothesis, we have created site-specifically modified oligodeoxynucleotides that contain either tetrapeptide (Lys-Trp-Lys-Lys) or dodecylpeptide (Lys-Phe-His-Glu-Lys-His-His-Ser-His-Arg-Gly-Tyr) γ-HOPdG-mediated cross-links (Fig. 1) and performed replication assays in vitro. Additionally, the DNA–peptide cross-links were introduced into a single-stranded DNA vector (pMS2) (40) and the consequences of replication past these lesions were investigated in both the wild type and pol IV-deficient E. coli strains.
Materials and Methods
DNA polymerases
Human pol κ was purchased from Enzymax Inc, Lexington, KY. The polymerase is full length and tagged at its N-termini with the calmodulin-binding peptide. Subunits of the pol III replicase were purified as described before (41, 42). Pol III core was reconstituted by mixing α,ε and θ subunits (1:1.5:3 molar ratio, respectively) and incubating for 30 min at 15 °C. Pol III core was resolved from unbound proteins by chromatography on MonoQ as described previously (43). The pol III replicase, containing two molecules of pol III core connected by the clamp loader, was reconstituted as follows. A mixture of γ and τ (2.5:1 molar ratio, respectively) was incubated for 90 min at 15 °C, followed by the addition of δ,δ',χ, and ψ (3-fold molar excess of each subunit over τ) followed by a further 1 hr incubation. The τ2γ1δδ'χψ complex was resolved from other species (τ3δδ'χψ, τ1γ2δδ'χψ and γ3δδ'χψ) using a 1-ml MonoS column by elution with a 0.0–0.5 M NaCl gradient as described before (44). The τ2γ1δδ'χψ clamp loader was incubated with a 4-fold molar excess of pol III core, and pol III replicase was purified from excess pol III core by MonoQ anion exchange chromatography as reported previously (42). E. coli pol II (45), pol IV (46, 47), pol V (48), and RecA (49) were purified as previously described.
Bacterial strains
The E.coli strains ZK126 (W3110 DlacU169 tna2) and its pol IV-deficient derivative (ZK126 dinB::Kan) (50) used in this study were the generous gift of Dr. Steven Finkel, University of Southern California, Los Angeles.
DNA substrates
DNA oligodeoxynucleotides containing a site-specific γ-HOPdG (Fig. 1A) were synthesized and purified as previously described (51). This 30-mer had the following sequence: GCTAGTACTCGTCGACAATTCCGTATCCAT, where the bolded G denotes the position of the γ-HOPdG. Procedures to convert γ-HOPdG into the permanently ring-opened adduct, reduced γ-HOPdG (Fig. 1B) were performed as previously described (31). The Lys-Trp-Lys-Lys tetrapeptide was synthesized by Sigma-Genosys. The Lys-Phe-His-Glu-Lys-His-His-Ser-His-Arg-Gly-Tyr dodecylpeptide was purchased from Sigma-Genosys. Generation of the DNA–peptide cross-links (Fig. 1C) and purification of the DNA–peptide cross-link-containing oligodeoxynucleotides were carried out according to a published protocol (31). These 30-mer oligodeoxynucleotides were used as templates in replication reactions in vitro and also for preparation of site-specifically modified vectors for cellular studies. All other oligodeoxynucleotides were obtained from the Molecular Microbiology and Immunology Research Core Facility, Oregon Health & Science University.
Replication assays in vitro
Preparations of primer-template DNA substrates were performed as previously described (52). Polymerase bypass assays with human pol κ, and E. coli pol II, pol III, and pol IV were carried out using 5 nM primer-template DNA substrates in the presence of 25 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 10% (v/v) glycerol, 10 mM NaCl, 0.1 mg/ml bovine serum albumin, and 5 mM dithiothreitol at 37 °C. Polymerase bypass assays with E. coli pol V were carried out using 5 nM primer-template DNA substrates in the presence of 20 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 4% (v/v) glycerol, 10 mM sodium glutamate, 5 mM dithiothreitol, 100 µM EDTA, 2 µM a single-stranded 36-mer oligodeoxynucleotide, 12 µM RecA, and 1 mM adenosine 5'[γ-thio]triphosphate. Prior to pol V-catalyzed reactions, single-stranded oligodeoxynucleotide and RecA were preincubated in reaction buffer in the presence of adenosine 5'[γ-thio]triphosphate at 37 °C for 3 min and then added to the primer-template DNA substrates. The reactions were initiated by addition of dNTPs and pol V and conducted at 37 °C. Protein concentrations, dNTPs concentrations, and incubation times are given in the figure legends. Polymerase reactions were terminated by the addition of an equal volume of a solution of 95% (v/v) formamide, 20 mM EDTA, 0.2% (w/v) bromphenol blue, and 0.2% (w/v) xylene cyanol. Products were resolved through a 15% (w/v) denaturing polyacrylamide gel in the presence of 8 M urea and visualized using a PhosphorImager screen (GE Healthcare).
Replication of DNAs containing site-specific DNA–peptide cross-links in E. coli
The pMS2 phagemid shuttle vector (40) was a gift from Dr. M. Moriya, State University of New York, Stony Brook. Single-stranded, site-specifically modified pMS2 vectors were constructed as in previous studies (17, 31, 40). Transformations of E. coli cells with these vectors and analyses of the progeny DNAs by differential hybridization were performed as previously described (17, 31). Individual DNAs were isolated using Qiagen plasmid minipreparation kits and tested by ScaI restriction endonuclease according to manufacturer’s recommendations.
