Abstract
The Brn-3 family of transcription factors play a critical role in regulating expression of genes that control cell fate, including the small heat shock protein Hsp27. The aim of this study was to investigate the relationship between Brn-3a and Brn-3b and Hsp27 expression in the developing rodent heart. Brn-3a and Brn-3b were detected from embryonic days 9.5–10.5 (E9.5–E10.5) in the mouse heart, with significant increases seen later during development. Two isoforms (long and short) of each protein were detected during embryogenesis and postnatally. Brn-3a messenger RNA (mRNA) and protein were localized by E13.0 to the atrio-ventricular (AV) valve cushions and leaflets, outflow tract (OFT), epicardium and cardiac ganglia. By E14.5, Brn-3a was also localised to the septa and compact ventricular myocardium. An increase in expression of the long Brn-3a(l) isoform between E17 and adult coincided with a decrease in expression of Brn-3b(l) and a marked increase in expression of Hsp27. Hearts from Brn-3a−/− mice displayed a partially penetrant phenotype marked by thickening of the endocardial cushions and AV valve leaflets and hypoplastic ventricular myocardium. Loss of Brn-3a was correlated with a compensatory increase in Brn-3b and GATA3 mRNA but no change in Hsp27 mRNA. Reporter assays in isolated cardiomyocytes demonstrated that both Brn-3a and Brn-3b activate the hsp27 promoter via a consensus Brn-3-binding site. Therefore, Brn-3 POU factors may play an important role in the development and maintenance of critical cell types and structures within the heart, in part via developmental regulation of myocardial Hsp27 expression. Furthermore, Brn-3a may be necessary for correct valve and myocardial remodelling and maturation.
Keywords: Brn-3a, Brn-3b, POU domain, Transcription factor, Heart, Development, Hsp27
Introduction
A hierarchy of transcription factors controls the temporo-spatial regulation of gene expression during cardiac development. Characterisation of their role during cardiogenesis is important to understand changes that may contribute to abnormal development or disease. The Pit-Oct-Unc (POU) transcription factors, Brn-3a and Brn-3b, are essential for determining the fate of cells that express them (Fedtsova and Turner 2001; Gan et al. 1999). Brn-3a was originally identified in brain complementary DNA (cDNA) whereas Brn-3b was isolated from neuroblastoma cDNA (Lillycrop et al. 1992; Turner et al. 1994). The expression and effects of these proteins have been widely studied in neurons of the central and peripheral nervous systems, for instance, in retinal, dorsal root, trigeminal and cranial ganglia. More recently, these factors have been detected in epithelial cells of non-neuronal tissues such as the reproductive tract (cervix, ovary, testes and breast epithelium) and the lung (Latchman 1999; Budhram-Mahadeo et al. 1999a, 2001, 2006; Ndisang et al. 2001). Brn-3a and Brn-3b are encoded by distinct genes, and each exists as two isoforms resulting from alternate promoter usage (Theil et al. 1995). The longer 46-kDa isoform of Brn-3a [Brn-3a(l)] and 43 kDa Brn-3b [Brn-3b(l)] have N-terminal domains encoded by exon 1 not found in the shorter 32- to 35-kDa Brn-3a(s) and Brn-3b(s) isoforms. However, exon 2, which encodes the DNA-binding POU domain, is common to both the long and short isoforms.
The gene regulatory effects of Brn-3a and Brn-3b are complex and largely depend on the cell types in which they are expressed. For instance, in neuroblastoma cells, Brn-3a and Brn-3b exhibit opposite expression patterns and antagonistic functions on the regulation of target genes. Thus, high levels of Brn-3b are associated with proliferation in these cells, whereas increasing Brn-3a enhances survival and differentiation. In contrast to the opposite expression patterns and antagonistic effects of Brn-3a and Brn-3b seen in neuronal cells, both Brn-3a and Brn-3b can transactivate the promoter of the small heat shock protein Hsp27 and increase protein expression (Farooqui-Kabir et al. 2004), suggesting that the effects of Brn-3a and Brn-3b are highly dependent on the specific target gene as well as cell type.
In this study, we report the novel observation that Brn-3a and Brn-3b are expressed in the developing and adult heart. Brn-3a is strongly expressed in the cardiac ganglia, AV cushions, valves, outflow tract (OFT), septa and the ventricular compact myocardium. The loss of Brn-3a resulted in a partially penetrant phenotype of endocardial cushion hyperplasia and failure of correct atrioventricular valve remodelling and maturation and hypoplastic/non-compacted ventricular myocardium in a few hearts. An isotype switch occurred at birth, with upregulation of Brn-3a and downregulation of Brn-3b, which was reversed in Brn-3a−/− hearts that may explain the variation in morphological phenotype. Furthermore, we have identified the hsp27 gene promoter as a target for positive regulation by both Brn-3a and Brn-3b in cardiomyocytes and suggesting non-redundant roles for these two proteins in the context of Hsp27 gene regulation and a mechanism for compensation by Brn-3b upon loss of Brn-3a.
Materials and methods
Cardiomyocyte culture
Primary cultures of cardiomyocytes were prepared from 2-day-old neonatal rats as previously described (Punn et al. 2000) and cultured on six-well plates (Greiner) at an initial density of 1 × 106 cells per well. Cardiomyocytes were >95% pure at time of plating as determined by immunofluorescence detection of the sarcomeric protein α-actinin.
Brn-3a-gene-targeted mice
Brn-3a knockout mice have been described previously (Gan et al. 1996). Heterozygous male/female breeding pairs were set up, which generated wild-type (+/+), knockout (−/−) and heterozygous (+/−) progeny. Genotypes were determined by polymerase chain reaction (PCR). Only male +/+ and −/− mice were used for the experiments. Animals were kept in accordance with UK Home Office guidelines and the study conformed to the NIH guide for the care and use of laboratory animals.
Plasmids, transfections and lucifersae assays
Transfections were performed using a non-viral integrin-targeting peptide as previously described (Hart et al. 1998; Obasanjo-Blackshire et al. 2006). Cells were transfected with pLTR (null vector), pLTR-Brn-3a(l), pLTR-Brn-3a(s), pLTR-Brn-3b(l), pGL2-hHsp27-luc (hHsp27 promoter) and pGL2-basic (null vector) as appropriate. To control for differences in transfection efficiency, 0.2 μg Renilla luciferase plasmid was also included in each transfection. A dual luciferase assay was performed (Promega) using a luminometer (TD 20/20, Turner Designs), and activity was normalised to Renilla activity. Brn-3a and Brn-3b expression vectors in pLTR have been described previously (Budhram-Mahadeo et al. 1995a). The construction of pGL2-hHsp27-luc has been described previously (Farooqui-Kabir et al. 2004).