Results
Preparation of DNA–Peptide Cross-links
Although there are multiple potential sites of adduction within both DNA and protein, we have chosen to model DNA–peptide cross-links via acrolein-modified DNA at N2-dG. Acrolein reacts with DNA primarily at N2-dG, yielding γ-HOPdG, a lesion that undergoes spontaneous ring-opening and closing (Fig. 1A) (21). In the presence of NaBH4, γ-HOPdG can be converted to its reduced derivative (Fig. 1B) that serves in our assays as a model for the permanently ring-opened form of the adduct. The free aldehyde of the natural, unreduced γ-HOPdG can react with primary and secondary amines in peptides and proteins (23, 24). When DNAs containing these lesions are incubated in the presence of peptides that contain both alpha and epsilon amino groups, the relative pKa of the alpha amino group dominates the course of the cross-linking reaction, forming a carbinolamine linkage between the N-terminal alpha amino group and the aldehyde of the γ-HOPdG adduct. The initial carbinolamine linkage can dehydrate to a Schiff base, that can be reductively trapped in the presence of a mild reducing agent such as NaCNBH3 to form an irreversible cross-link (Fig. 1C) (23). Thus, DNAs incubated as described above have stable peptide cross-links that are directly amenable to replication bypass analyses. These linkages through the alpha amino group are common intermediates for a variety of DNA glycosylase/abasic site lyases (53) and thus, our DNA–peptide cross-links model biologically relevant lesions.
The 30-mer oligodeoxynucleotides containing DNA–peptide cross-links were prepared as previously described (31) and tested for purity using denaturing (8M urea) polyacrylamide gel. No bands were detected that would suggest either the presence of DNAs without cross-link or decomposition of oligodeoxynucleotides (data not shown).
Bypass of DNA–peptide cross-links by human pol κ
Our previous studies had indicated that both the γ-HOPdG adduct and its reduced derivative posed a severe block to DNA synthesis by eukaryotic replicative polymerases, pol δ and pol ε (17, 54). Based on these observations, we considered it unlikely that either of them would be able to replicate past bulky γ-HOPdG-mediated DNA–peptide cross-links. In contrast to replicative polymerases, human pol κ could efficiently incorporate the correct nucleotide, C, opposite the ring-opened, reduced γ-HOPdG (54, 55). Further, pol κ will efficiently extend a 0 primer if the correct C has been placed opposite either γ-HOPdG or its reduced derivative (54, 55). In the current study, we compared the ability of pol κ to carry out translesion synthesis past tetra- and dodecylpeptides linked to DNA via the ring-opened γ-HOPdG adduct versus control (unadducted) DNAs (Fig 1C).
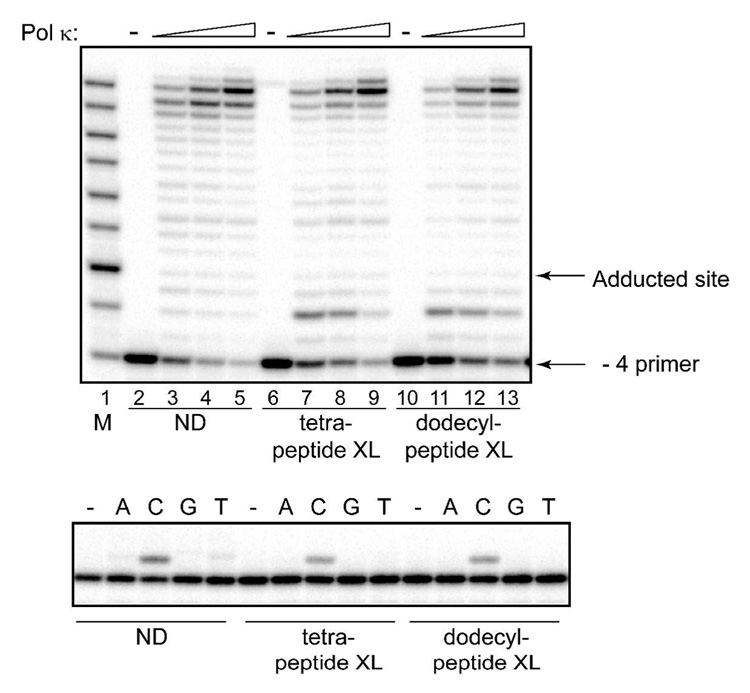
Using a - 4 primer, pol κ showed a significant processivity on nondamaged DNA, with a large percentage of primers being extended to near full length (Fig. 2, upper panel, lanes 2–5). Surprisingly, neither of the peptide-modified γ-HOPdG substrates blocked the progression of pol κ (Fig. 2, upper panel, lanes 6–13). Although there was a minor pause site, two nucleotides prior to the adducted site, there was no evidence of even a minor replication pause at the site of the lesion, and primers could be extended to full length comparable with the control DNAs.
Fig. 2.
Replication bypass of DNA–peptide cross-links by human pol κ. Upper panel: Primer extensions by pol κ were carried out for 20 min in the presence of 100 µM dNTPs at 1.25 nM (lanes 3, 7, and 11), 2.5 nM (lanes 4, 8, and 12) or 5 nM (lanes 5, 9, and 13) protein concentrations. Lower panel: Single nucleotide incorporations by pol κ (1 nM) were carried out for 5 min in the presence of 20 µM individual dNTPs. M – oligodeoxynucleotide sizing markers; ND – non-damaged template; XL – cross-link-containing template.
Since pol κ incorporated a nucleotide opposite both lesions, the relative fidelity of the reactions was examined by carrying out single nucleotide incorporation assays using a −1 primer (Fig 2, lower panel). These assays revealed that in addition to these DNA–peptide cross-links not posing a block, pol κ utilized dCTP with a very strong preference over any other of the dNTPs (Fig. 2, lower panel). Taken together, these data indicate that pol κ has the capacity to accurately bypass the complex N2-dG acrolein-derived peptide adducts.
Replication of DNAs containing DNA–peptide cross-links by E. coli DNA polymerases
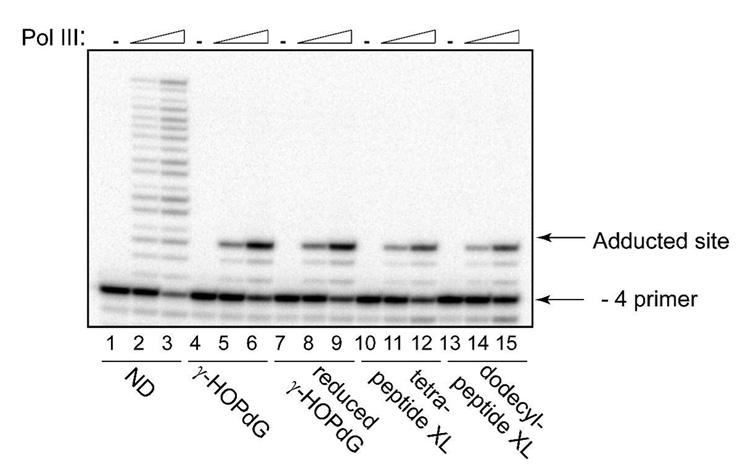
Whereas the blocking effect of γ-HOPdG and reduced γ-HOPdG has been shown for eukaryotic replicative polymerases (17, 54), the replicative polymerase in E. coli cells, pol III, has not been previously tested with any of these adducts. Here, the primer extensions by pol III were conducted with – 4 primers that were annealed with DNA templates containing either γ-HOPdG, reduced γ-HOPdG, or γ-HOPdG-derived DNA–peptide cross-links (Fig. 3). The form of the multi-subunit pol III replicase employed here is the dimeric replicase that is utilized for chromosome replication; it contains two molecules of pol III core that attach to two τ subunits of the clamp loading assembly. The observed results reveal that while the pol III replicase could efficiently catalyze DNA synthesis utilizing the nondamaged substrate (Fig. 3, lanes 2 and 3), no bypass was detected in reactions containing any of the adducted templates; all primer extensions were terminated one nucleotide prior to the lesion even at the highest enzyme concentrations (Fig. 4, lanes 6, 9, 12, and 15). Previously, it has been shown that the exonuclease-deficient Klenow fragment of pol I, which is another major DNA polymerase in E. coli, catalyzed very inefficient and extremely error-prone DNA synthesis past the γ-HOPdG adduct (56, 57). Collectively, these data suggest that similar to mammalian cells, DNA synthesis past γ-HOPdG-derived lesions in bacteria is likely to utilize specialized bypass polymerases.
Fig. 3.
Replication bypass of γ-HOPdG, reduced γ-HOPdG, and DNA–peptide cross-links by E. coli pol III. Primer extensions by the pol III replicase were carried out for 20 min in the presence of 200 µM dNTPs at 0.01 nM (lanes 2, 5, 8, 11, and 14), or 0.05 nM (lanes 3, 6, 9, 12, and 15) protein concentrations. ND – non-damaged template; XL – cross-link-containing template.
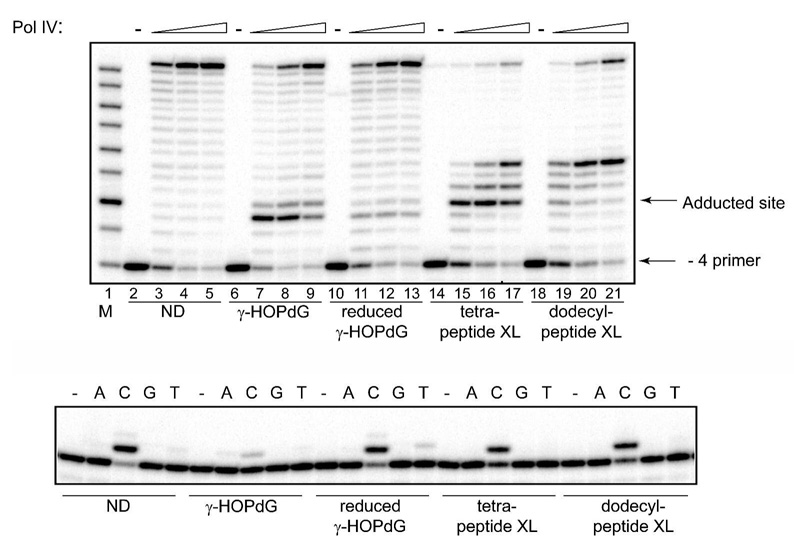
Fig. 4.
Replication bypass of γ-HOPdG, reduced γ-HOPdG, and DNA–peptide cross-links by E. coli pol IV. Upper panel: Primer extensions by pol IV were carried out for 20 min in the presence of 100 µM dNTPs at 1.25 nM (lanes 3, 7, 11, 15, and 19), 2.5 nM (lanes 4, 8, 12, 16, and 20) or 5 nM (lanes 5, 9, 13, 17, and 21) protein concentrations. Lower panel: Single nucleotide incorporations by pol IV (1 nM) were carried out for 20 min in the presence of 20 µM individual dNTPs. M – oligodeoxynucleotide sizing markers; ND – non-damaged template; XL – cross-link-containing template.
Given that human pol κ could readily bypass these lesions, it was hypothesized that E. coli pol IV, which is an orthologue of mammalian pol κ (58), might be capable of catalyzing translesion synthesis past these adducts. As anticipated, pol IV extended a - 4 primer annealed with an undamaged template to full length product (Fig. 4, upper panel, lanes 2–5). In contrast, extension of the - 4 primer annealed to the DNA template containing γ-HOPdG was significantly blocked one nucleotide prior to the lesion and modestly opposite the lesion (Fig. 4, upper panel, lanes 6–9). However, given a sufficient number of enzyme-DNA encounters, extension could proceed to full length. Similar reactions, but using the reduced γ-HOPdG showed only a modest pause site one nucleotide prior to the lesion (Fig. 3, upper panel, lanes 10–13). Thus, the permanently ring-opened analog of the lesion affords efficient bypass.