Protein isolation and immunoblotting
Hearts from embryos from E9.5 to E18.5 and neonates from P1 to P10 were isolated by dissection. Hearts were rinsed in ice-cold phosphate-buffered saline (PBS), snap frozen in N2(l) and stored at −80°C until use. Fifty to 100 mg of frozen tissue were powdered in N2(l) then homogenised in 250 μl of sample loading buffer and heated to 100°C for 5 min. Electrophoresis and Western blotting was performed essentially as described previously (Farooqui-Kabir et al. 2004). Sample loadings were equalised after Coomassie Blue staining of test gels and densitometry. Proteins were transferred to nitrocellulose membranes (Hybond-C, Amersham). After blocking in non-fat milk (Marvel), membranes were probed with primary antibodies at 1:1,000 dilution. These were monoclonal anti-Brn-3a (Chemicon International), polyclonal anti-Brn-3b (Santa Cruz), polyclonal goat anti-mouse Hsp27 C-terminal (Santa Cruz), polyclonal anti-pan-myosin light chain (Sigma), polyclonal anti-troponin-T (Sigma) and polyclonal anti-actin (Santa Cruz). Secondary antibodies were horseradish peroxidase-conjugated pig-anti-rabbit, rabbit-anti-mouse or rabbit-anti-goat (DAKO) used at 1:2,500 dilution. Antibody–antigen complexes were visualised by enhanced chemiluminescence (ECL, Amersham).
Dissection and fixation of embryos for in situ hybridisation
Hearts were dissected from embryos at specific time points for RNA and protein isolation. Isolated hearts were rinsed in diethyl pyrocarbonate (DEPC)-treated PBS, snap frozen in liquid nitrogen and stored at −80°C until use. The neonatal and adult hearts were isolated in a similar manner to that of the older embryos; however, care was taken to squeeze the hearts gently with forceps in the DEPC-treated PBS to ensure that blood contamination was kept to a minimum. For immunostaining and in situ hybridization, hearts were carefully dissected from embryos between E12.5–E16.5, washed and either frozen in optimal cutting temperature (OCT) compound or fixed in 4% paraformaldehyde for mounting in paraffin. After fixation, the embryos were washed twice for 5 min with DEPC-treated PBS on ice and serially dehydrated through 30%, 50% and 70% ethanol in DEPC-treated MilliRO water. The length of time used for each wash depended upon the size of the embryo. E12.5 and E13.5 embryos were washed at each stage for 60 min; however, embryos of E14.5 and older were washed for 90 min. Embryos were then embedded in paraffin wax for sectioning.
Immunostaining and microscopy
For Brn-3a and Brn-3b localisation in adult rat myocardium, adult rat hearts were washed free of blood using ice-cold PBS, frozen in OCT compound and sectioned using a crystat. Frozen sections were fixed, permeabilised and incubated with monoclonal anti-Brn-3a (Chemicon) or polyclonal anti-Brn-3b (BAbCo) overnight at 4°C, followed by horseradish peroxidase-conjugated rabbit-anti-mouse or pig-anti-rabbit secondary antibodies (DAKO) as appropriate, for 1 h at room temperature. Signals were developed using diaminobenzidine and photographed at ×40 using a 35-mm camera mounted on a Nikon Diaphot microscope. For immunohistochemical analysis of protein expression during embryonal development, embryos were dissected and fixed as described above. Slides were incubated with primary antibody (i.e., monoclonal anti-Brn3a, Chemicon) at 1:50 dilution in 2% goat serum in Tris-buffered saline (TBS) at room temperature for 1 h. After washing five times for 5 min each with TBS, 2% goat serum, 0.1% Triton X-100 and 0.15M NaCl, the slides were incubated with biotinylated secondary antibody (goat anti-mouse Ig, DAKO) at 1:1,000 dilution for 30 min at room temperature. Following this, the slides were washed five times for 5 min each then overlaid with ABComplex/horseradish peroxidase (DAKO) prepared according to the manufacturers instructions (50 μl of biotinylated horseradish peroxidase to 50 μl of streptavidin in 5 ml of TBS) and left to incubate for 30 min. After this, the washing steps were then repeated as above, and the antibody signal was visualised by addition of metal enhanced-3,3-diaminobenzidine tetrahydrochloride prepared according to the manufacturer’s instructions. Two percent goat serum was used instead of the primary antibody to provide negative controls. Images were captured using a digital camera attached to a Zeiss Axiophot II microscope and processed using Adobe Photoshop (v 7.0) and Microsoft PowerPoint.
Slide in situ hybridisation
Slide in situ hydridisation was based on (Breitschopf et al. 1992). Parafin wax-embedded embryo sections were mounted on Tespa-coated slides, de-waxed, rehydrated through a graded ethanol series, washed, treated with proteinase-K followed by triethanolamine/acetic anhydride treatment to reduced background and hybridised with the appropriate digoxygenin (DIG)-labelled ribonucleotide probes overnight at 55°C. After high-stringency washing, the DIG label was detected using alkaline phosphatase-conjugated anti-DIG Fab fragments (Boehringer Mannheim). Antibody visualisation was achieved using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate solution (Roche). Riboprobes were specific for Brn-3a or Brn-3b but recognised both long and short isoforms of each mRNA.
RNA isolation and PCR
Total RNA was isolated from tissues using the TRIzol RNA isolator kit (Gibco-BRL). To remove any contaminating genomic DNA, the isolated RNA was treated with RQ1 RNase-free DNase I (Promega) for 1 h, incubated at 37°C, followed by phenol/chloroform extraction and ethanol precipitation. One to 2 μg of total RNA was used for cDNA synthesis using AMV reverse transctriptase (Promega) in 50µl volume. The resulting cDNA was aliquoted and stored at −20°C until ready for use. Quantitative real-time PCR (qPCR) was carried out to measure changes in the Brn-3b mRNA expressed in Brn-3a null hearts compared with wild-type control littermates using RNA isolated from hearts of mouse embryos at E14.5 and E15.5, and also for Brn-3a(l) mRNA from E13.5 to P19.5 hearts. For Brn-3a(l) mRNA, specific primers were designed that spanned exon-1 and the exon-1/exon-2 boundary, exon-1 being specific for the long form of Brn-3a (Brn-3a(l). After cDNA synthesis (see above), q-RT-PCR was carried out as described (Hudson et al. 2004) using Brn-3b-specific primers for amplification using QuantiTect SYBR I Green (Qiagen). Housekeeping gene, GAPDH was used as an internal control. All qPCR reactions were carried out using the DNA Engine Opticon system (MJ Research). To quantify the mRNA for each amplified target gene, the standard curve method was used as this allowed PCR products to be expressed as femtograms (fg) of RNA. Standards were constructed using serial dilutions of purified product. Once tested, sets of standards (of different concentrations, fg) were stored as aliquots that could be used in all subsequent studies and thus allowed for comparisons between different experiments.