In contrast to these data, replication of the templates containing the γ-HOPdG adducts with tetra- and dodecylpeptide revealed different patterns of inhibition. For the DNA modified with tetrapeptide, the dominant pause site occurred following incorporation of a nucleotide opposite the lesion (Fig. 4, upper panel, lanes 14–17). However, using increasing enzyme concentrations, blockage opposite the lesion could be overcome, only to encounter replication blockage one and three nucleotides beyond the lesion (Fig. 4, upper panel, lanes 16, 17). The block at the third position beyond the lesion was very poorly extended, suggesting that there are structural impediments to the passage of the bulky adducted duplex DNA through the backside of pol IV. Comparable analyses using the dodecylpeptide-containing substrate revealed a less significant blockage opposite the lesion, but an equivalent pause site at the third position beyond the lesion (Fig. 4, upper panel, lanes 18–21). These data confirm that the presence of N2-dG-linked peptide inhibits pol IV-catalyzed processive synthesis several nucleotides past the site of the lesion. However, since the magnitude and site of the pause was not affected by the increased bulk of the larger peptide, these data suggest that the impeding interactions are localized to the first few amino acids of the adducted peptides.
Single nucleotide incorporation analyses revealed that pol IV preferentially incorporated the correct nucleotide, C, opposite all of these lesions; however, misincorporations of A and T opposite the γ-HOPdG and reduced γ-HOPdG adducts were also observed (Fig. 4, lower panel).
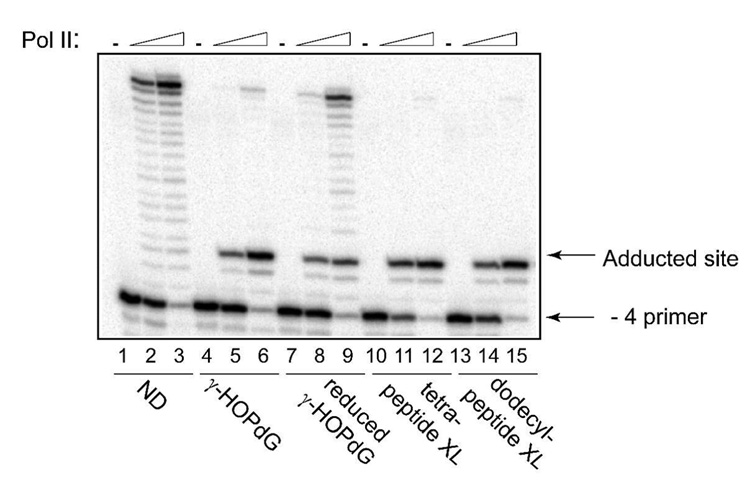
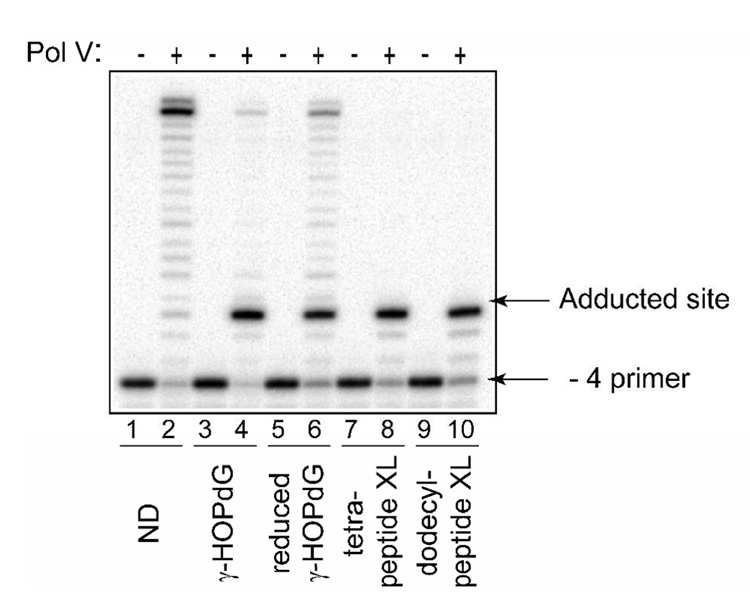
In addition to pol IV, E. coli possesses two other DNA damage-induced polymerases, pol II and pol V (59). Here, the abilities of pol II and pol V to synthesize DNA past γ-HOPdG, reduced γ-HOPdG, and γ-HOPdG-derived DNA–peptide cross-links have been tested by conducting primer extension experiments similar to those described above. The data revealed that pol II was strongly inhibited by all these lesions one nucleotide prior to the site of modification (Fig. 5). The blockage was less severe on the DNA substrate containing the ring-opened, reduced γ-HOPdG adduct with a significant percentage of primers being extended to the end of the template (Fig. 5, lanes 8 and 9). In contrast, almost no full-length products were detected in reactions using substrates that contained either γ-HOPdG (Fig. 5, lanes 5 and 6) or DNA–peptide cross-links (Fig. 5, lanes 11, 12, 14, and 15). When pol V was examined with the same set of modified DNAs (Fig. 6), patterns of inhibition of DNA synthesis appeared to be very similar to that observed for pol II, with the only exception that in pol V-catalyzed reactions, no bypass products were detected on substrates adducted with DNA–peptide cross-links (Fig. 6, lanes 8 and 10). Thus, among E. coli DNA polymerases, pol IV was uniquely able to tolerate N2-dG linked DNA–peptide cross-links in vitro.
Fig. 5.
Replication bypass of γ-HOPdG, reduced γ-HOPdG, and DNA–peptide cross-links by E. coli pol II. Primer extensions by pol II were carried out for 20 min in the presence of 250 µM dNTPs at 0.5 nM (lanes 2, 5, 8, 11, and 14), or 5.0 nM (lanes 3, 6, 9, 12, and 15) protein concentrations. ND – non-damaged template; XL – cross-link-containing template.
Fig. 6.
Replication bypass of γ-HOPdG, reduced γ-HOPdG, and DNA–peptide cross-links by E. coli pol V. Primer extensions by pol V (200 nM) were carried out for 20 min in the presence of 500 µM dNTPs. ND – non-damaged template; XL – cross-link-containing template.
Replication of plasmid DNA containing DNA–peptide cross-links in E. coli
Having observed replication bypass of the γ-HOPdG-derived DNA–peptide cross-links by pol IV in vitro, we hypothesized that this polymerase may function in such a process intracellularly. To test this hypothesis, 30-mer DNA oligodeoxynucleotides adducted with either tetrapeptide or dodecylpeptide or containing no damage, were individually inserted into a single-stranded pMS2 vector, and these vectors were used to transform the wild type and pol IV-deficient E. coli cells. Following overnight selection on ampicillin-containing plates, colonies were counted, and the relative efficiency of transformation was calculated as the ratio of the number of clones originating from the adducted vectors to the number of clones originating from the nondamaged control. The average relative efficiencies with their standard deviations obtained from three independent experiments are given in Table 1, third column. These data suggested that DNA–peptide cross-links represented more severe block for replication in pol IV-deficient mutant than in wild-type cells. However, based on our previous experience (31), it was possible that a substantial percentage of the progeny DNAs originated from ligated pMS2 vectors did not contain the insert sequences, and therefore, they should be excluded from the analyses. To evaluate the number of progeny DNAs having specific insert sequences, the following approach was utilized. A subpopulation of clones were individually grown in the 96-well plates, transferred onto Whatman 541 paper, and subjected to differential hybridization, utilizing the 5′-radioactively labeled oligodeoxynucleotide probe G. This probe was designed to be complementary to the insert sequence, assuming no mutations were introduced. Additional 20-mer oligodeoxynucleotide probes, L and R were designed to be complementary to the vector sequences located 60 and 52 nucleotides from the site of insertion. Hybridization with these probes allowed for the exclusion of the progeny DNAs with deletions involving the region of interest. Individual DNAs that hybridized with a mixture of L and R probes, but did not hybridize with G probe, were isolated and analyzed further. Specifically, they were tested by cleavage with ScaI endonuclease. Since both the original pMS2 vector and the inserted DNAs contain ScaI recognition sites, cleavage of progeny DNAs containing the insert sequences can be easily identified by the generation of DNA fragments, ~1.9 and ~3.2 kb in length. Thus, the combination of differential hybridization and analyses of isolated plasmids by cleavage with ScaI facilitated the determination of the percentage of the progeny DNAs having inserted sequences (Table 1, fifth column). Subsequently, these values were used to re-evaluate the relative efficiencies of transformation, taking into account only the transformants in which vectors with inserts, but not other products of ligation and/or intracellular DNA rearrangements, were replicated. These data indicated that relative to wild type cells, the pol IV-deficient mutant was very inefficient in replicating the adducted vectors, thus, confirming the intracellular role for pol IV in bypass of γ-HOPdG-derived DNA–peptide cross-links.
Table 1.
Efficiency of E. coli transformation using vector DNAs containing DNA–peptide cross-links
| E. coli strain | Insert | Relative efficiency of transformation | Number of colonies tested | Colonies with inserts, % | Relative efficiency of transformation adjusted for number of colonies with inserts | Efficiency of transformation of pol IV-deficient mutant relative to wild type |
|---|---|---|---|---|---|---|
| Wild type | Non-damaged | 1 | 106 | 97 | 1 | |
| Tetra-peptide cross-link | 0.45 ± 0.04 | 136 | 85 | 0.39 ± 0.035 | 1 | |
| Dodecyl-peptide cross-link | 0.39 ± 0.15 | 128 | 93 | 0.37 ± 0.14 | 1 | |
| Pol IV− | Non-damaged | 1 | 136 | 98.5 | 1 | |
| Tetra-peptide cross-link | 0.05 ± 0.01 | 67 | 37 | 0.02 ± 0.004 | 0.05 | |
| Dodecyl-peptide cross-link | 0.12 ± 0.07 | 80 | 56 | 0.07 ± 0.04 | 0.19 |
Mutagenic properties of γ-HOPdG-mediated DNA–peptide cross-links in E. coli
Earlier reports (17, 56, 57) indicated that translesion synthesis past the γ-HOPdG adduct was essentially error-free in E. coli. Although these studies assayed bacterial strains with various genetic backgrounds and utilized different vector systems, mutations (G to T transversions) were consistently observed at less than 1 % frequencies or not detected at all. In this report, we examined the potential the γ-HOPdG-mediated DNA–peptide cross-links to cause mutations during extrachromosomal replication in E. coli. As described above, following intracellular replication of site-specifically modified pMS2 vectors, individual clones were tested by differential hybridization using a probe that hybridized only when nonmutagenic replication bypass had occurred. Using this method, the progeny DNAs containing no mutations were readily identified. In order to verify the accuracy of hybridization procedure, approximately 10 % of such clones were analyzed by DNA sequencing using oligodeoxynucleotide R as a primer. Among these DNAs, no clone had any sequence alteration. However, a few clones were identified that did not hybridize with probe G, but according to the ScaI cleavage analyses, they contained inserts. Targeted mutations were detected in these clones by DNA sequencing indicating that in both the wild type and pol IV-deficient cells the γ-HOPdG-mediated DNA–peptide cross-links were only marginally miscoding. Specifically, two mutations were observed in the wild type cells: one G to A transition (caused by the tetrapeptide cross-link) and one G to T transversion (caused by the dodecylpeptide cross-link). In pol IV-deficient mutant, the only mutation found was G to C transversion (caused by the tetrapeptide cross-link).