Mouse genotyping
A standard PCR protocol was used to amplify Brn-3a mRNA and the neomycin gene (Xiang et al. 1996) from littermates of Brn-3a−/− crosses. Briefly, genomic DNA was isolated from mouse tails or other embryonic tissue using the DNeasy tissue kit (Qiagen). The isolated DNA was used for standard PCR.
Statistical analysis
For experiments on isolated cardiomyocytes, at least three independent cell preparations were used (unless otherwise stated) and treatments performed in triplicate wells for each preparation. Data was pooled and expressed as mean ± SD (n = 3). Groups were compared by one-way analysis of variance (ANOVA) with Tukey’s post hoc test using Graphpad Prism software.
Results
Expression of Brn-3a and Brn-3b in cardiomyocytes
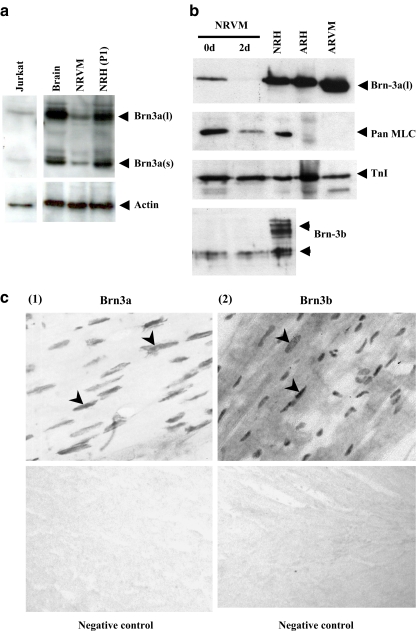
The expression of Brn-3a was analysed in neonatal hearts isolated from 1-day-old rats (P1) and in isolated primary neonatal rat ventricular cardiomyocytes (NRVM). As shown in Fig. 1a, strong expression of both Brn-3a(l) and Brn-3a(s) isoforms in heart or NRVM corresponded to the pattern observed in both brain and Jurkat cells. The antibody was specific for Brn-3a, as determined using a positive control from cells expressing the long form of Brn-3a from a transfected cDNA. Nevertheless, two minor bands were detected running slower than both the main Brn-3a(l) and Brn-3a(s) bands. These are believed to be due to post-translational phosphorylation of Brn-3a, as ongoing work suggests that several sites within both the N-terminal and POU domains are phosphorylated (see for instance Calissano et al. 2005). Brn-3a(l) was also expressed in adult rat heart or freshly isolated adult cardiomyocytes (Fig. 1b). After 2 days in culture, Brn-3a(l) was downregulated in cultured NRVM (Fig. 1b). Analysis of cardiomyocyte-specific contractile protein markers showed that the expression of myosin light chain also decreased after 2 days, whereas troponin I levels remained unchanged. This may be indicative of sarcomere remodelling at 2 days in culture, although gross sarcomere structure and spontaneous contractile activity were maintained. Brn-3b(l) and Brn-3b(s) isoforms were also expressed in the intact heart (Fig. 1b), although only the short form of Brn-3b appeared to be expressed in isolated NRVM. Immunohistochemical staining of sections of adult rat heart using Brn-3a and Brn-3b-specific antibodies shows that both Brn-3a and Brn-3b expression were localised to the nucleus of cardiomyocytes [Fig. 1c(1) and (2)].
Fig. 1.
Analysis of Brn-3a protein expression in heart and isolated cardiomyocytes. a immunoblot of Brn-3a protein in cultured neonatal rat ventricular cardiomyocytes (NRVM) and neonatal rat heart (NRH). Lane 1 Purified protein from Jurkat cells (lane 2), rat brain tissue, NRVM (lane 3) and NRH (lane 4) at stage P1 were blotted and probed with a monoclonal anti-Brn-3a antibody. The samples were also probed with a polyclonal anti-actin antibody to assess variation in protein loading. b NRVM cultured for 2 days, and Brn-3a(l) and Brn-3b expression assessed by Western blotting. NRVM at day 0 (lane 1) was compared to NRVM after 2 days in culture (lane 2), NRH at P1, adult rat heart (ARH) and isolated adult rat ventricular cadiomyocytes (ARVM). Blots were also probed for pan-myosin light chain (MLC) and troponin I (TnI) for comparison. C Immunohistochemical analysis of Brn-3a (1) and Brn-3b (2) in sections of adult rat heart
Temporal expression profile of Brn-3a and Brn-3b in the developing heart
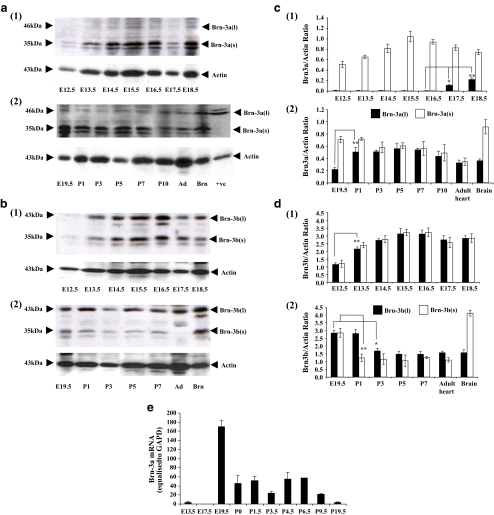
Since both Brn-3a and Brn-3b were detected in the neonatal and adult rat heart, we next analysed the temporal expression of these factors in the developing mouse heart. The levels of Brn-3a and Brn-3b protein and mRNA were analysed by Western blotting and qPCR, respectively, at different time points during embryonal and postnatal development. Analysis of Brn-3a and Brn-3b protein levels by Western blotting facilitated the detection of the different isoforms of these proteins. The shorter Brn3a(s) isoform was the major species expressed and could be detected as early as E12 and increased steadily from E12 to E15 [Fig. 2a(1) and c(1)], then decreased between P10 to adult [Fig. 2a(2) and c(2)]. Interestingly, expression of the longer Brn-3a(l) isoform was very low prior to E17 but increased immediately before birth (E18 to P1) and then remained elevated into adulthood [Fig. 2a(2) and c(2)]. However, in contrast to Brn-3a, both the long and short isoforms of Brn-3b were expressed at similar levels (Fig. 2b and d). An increase was observed between E12 and E16 and levels then remained stable until after birth, where a fall in levels of Brn-3b (particularly the short isoform) was observed at P1-P3, concomitant with the increase in levels of Brn-3a(l) [Fig. 2b(ii) and d(ii)]. The antibodies used were specific for Brn-3a and Brn-3b, as these were the major bands detected as confirmed by blotting against positive controls expressed from transfected cDNAs. However, there were other minor bands detected which may be non-specific. As mentioned above, some slower migrating phosphorylated forms of Brn-3a were detected. Furthermore, the most prominent minor band running slightly faster than the main Brn-3a(s), Brn-3b(l) and Brn-3b(s) bands are likely to be proteolytically derived, but it is not certain whether or not these are generated by post-translational processing. Both Brn-3a and Brn-3b mRNAs were detected as early as E9.5 (the earliest time point studied due to the limited amount of tissue-results not shown). Figure 2e shows that Brn-3a (l) mRNA expression was low between E13.5-E17.5 but peaked sharply at E18.5-E19.5 prior to the increase in Brn-3a(l) protein. From P0 to P6.5 Brn-3a mRNA was maintained at a lower level and then decreased from P9.5 to adult. Brn-3b mRNA increased gradually up to E13.5 and was maintained until birth, decreasing postnatally between P1–P3 in keeping with Brn-3b protein (results not shown).