Discussion
Previous investigations have provided insights into the mechanisms by which DNA–protein cross-links are processed and removed (1). Although there have been inconsistent reports in the literature on the role of nucleotide excision repair in the removal of these adducts, these apparent contradictions may potentially be explained by differences in both the damage-inducing agents and the assays used to assess repair capacity. With the advent of technologies to create site-specific DNA–protein and DNA–peptide adducts, a much clearer picture has emerged for the role of NER in processing of these lesions. Specifically, data in both bacterial and mammalian systems indicate that relative to DNA–protein cross-links, DNA–peptide cross-links are preferable substrates for the initiation of repair by NER proteins (25–28, 30). These data are highly consistent with a role for proteolytic degradation of cross-linked proteins occurring prior to repair, since proteasomal inhibitors significantly reduce repair (29).
Since proteolytic processing of DNA–protein cross-links appears to be integral to the ultimate repair, these data raise the possibility that replication forks could encounter these intermediates prior to nucleotide excision repair. Therefore, it is likely that replicative polymerases, such as pol δ and pol ε in mammalian cells and pol III in bacteria, would be blocked. In such a scenario, specialized translesion polymerases could be recruited to these sites. Germane to this hypothesis, the study presented herein shows that pol κ was able to catalyze high fidelity efficient bypass of DNA–peptide cross-links, where the linkage of the peptide to DNA was via an acrolein-mediated bridge to N2-dG. These biochemical data suggest that pol κ contributes to the non-mutagenic bypass events during intracellular replication past N2-dG DNA–peptide cross-links. Thus, the relatively accurate replication past the γ-HOPdG-mediated tetrapeptide cross-link that previously was observed in mammalian cells (31) maybe be provided, at least partially, by pol κ. Pol κ has been shown to catalyze high fidelity bypass of benzo[a]pyrene and other bulky N2-dG adducts (32–38). Recently, we have extended the substrate repertoire of pol κ to include replication bypass of DNA interstrand cross-links (60). In this study, DNAs were utilized containing site-specifically engineered N2-N2-dG interstrand cross-links that model DNA repair intermediates, hypothesized to be in a repair pathway not requiring homologous recombination. We demonstrated that pol could catalyze bypass of cross-linked substrates in which nucleotides 3' to the cross-link had been removed. Data were also obtained which demonstrated that loss or even reduction in cellular levels of pol κ, resulted in increased cytotoxicity and chromosomal damage when cells were exposed to mitomycin C, an N2-N2-dG cross-link-inducing agent.
Collectively, these data reveal a consistent pattern of pol κ being an efficient and high fidelity enzyme in the bypass of extremely bulky N2-dG adducts. Relative to these and other catalytic bypass substrates, it is of interest that the co-crystal structure has been solved for the catalytic core of pol κ with a primer-template complex and an incoming nucleotide (61). This structure clearly shows pol κ completely encircling the duplex DNA, using an N-terminal clasp. It was hypothesized that this structure would enhance the thermodynamic stability of the core complex, thus facilitating replication bypass. Given this tight-fitting encircling of the DNA, we hypothesized that there must be considerable flexibility in the hinge domains to allow such bulky lesions to pass through the core of the complex. Thus, it is of considerable interest to determine the structures of additional complexes to reveal the conformational changes that must take place to permit these error-free reactions.
Our data also demonstrated that the E. coli pol III, pol II, and pol V were all incapable of replicating past DNA–peptide cross-links as assayed herein, but that pol IV was modestly efficient at the incorporation and extension steps and that incorporation of the correct nucleotide opposite the lesions was highly preferred. However, unexpectedly, processive synthesis of pol IV was strongly inhibited beyond the site of the lesions. Such blocks were not observed with the γ-HOPdG, reduced γ-HOPdG (this study) or furfuryl N2-dG adducts (34). These data suggest that the ability of pol IV to continue processive synthesis is blocked via a physical obstruction of the peptide cross-link with a portion of pol IV in a location where the newly synthesized DNA exits the polymerase. Such downstream blockages have been previously observed with the HIV reverse transcriptase replicating N2-dG styrene oxide-modified DNAs (62, 63). This blockage could be attributed directly to the interaction of α-helix H and I, tracking in the minor groove of the newly synthesized duplex DNA, such that site-directed mutagenesis of selected amino acid residues in the tracking face of α-helix H modulated the downstream pause sites (62, 64). We hypothesize that similar “reading heads” of pol IV may monitor the newly synthesized duplex DNA. Insights into this mechanism await crystal and co-crystal structures of pol IV.
Although DNA synthesis past the γ-HOPdG-mediated DNA–peptide cross-links by pol IV in vitro was partially inhibited relative to the non-damaged DNA, our cellular studies clearly implicate pol IV as a major contributor to the replication bypass of these lesions in vivo. In its absence, we observed ~ 5- and 20-fold reduction in recovery of plasmids when the dodecylpeptide or tetrapeptide, respectively, was site-specifically linked to the single-stranded vector DNAs, and these DNAs replicated in E. coli cells. Surprisingly, such cross-links appeared to be only marginally miscoding in both the wild-type and pol IV-deficient strains. In wild type E. coli, mutations were observed at overall frequencies of ~ 1 %, which is significantly less than the frequency of mutations caused by the identical tetrapeptide cross-link in mammalian cells, in which ~ 8 % of bypass events resulted in targeted mutations (31). Interestingly, the γ-HOPdG adduct is also less miscoding in E. coli relative to mammalian system (17, 56, 57). Overall, these data give strong support for the role of DinB family polymerases in the bypass of very bulky N2-dG adducts.
Acknowledgments
This work was supported by NIH grants CA 106858 (R.S.L.), ES 05355 (R.S.L and C.J.R.), GM38839 (M.O.D.), R37 GM21244 (M.F.G), ES012259 (M.F.G.) and center grant P30 ES000267 (C.J.R). We are grateful to Dr. M. Moriya for a generous gift of the pMS2 vector and Dr. S. Finkel for providing the E. coli strains. We thank Ms. L. Earley for purification of single-stranded pMS2 vector.
Footnotes
Abbreviations: dG, deoxyguanosine; γ-HOPdG, γ-hydroxypropano-deoxyguanosine; NER, nucleotide excision repair; pol, polymerase; E. coli, Escherichia coli.