Fig. 2.
Levels of total Brn-3a and Brn-3b protein and mRNA in the heart during development. Immunoblots of both the long and short isoforms of Brn-3a and Brn-3b in embryonic (1) and neonatal (2) heart tissue samples are shown in a and b, respectively. Protein levels were quantified using laser densitometry and normalised to that of actin (c and d). Positive controls were brain (Brn) and expression of the long form of Brn-3a expressed in cells from a transfected cDNA (+ve). Values were obtained from three experiments carried out using independent heart samples from separate litters, and data is presented as mean ± SD. Brn-3a(l) mRNA levels were quantified by qPCR using primers specific for exon 1 and normalised to that of GAPDH (en = 5 hearts per time point). Statistical analysis was carried out using the students t test, and significance is expressed as asterisks where **p < 0.005 and where *p < 0.05
Localisation of Brn-3a expression in the heart
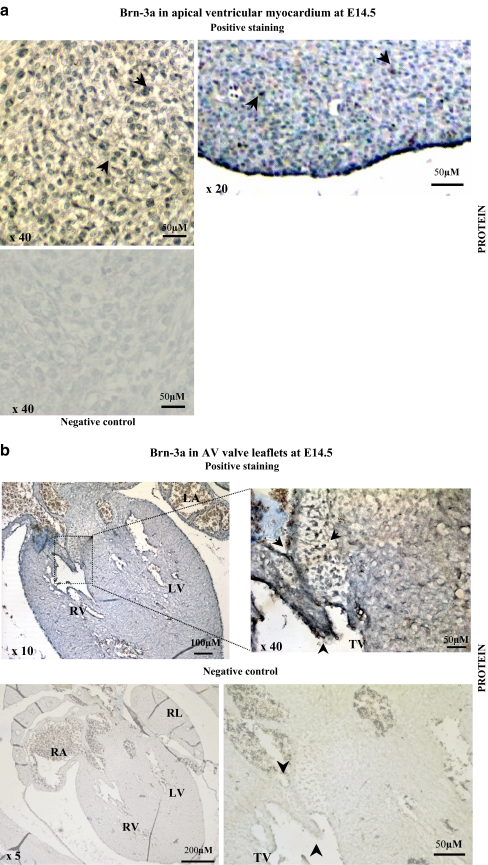
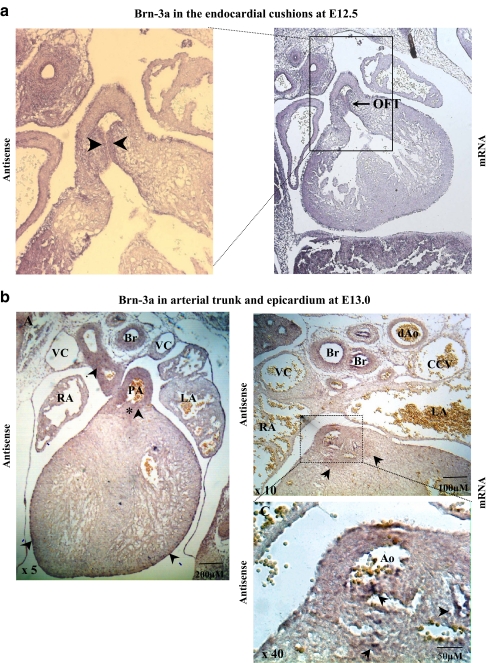
Immunohistochemical analysis and in situ hydridisation were used to determine the localisation of Brn-3a protein and mRNA expression between E11.5 to E14.5 when the major cardiac structures are formed and remodelled. Brn-3a protein (Fig. 3a and b) and mRNA (Figs. 3d and 4) were detectable from E12.5 with prominent expression observed in the forming valve structures of the outflow tract. By E13.0, Brn-3a protein and mRNA were also detected in the epicardium, aorta and semilunar valve leaflets (Figs. 3b,e and 4b). By E14.5, Brn-3a protein and mRNA expression were strong in the compact zone myocardium near the apex of the heart (Fig. 3a and c) and in the lining of the atrioventricular (AV) valve leaflets (Fig. 3b and d), where it was restricted to the endocardial cell layer and to a distinct subendocardial cell layer. Figure 4b shows that the walls of the great arteries stain very strongly for Brn-3a mRNA. Consistent with previously reported expression in the lung (Budhram-Mahadeo et al. 2001), strong staining of the epithelial cell layers of the trachea and bronchioles was observed. Figures 3f and 4d show Brn-3a mRNA expression in the cardiac pre-ganglionic nerves and ganglia that may suggest that it plays a role in the survival and differentiation of cardiac sensory neurons in a similar manner to sensory neurons of the peripheral nervous system. Neurofilament protein staining is shown in Fig. 3f and was used to confirm the sensory neuronal tissue type of these structures. Thus Brn-3a protein and mRNA have been detected in a number of structures within the developing heart, which undergo dynamic changes during embryogenesis. These include the myocardium, endocardial cushions, valve leaflets and the cardiac ganglia.
Fig. 3.
Expression of Brn-3a protein and mRNA in different cardiac structures at E14.5. Immunohistochemistry was carried out using a monoclonal Brn-3a antibody on transverse sections (8 μm) of E14.5 embryos to show sites of expression. a Nuclear Brn-3a expression is visible in isolated cells of the ventricular myocardium (black arrowheads) and epicardium. The lower panel shows negative control with secondary antibody alone. Expression of Brn-3a protein in the endocardium surrounding the mitral and tricuspid valves (arrowheads, b) and in a layer of cells beneath the endocardial layer. The lower panels show negative control. Brn-3a expression is also noted in the semilunar valve leaflets (arrowheads, e). Ao aorta. Brn-3a expression in the cardiac innervation was compared with the distribution of Neurofilament protein in the same region to confirm Brn-3a expression in the cardiac ganglia. f Prominent Brn-3a protein expression is seen in the cardiac ganglia (black arrowheads), as seen with the mRNA expression; the DRG’s exhibit strong Brn-3a protein expression. The lower panel shows negative control carried out with secondary antibody alone. Strong expression of neurofilament protein is apparent in the neuronal processes of the cardiac innervation and in the DRG (arrowheads) and neurons in the spinal cord. Br bronchus, Oe oesophagus. Slide in situ hybridisation using Brn-3a-specific DIG-labelled riboprobes on transverse sections (8 μm) of E12.5–E14.5 embryos. Expression of Brn-3a mRNA was observed in the ventricular myocardium of the apex at E14.5, the muscular interventricular septum and myocytes adjacent to the epicardium (c), the endocardium surrounding the mitral valve (left), tricuspid valve (right) and a layer of cells beneath the endocardial layer (arrowheads, d). Negative (sense) controls for the corresponding sections of the heart are shown. LA left atrium, MV mitral valve, TV tricuspid valve
Fig. 4.
Expression of Brn-3a mRNA in different cardiac structures. Slide in situ hybridisation using Brn-3a-specific DIG-labelled riboprobes on transverse sections (8 μm) of E12.5–E14.5 embryos. Expression of Brn-3a mRNA was observed in the developing endocardial cushions (arrowheads) at E12.5 (a), the epicardium and the semilunar valve leaflets (arrowheads, b) at E13.0, the aorta and pulmonary trunk and pulmonary valve leaflets (arrowheads, c) at E14.5, the individual preganglionic nerves and ganglia (arrowheads, d) at E14.5. Negative (sense) controls for the corresponding sections of the heart are shown. Ao aorta, dAo descending aorta, LA left atrium, RA right atrium, CCV common cardinal vein, PT pulmonary trunk, LL left lung, RL right lung, IVS interventricular septum, LV left ventricle, RV right ventricle, VC vena cava, Br and asterisk bronchioles, T trachea, MV mitral valve, TV tricuspid valve
Temporal expression profile of Hsp27 in the developing heart
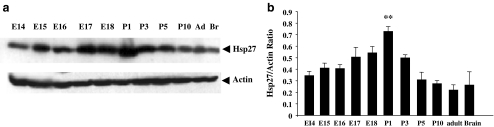
We have previously identified the small heat shock protein hsp27 gene as a target for regulation by Brn-3 in sensory neuronal derived ND7 cells (Farooqui-Kabir et al. 2004) and breast-cancer-derived MCF7 cells (Lee et al. 2005). Hsp27 is also an important factor for differentiation and survival of post-mitotic cardiomyocytes (Davidson and Morange 2000); however, its temporal expression and mechanisms of regulation in the heart have not been determined. Therefore, we investigated the possible regulation of Hsp27 expression by Brn-3 in cardiomyocytes. Thus, expression of Hsp27 (mHsp25) protein during cardiac development was determined by Western blotting. Figure 5 shows that Hsp27 was expressed from at least E14.5. Expression increased sharply around birth, between E17 and P1, concomitant with the increase in Brn-3a(l) expression. Hsp27 expression then decreased from P3 onwards, although a modest level was maintained into adulthood. This pattern of Hsp27 expression concords with previous reports that Hsp27 levels are high in the early neonatal heart and decrease in the adult heart (Martin et al. 1999).
Fig. 5.
Analysis of Hsp27 protein levels in hearts during development. a Representative mmunoblots of Hsp27 and actin protein in heart samples during development. b Band densities were obtained by laser densitometry from three independent sets of protein samples and the Hsp27 values normalized to actin. Hsp27/actin ratios are expressed as mean ± SD, and statistical analysis was conducted using the t test and is expressed as asterisks where **p < 0.005 versus E14
Reciprocal expression of Brn-3a and Brn-3b in the heart
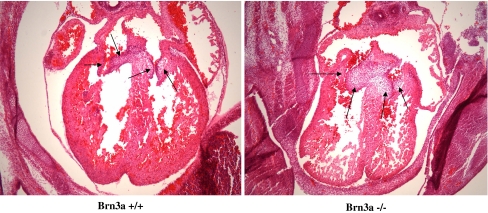
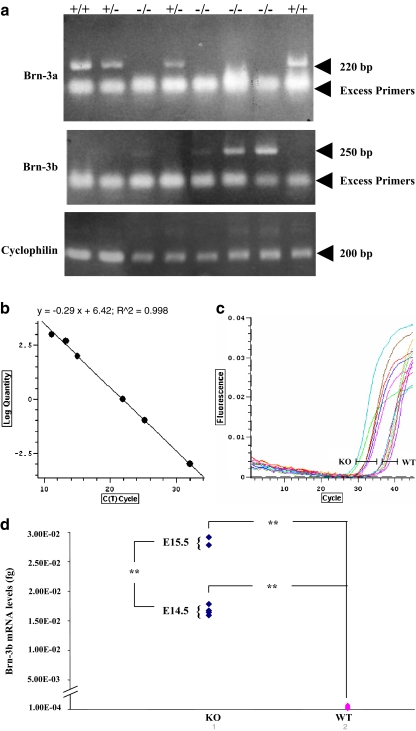
Brn-3a knockout (KO) mice die in the first day after birth. This is considered to be due to the loss of sensory neuronal cells (Xiang et al. 1996), and as such, no analysis of Brn-3a−/− hearts has been undertaken. We therefore analysed hearts taken from Brn-3a−/− embryos and compared them to wild-type littermate controls. Approximately 30% of hearts displayed evidence of thickened endocardial cushion tissue and valve leaflets (a representative example is shown in Fig. 6), which was associated with an increased cell density and thus may be due to cushion cell hyperplasia and failure to differentiate. Furthermore, these hearts also displayed hypoplastic ventricular myocardium and less compaction. As we have observed reciprocal expression patterns for Brn-3a and Brn-3b in sensory neuronal cells (Budhram-Mahadeo et al. 1995b), we analysed Brn-3a and Brn-3b mRNA expression in hearts isolated from wild-type and KO mice at 1 day after birth. As expected, Brn-3a mRNA was not expressed in hearts isolated from Brn-3a−/− mice. However, Brn-3b mRNA was upregulated to different extents in all Brn-3a−/− hearts (Fig. 7a), suggesting a compensatory regulatory mechanism whereby upregulation of Brn-3b is able to compensate for loss of Brn-3a. To confirm this result quantitatively, real-time qRT-PCR was used to quantitate Brn-3b mRNA in wild-type and Brn-3a−/− hearts at two different time points, E14.5 and E15.5. The results shown in Fig. 7b–d clearly demonstrate that Brn-3b mRNA is consistently and significantly upregulated in Brn-3a−/− hearts.
Fig. 6.