References
- 1.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Grafstrom RC, Dypbukt JM, Willey JC, Sundqvist K, Edman C, Atzori L, Harris CC. Pathobiological effects of acrolein in cultured human bronchial epithelial cells. Cancer Res. 1988;48:1717–1721. [PubMed] [Google Scholar]
- 3.Costa M, Zhitkovich A, Harris M, Paustenbach D, Gargas M. DNA-protein cross-links produced by various chemicals in cultured human lymphoma cells. J. Toxicol. Environ. Health. 1997;50:433–449. doi: 10.1080/00984109708984000. [DOI] [PubMed] [Google Scholar]
- 4.Kuykendall JR, Bogdanffy MS. Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat. Res. 1992;283:131–136. doi: 10.1016/0165-7992(92)90145-8. [DOI] [PubMed] [Google Scholar]
- 5.Curren RD, Yang LL, Conklin PM, Grafstrom RC, Harris CC. Mutagenesis of xeroderma pigmentosum fibroblasts by acrolein. Mutat. Res. 1988;209:17–22. doi: 10.1016/0165-7992(88)90104-2. [DOI] [PubMed] [Google Scholar]
- 6.Marnett LJ, Hurd HK, Hollstein MC, Levin DE, Esterbauer H, Ames BN. Naturally occurring carbonyl compounds are mutagens in Salmonella tester strain TA104. Mutat. Res. 1985;148:25–34. doi: 10.1016/0027-5107(85)90204-0. [DOI] [PubMed] [Google Scholar]
- 7.Smith RA, Cohen SM, Lawson TA. Acrolein mutagenicity in the V79 assay. Carcinogenesis. 1990;11:497–498. doi: 10.1093/carcin/11.3.497. [DOI] [PubMed] [Google Scholar]
- 8.Cohen SM, Garland EM, St John M, Okamura T, Smith RA. Acrolein initiates rat urinary bladder carcinogenesis. Cancer Res. 1992;52:3577–3581. [PubMed] [Google Scholar]
- 9.Uchida K, Kanematsu M, Morimitsu Y, Osawa T, Noguchi N, Niki E. Acrolein is a product of lipid peroxidation reaction. Formation of free acrolein and its conjugate with lysine residues in oxidized low density lipoproteins. J. Biol. Chem. 1998;273:16058–16066. doi: 10.1074/jbc.273.26.16058. [DOI] [PubMed] [Google Scholar]
- 10.Chung FL, Nath RG, Nagao M, Nishikawa A, Zhou GD, Randerath K. Endogenous formation and significance of 1, N2-propanodeoxyguanosine adducts. Mutat. Res. 1999;424:71–81. doi: 10.1016/s0027-5107(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 11.Chung FL, Pan J, Choudhury S, Roy R, Hu W, Tang MS. Formation of trans-4-hydroxy-2-nonenal- and other enal-derived cyclic DNA adducts from ω-3 and ω-6 polyunsaturated fatty acids and their roles in DNA repair and human p53 gene mutation. Mutat. Res. 2003;531:25–36. doi: 10.1016/j.mrfmmm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Chung FL, Young R, Hecht SS. Formation of cyclic 1, N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 13.Pawlowicz AJ, Munter T, Zhao Y, Kronberg L. Formation of acrolein adducts with 2'-deoxyadenosine in calf thymus DNA. Chem. Res. Toxicol. 2006;19:571–576. doi: 10.1021/tx0503496. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro R, Sodum RS, Everett DW, Kundu SK. Reactions of nucleosides with glyoxal and acrolein. IARC Sci. Publ. 1986. pp. 165–173. [PubMed]
- 15.Smith RA, Williamson DS, Cohen SM. Identification of 3, N4-propanodeoxycytidine 5'-monophosphate formed by the reaction of acrolein with deoxycytidine 5'-monophosphate. Chem. Res. Toxicol. 1989;2:267–271. doi: 10.1021/tx00010a009. [DOI] [PubMed] [Google Scholar]
- 16.Smith RA, Williamson DS, Cerny RL, Cohen SM. Detection of 1, N6-propanodeoxyadenosine in acrolein-modified polydeoxyadenylic acid and DNA by 32P postlabeling. Cancer Res. 1990;50:3005–3012. [PubMed] [Google Scholar]
- 17.Kanuri M, Minko IG, Nechev LV, Harris TM, Harris CM, Lloyd RS. Error prone translesion synthesis past γ-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J. Biol. Chem. 2002;277:18257–18265. doi: 10.1074/jbc.M112419200. [DOI] [PubMed] [Google Scholar]
- 18.Nath RG, Chung FL. Detection of exocyclic 1, N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7491–7495. doi: 10.1073/pnas.91.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nath RG, Ocando JE, Chung FL. Detection of 1, N2-propanodeoxyguanosine adducts as potential endogenous DNA lesions in rodent and human tissues. Cancer Res. 1996;56:452–456. [PubMed] [Google Scholar]
- 20.Penn A, Nath R, Pan J, Chen L, Widmer K, Henk W, Chung FL. 1, N2-propanodeoxyguanosine adduct formation in aortic DNA following inhalation of acrolein. Environ. Health Perspect. 2001;109:219–224. doi: 10.1289/ehp.01109219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de los Santos C, Zaliznyak T, Johnson F. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct γ-OH-1,-N2-propano-2'-deoxyguanosine. J. Biol. Chem. 2001;276:9077–9082. doi: 10.1074/jbc.M009028200. [DOI] [PubMed] [Google Scholar]
- 22.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Interstrand DNA cross-links induced by, αβ-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc. Chem. Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurtz AJ, Lloyd RS. 1, N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J. Biol. Chem. 2003;278:5970–5976. doi: 10.1074/jbc.M212012200. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived α-HOPdG and γ-HOPdG regioisomeric deoxyguanosine adducts. Chem. Res. Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 25.Minko IG, Zou Y, Lloyd RS. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4056–4061. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reardon JT, Cheng Y, Sancar A. Repair of DNA-protein cross-links in mammalian cells. Cell Cycle. 2006;5:1366–1370. doi: 10.4161/cc.5.13.2892. [DOI] [PubMed] [Google Scholar]
- 28.Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, O'Connor TR. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 29.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–1580. [PubMed] [Google Scholar]