Comparison of the hearts of Brn-3a null and wild-type mice at E14.5 (abnormal phenotype). Brn-3a null and wild-type E14.5 embryos were sectioned (8 μm) transversely and subjected to H & E staining to examine the morphology of their hearts. Wild-type (+/+) mice are shown in the left panel and Brn-3a null (−/−) mice are shown in the right panel. The position of the endocardial cushions are indicated by arrows. In this example, the Brn-3a null mouse exhibits thickened endocardial cushions with increased cell density
Fig. 7.
Comparison of Brn-3b mRNA levels in Brn-3a KO and wild-type hearts. RT-PCR (a) and quantitative real-time PCR (qRT-PCR; b–d) of hearts isolated from wild-type or Brn-3a null mice at E14.5 and E15. Levels of cyclophilin (a) or GAPDH (b–d) mRNA were measured to control for variations in cDNA levels between samples. b A typical Brn-3b mRNA standard curve used to quantify the Brn-3b mRNA. c Typical cycle curves, with the Brn-3b mRNA levels being measured during the linear phase of the reaction at the point marked on the graph. d The mean values in femtograms (fg) for each sample normalized to GAPDH. Values were obtained in triplicate for each heart with an n = 4 hearts for E14.5 and n = 2 hearts for E15.5. Data is expressed as mean ± SD, and statistical analysis was conducted using the Student’s t test and is expressed as **p < 0.005
Hsp27 target gene regulation by Brn-3 in cardiomyocytes
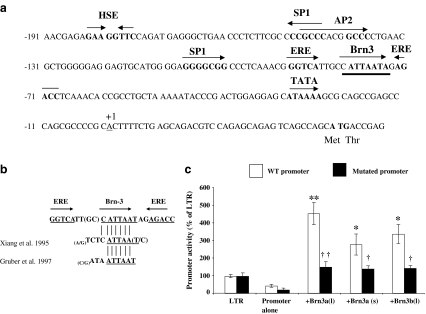
Since the temporal expression patterns of Brn-3a(l) and Hsp27/mHsp25 in the heart were coincident, we next tested whether or not Brn-3a was able to activate the hsp27 proximal promoter in isolated cardiomyocytes. The sequence of the proximal hsp27 promoter containing the putative Brn-3a-binding site is depicted in Fig. 8a. The putative Brn-3-binding site, which has high homology to previously described consensus Brn-3a-binding sequences (Xiang et al. 1995; Gruber et al. 1997) is shown in Fig. 8b. Co-transfection of Brn-3a(l), Brn-3a(s) or Brn-3b(l) expression vectors with a reporter construct comprising the proximal human hsp27 promoter driving expression of the luciferase reporter gene (Farooqui-Kabir et al. 2004; Lee et al. 2005) is shown in Fig. 7c. Brn-3a(l), Brn-3a(s) and Brn-3b(l) were all able to transactivate the hsp27 promoter, with Brn-3a(l) being slightly more effective. This demonstrates that the N-terminal domain of Brn-3a may be necessary for maximal activation. Furthermore, Brn-3a and Brn-3b may play functionally redundant roles in regulating Hsp27. Mutation of the Brn-3-binding site in the context of the otherwise intact hsp27 promoter resulted in a loss of activation by either Brn-3a or Brn-3b, confirming that this site is necessary and sufficient for activation by the Brn-3 proteins in cardiomyocytes. This is supported by previous findings that Brn-3a and Brn-3b specifically bind this site in vitro and in vivo (Farooqui-Kabir et al. 2004; Lee et al. 2005).
Fig. 8.
Mutation of the putative Brn3-binding site results in a loss of activation of the Hsp27 promoter in cardiomyocytes. a The 200-bp proximal region of the Hsp27 promoter showing the major transcriptional regulatory elements including the TATA, HSE and ERE, and the putative Brn-3-binding site. b Comparison of the Brn-3-binding site found in the Hsp27 promoter with published Brn-3 consensus binding sites. c Activation of the wild-type Hsp27-luciferase reporter by Brn-3a(l), Brn-3a(s) and Brn-3b(l) (open bars) and the loss of activation following mutation of the putative Brn-3-binding site (filled bars). The Hsp27 promoter alone and empty pLTR vectors were used as controls. Luciferase activation was expressed as percent of pLTR, which was taken as 100%. Luminescence was determined from three independent experiments carried out in triplicate, and results are expressed as mean ± SD. Statistical analysis was carried out using the one-way ANOVA and is expressed as **p < 0.005 and as *p < 0.05 versus pLTR, and as ††p < 0.005 and as †p < 0.05 versus the WT promoter
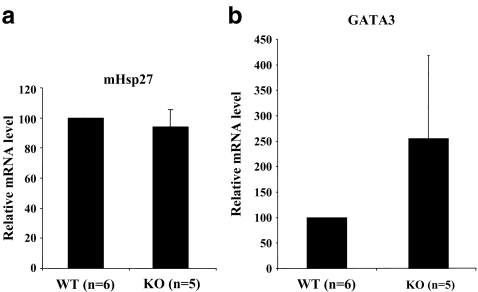
Because Brn-3b mRNA is elevated in Brn-3a−/− hearts and Brn-3b is also able to activate the Hsp27 promoter, we analysed Hsp27 mRNA levels by qPCR in wild-type and Brn-3a−/− hearts. Hsp27 mRNA was not different between genotypes (see Fig. 9a), suggesting that the compensatory increase in Brn-3b may ensure that Hsp27 levels are maintained in Brn-3a−/− hearts. As a positive control, we analysed the expression of GATA3 mRNA, as this has been shown to increase in sensory neurons in Brn-3a−/− mice (Eng et al. 2004) and is also a marker of cardiomyocyte growth and differentiation (Crawford et al. 2002). Figure 9b shows that GATA3 mRNA is also increased in hearts from Brn-3a−/− mice compared to wild type.
Fig. 9.
Levels of Hsp27 and GATA3 mRNA in wild-type and Brn-3a−/− hearts at P0. Hsp27 (a) and GATA3 (b) mRNA levels were quantified by qPCR and normalised to that of GAPDH (en = 5 hearts for each genotype)
Discussion
This study demonstrates that the POU domain transcription factors Brn-3a and Brn-3b are expressed in the heart throughout development with low levels detected as early as E9.5. Expression significantly increases at critical periods such as during septation, chamber formation and development of the valves and outflow tract, indicative of its likely role in specific processes critical for cardiac morphogenesis. For instance, Brn-3a mRNA is markedly expressed in the developing endocardial cushions of the AV valves and outflow tract (OFT) at E12.5 and, later, is highly expressed in the endocardial cell layer of the valve leaflets at E14.5. A similar pattern was observed in the vascular wall of the aorta and semi-lunar valves. Interestingly, at E14.5, there is also staining of a distinct cell layer beneath the endocardial layer and also of cells deeper within the valve leaflets. Furthermore, in the myocardium, strong staining is first seen in the epicardial cell layer and later in the compact zone myocardium (i.e., around E14.5), particularly near the apex of the heart.
The reciprocal expression of Brn-3b and Brn-3a in the heart would suggest that Brn-3b may be able to compensate for Brn-3a in certain cardiac cell types. This is particularly apparent on the hsp27 promoter, which is activated by both Brn-3a and Brn-3b in cardiac myocytes. Both Brn-3a and Brn-3b are expressed in the myocardium at a time when terminal differentiation of cardiomyocytes occurs, and expression persists into the adult heart. Interestingly, expression of the long isoform of Brn-3a [Brn-3a(l)] protein increased around birth, concomitant with a decrease in Brn-3b protein. This reciprocal expression pattern of Brn-3a and Brn-3b is reminiscent of that which occurs upon cAMP-induced differentiation of sensory neuronal-derived ND7 cells (Budhram-Mahadeo et al. 1995b). Thus, Brn-3b is highly expressed in proliferating cells and is downregulated when cells exit the cell cycle, whereas low levels of Brn-3a expressed in proliferating cells are significantly increased upon differentiation. Brn-3a(l) mRNA peaked at E19.5, and protein between P1–P7, during which time myocytes are under most stress, exit the cell cycle and undergo hypertrophy, particularly in the left ventricle. As Brn-3a is a known survival factor that regulates the expression of anti-apoptotic pro-survival genes, it is likely that Brn-3a enhances cardiomyocyte survival during this critical period.
Thus, in non-cardiac cell types, the expression of Brn-3a is increased during growth arrest and supports survival and differentiation by upregulating the expression of anti-apoptotic genes such as Bcl-2 (Budhram-Mahadeo et al. 1999b), Bcl-XL (Sugars et al. 2001) and Hsp27 (Farooqui-Kabir et al. 2004), and suppressing the expression of pro-apoptotic genes such as Bax and Noxa (Hudson et al. 2005). Interestingly, the reciprocal expression of Brn-3a and Brn-3b that we observed in mouse heart suggests a regulatory loop.
Hsp27 (and the orthologous mHsp25) are members of the small heat shock protein/molecular chaperone family that contain conserved crystalin domains, thus are related to the lens proteins αA and αB crystalin (Sun and MacRae 2005). Both Hsp27/mHsp25 and αB crystalin are highly expressed in the heart and the skeletal muscle, and have been proposed to play an important protective role in cardiac myocytes (Martin et al. 1997; Sakamoto et al. 2000). A relatively high level of endogenous Hsp27 expression has been shown in the heart throughout development in several species such as zebrafish (Mao and Shelden 2006), pig (Tallot et al. 2003), mouse, rat and human (Lutsch et al. 1997), suggesting that Hsp27 may play an important role, possibly in protecting specific cell types within this organ or supporting proliferation and/or differentiation events. In the mouse, Hsp27 expression is detected early during heart development (E10.5) and is detected in the presumptive atrial and ventricular myocardium. Subsequently, expression is limited to cells of a myogenic lineage (Davidson et al. 2002). Indeed, Davidson and Morange (2000) suggested that Hsp27 is important for terminal differentiation of cardiomyocytes (Davidson and Morange 2000). However, a recent study using anti-sense morpholinos in Xenopus embryos demonstrated a crucial role for Hsp27 in the fusion of cardiac progenitors and closure of the linear heart tube (Brown et al. 2007). Although cardiac myocytes were specified and differentiated normally, Hsp27 knockdown resulted in actin filament and myofibrillar disorganization, as well as cardiac bifida, further supporting a critical role for Hsp27 in actin assembly, cell polarisation and migration. The regulation of hsp27 gene expression by Brn-3 may be temporally and/or cell-type restricted because Brn-3a and Hsp27 are co-expressed in cardiomyocytes but not endocardial cushion or valve tissue. Furthermore, high expression of Brn-3a in AV valve endocardium occurs early (E14.5), whereas Brn-3a/Hsp27 co-expression in myocardium occurs late (around birth).
The regulation of Hsp27 expression during development has been suggested to be at least partially dependent on heat shock factor-2 (HSF-2; Loones et al. 1997). However, the possible involvement of other transcription factors, particularly in the heart, has not been addressed. Therefore, this paper represents the first to demonstrate that other factors, specifically the POU domain transcription factors Brn-3a and Brn-3b, can activate the Hsp27 gene promoter and that induction of high levels of Hsp25 in mouse hearts in vivo correlates with the elevated expression of the long isoform of Brn-3a [Brn-3a(l)] between E17 and P3. Thus, we demonstrated that the Hsp27 proximal promoter was transactivated by Brn-3 in isolated cardiomyocytes, and this promoter required a conserved Brn-3 consensus binding site. This site was both necessary and sufficient for Brn-3-dependent activation, as mutation of this site abolished activation. This agrees with our recent findings that Hsp27 expression is also regulated by Brn-3 in other cell types where Brn-3a and Brn-3b are expressed, such as sensory neuron-derived cells and breast epithelial cells (Farooqui-Kabir et al. 2004; Lee et al. 2005). Hsp27 is also important for the survival of these cells under stress conditions, such as serum withdrawal (Farooqui-Kabir et al. 2004). Therefore, Hsp27 may represent an important transcriptional target for Brn-3-induced survival and differentiation and may be complementary to the roles of anti-apoptotic genes such as Bcl-2 and Bcl-XL.
In conclusion, the class IV subfamily POU factors Brn-3a and Brn-3b are expressed in the developing heart. Furthermore, they appear to be able to compensate for each other at least in cardiomyocytes where both proteins are expressed. In this regard, both Brn-3a and Brn-3b can transactivate the hsp27 promoter in isolated cardiomyocytes. It is interesting that Brn-3a(l), which can strongly activate this promoter, shows a reciprocal expression to Brn-3b. This points to a complex mechanism by which this family of transcription factors may act in the heart. Given their role in other cell types, it is likely that Brn-3a and Brn-3b may act together to control the fate of specific cells during cardiogenesis.
Acknowledgement
The work described in this manuscript was supported by the British Heart Foundation PhD Studentship FS99084 to S.R.F.