- 30.Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- 31.Minko IG, Kozekov ID, Kozekova A, Harris TM, Rizzo CJ, Lloyd RS. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat. Res. 2008;637:161–172. doi: 10.1016/j.mrfmmm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase κ. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 33.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 34.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 35.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Pol κ protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z. Activities of human DNA polymerase κ in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair (Amst.) 2002;1:559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki N, Ohashi E, Kolbanovskiy A, Geacintov NE, Grollman AP, Ohmori H, Shibutani S. Translesion synthesis by human DNA polymerase κ on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-N2-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- 38.Huang X, Kolbanovskiy A, Wu X, Zhang Y, Wang Z, Zhuang P, Amin S, Geacintov NE. Effects of base sequence context on translesion synthesis past a bulky (+)-trans-anti-B[a]P-N2-dG lesion catalyzed by the Y-family polymerase pol κ. Biochemistry. 2003;42:2456–2466. doi: 10.1021/bi026912q. [DOI] [PubMed] [Google Scholar]
- 39.Yuan B, Cao H, Jiang Y, Hong H, Wang Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2'-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8679–8684. doi: 10.1073/pnas.0711546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C--> T.A transversions in simian kidney cells. Proc. Natl. Acad. Sci. U. S. A. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 42.Onrust R, Finkelstein J, Naktinis V, Turner J, Fang L, O'Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J. Biol. Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 43.Stukenberg PT, Studwell-Vaughan PS, O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 44.Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of deltadelta' with DnaX(4) forms DnaX(3)deltadelta'. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai H, Yu H, McEntee K, Goodman MF. Purification and properties of DNA polymerase II from Escherichia coli. Methods Enzymol. 1995;262:13–21. doi: 10.1016/0076-6879(95)62004-4. [DOI] [PubMed] [Google Scholar]
- 46.Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi S, Valentine MR, Pham P, O'Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 2002;277:34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- 48.Bruck I, Woodgate R, McEntee K, Goodman MF. Purification of a soluble UmuD'C complex from Escherichia coli. Cooperative binding of UmuD'C to single-stranded DNA. J. Biol. Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 49.Cox MM, McEntee K, Lehman IR. A simple and rapid procedure for the large scale purification of the recA protein of Escherichia coli. J. Biol. Chem. 1981;256:4676–4678. [PubMed] [Google Scholar]
- 50.Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nechev LV, Harris CM, Harris TM. Synthesis of nucleosides and oligonucleotides containing adducts of acrolein and vinyl chloride. Chem. Res. Toxicol. 2000;13:421–429. doi: 10.1021/tx990167+. [DOI] [PubMed] [Google Scholar]
- 52.Minko IG, Washington MT, Kanuri M, Prakash L, Prakash S, Lloyd RS. Translesion synthesis past acrolein-derived DNA adduct, γ-hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase η. J. Biol. Chem. 2003;278:784–790. doi: 10.1074/jbc.M207774200. [DOI] [PubMed] [Google Scholar]
- 53.Dodson ML, Lloyd RS. Mechanistic comparisons among base excision repair glycosylases. Free Radic. Biol. Med. 2002;32:678–682. doi: 10.1016/s0891-5849(02)00767-0. [DOI] [PubMed] [Google Scholar]
- 54.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Human DNA polymerase ι promotes replication through a ring-closed minor-groove adduct that adopts a syn conformation in DNA. Mol. Cell. Biol. 2005;25:8748–8754. doi: 10.1128/MCB.25.19.8748-8754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell. Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VanderVeen LA, Hashim MF, Nechev LV, Harris TM, Harris CM, Marnett LJ. Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J. Biol. Chem. 2001;276:9066–9070. doi: 10.1074/jbc.M008900200. [DOI] [PubMed] [Google Scholar]
- 57.Yang IY, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J. Biol. Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- 58.Gerlach VL, Feaver WJ, Fischhaber PL, Friedberg EC. Purification and characterization of pol κ, a DNA polymerase encoded by the human DINB 1 gene. J. Biol. Chem. 2001;276:92–98. doi: 10.1074/jbc.M004413200. [DOI] [PubMed] [Google Scholar]
- 59.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 60.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase κ in the processing of N2-N2-guanine interstrand crosslinks. J. Biol. Chem. 2008;283:17075–17081. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 62.Forgacs E, Latham G, Beard WA, Prasad R, Bebenek K, Kunkel TA, Wilson SH, Lloyd RS. Probing structure/function relationships of HIV-1 reverse transcriptase with styrene oxide N2-guanine adducts. J. Biol. Chem. 1997;272:8525–8530. doi: 10.1074/jbc.272.13.8525. [DOI] [PubMed] [Google Scholar]
- 63.Latham GJ, Lloyd RS. Deoxynucleotide polymerization by HIV-1 reverse transcriptase is terminated by site-specific styrene oxide adducts after translesion synthesis. J. Biol. Chem. 1994;269:28527–28530. [PubMed] [Google Scholar]
- 64.Latham GJ, Forgacs E, Beard WA, Prasad R, Bebenek K, Kunkel TA, Wilson SH, Lloyd RS. Vertical-scanning mutagenesis of a critical tryptophan in the "minor groove binding track" of HIV-1 reverse transcriptase. Major groove DNA adducts identify specific protein interactions in the minor groove. J. Biol. Chem. 2000;275:15025–15033. doi: 10.1074/jbc.M000279200. [DOI] [PubMed] [Google Scholar]