References
- Breitschopf H, Suchanek G, Gould RM, Colman DR, Lassmann H (1992) In situ hybridization with digoxigenin-labeled probes: sensitive and reliable detection method applied to myelinating rat brain. Acta Neuropathol (Berl) 84:581–587 [DOI] [PubMed]
- Brown DD, Christine KS, Showell C, Conlon FL (2007) Small heat shock protein Hsp27 is required for proper heart tube formation. Genesis 45:667–678 [DOI] [PMC free article] [PubMed]
- Budhram-Mahadeo V, Morris PJ, Lakin ND, Theil T, Ching GY, Lillycrop KA, Moroy T, Liem RK, Latchman DS (1995a) Activation of the alpha-internexin promoter by the Brn-3a transcription factor is dependent on the N-terminal region of the protein. J Biol Chem 270:2853–2858 [DOI] [PubMed]
- Budhram-Mahadeo V, Lillycrop KA, Latchman DS (1995b) The levels of the antagonistic POU family transcription factors Brn-3a and Brn-3b in neuronal cells are regulated in opposite directions by serum growth factors. Neurosci Lett 185:48–51 [DOI] [PubMed]
- Budhram-Mahadeo V, Ndisang D, Ward T, Weber BL, Latchman DS (1999a) The Brn-3b POU family transcription factor represses expression of the BRCA-1 anti-oncogene in breast cancer cells. Oncogene 18:6684–6691 [DOI] [PubMed]
- Budhram-Mahadeo V, Morris PJ, Smith MD, Midgley CA, Boxer LM, Latchman DS (1999b) p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J Biol Chem 274:15237–15244 [DOI] [PubMed]
- Budhram-Mahadeo V, Moore A, Morris PJ, Ward T, Weber B, Sassone-Corsi P, Latchman DS (2001) The closely related POU family transcription factors Brn-3a and Brn-3b are expressed in distinct cell types in the testis. Int J Biochem Cell Biol 33:1027–1039 [DOI] [PubMed]
- Budhram-Mahadeo VS, Bowen S, Lee S, Perez-Sanchez C, Ensor E, Morris PJ, Latchman DS (2006) Brn-3b enhances the pro-apoptotic effects of p53 but not its induction of cell cycle arrest by cooperating in trans-activation of bax expression. Nucleic Acids Res 34:6640–6652 [DOI] [PMC free article] [PubMed]
- Calissano M, Faulkes D, Latchman DS (2005) Phosphorylation of the Brn-3a transcription factor is modulated during differentiation and regulates its functional activity. Brain Res Mol Brain Res 141:10–18 [DOI] [PubMed]
- Crawford SE, Qi C, Misra P, Stellmach V, Rao MS, Engel JD, Zhu Y, Reddy JK (2002) Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem 277:3585–3592 [DOI] [PubMed]
- Davidson SM, Morange M (2000) Hsp25 and the p38 MAPK pathway are involved in differentiation of cardiomyocytes. Dev Biol 218:146–160 [DOI] [PubMed]
- Davidson SM, Loones MT, Duverger O, Morange M (2002) The developmental expression of small HSP. Prog Mol Subcell Biol 28:103–128 [DOI] [PubMed]
- Eng SR, Lanier J, Fedtsova N, Turner EE (2004) Coordinated regulation of gene expression by Brn3a in developing sensory ganglia. Development 131:3859–3870 [DOI] [PubMed]
- Farooqui-Kabir SR, Budhram-Mahadeo V, Lewis H, Latchman DS, Marber MS, Heads RJ (2004) Regulation of Hsp27 expression and cell survival by the POU transcription factor Brn3a. Cell Death Differ 11:1242–1244 [DOI] [PubMed]
- Fedtsova N, Turner EE (2001) Signals from the ventral midline and isthmus regulate the development of Brn3.0-expressing neurons in the midbrain. Mech Dev 105:129–144 [DOI] [PubMed]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J (1996) POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci USA 93:3920–3925 [DOI] [PMC free article] [PubMed]
- Gan L, Wang SW, Huang Z, Klein WH (1999) POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev Biol 210:469–480 [DOI] [PubMed]
- Gruber CA, Rhee JM, Gleiberman A, Turner EE (1997) POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Mol Cell Biol 17:2391–2400 [DOI] [PMC free article] [PubMed]
- Hart SL, rancibia-Carcamo CV, Wolfert MA, Mailhos C, O’Reilly NJ, Ali RR, Coutelle C, George AJ, Harbottle RP, Knight AM, Larkin DF, Levinsky RJ, Seymour LW, Thrasher AJ, Kinnon C (1998) Lipid-mediated enhancement of transfection by a nonviral integrin-targeting vector. Hum Gene Ther 9:575–585 [DOI] [PubMed]
- Hudson CD, Podesta J, Henderson D, Latchman DS, Budhram-Mahadeo V (2004) Coexpression of Brn-3a POU protein with p53 in a population of neuronal progenitor cells is associated with differentiation and protection against apoptosis. J Neurosci Res 78:803–814 [DOI] [PubMed]
- Hudson CD, Morris PJ, Latchman DS, Budhram-Mahadeo VS (2005) Brn-3a transcription factor blocks p53-mediated activation of proapoptotic target genes Noxa and Bax in vitro and in vivo to determine cell fate. J Biol Chem 280:11851–11858 [DOI] [PubMed]
- Latchman DS (1999) POU family transcription factors in the nervous system. J Cell Physiol 179:126–133 [DOI] [PubMed]
- Lee SA, Ndisang D, Patel C, Dennis JH, Faulkes DJ, D’Arrigo C, Samady L, Farooqui-Kabir S, Heads RJ, Latchman DS, Budhram-Mahadeo VS (2005) Expression of the Brn-3b transcription factor correlates with expression of HSP-27 in breast cancer biopsies and is required for maximal activation of the HSP-27 promoter. Cancer Res 65:3072–3080 [DOI] [PubMed]
- Lillycrop KA, Budrahan VS, Lakin ND, Terrenghi G, Wood JN, Polak JM, Latchman DS (1992) A novel POU family transcription factor is closely related to Brn-3 but has a distinct expression pattern in neuronal cells. Nucleic Acids Res 20:5093–5096 [DOI] [PMC free article] [PubMed]
- Loones MT, Rallu M, Mezger V, Morange M (1997) HSP gene expression and HSF2 in mouse development. Cell Mol Life Sci 53:179–190 [DOI] [PMC free article] [PubMed]
- Lutsch G, Vetter R, Offhauss U, Wieske M, Grone HJ, Klemenz R, Schimke I, Stahl J, Benndorf R (1997) Abundance and location of the small heat shock proteins HSP25 and alphaB-crystallin in rat and human heart. Circulation 96:3466–3476 [DOI] [PubMed]
- Mao L, Shelden EA (2006) Developmentally regulated gene expression of the small heat shock protein Hsp27 in zebrafish embryos. Gene Expr Patterns 6:127–133 [DOI] [PubMed]
- Martin JL, Mestril R, Hilal-Dandan R, Brunton LL, Dillmann WH (1997) Small heat shock proteins and protection against ischemic injury in cardiac myocytes. Circulation 96:4343–4348 [DOI] [PubMed]
- Martin JL, Hickey E, Weber LA, Dillmann WH, Mestril R (1999) Influence of phosphorylation and oligomerization on the protective role of the small heat shock protein 27 in rat adult cardiomyocytes. Gene Expr 7:349–355 [PMC free article] [PubMed]
- Ndisang D, Budhram-Mahadeo V, Pedley B, Latchman DS (2001) The Brn-3a transcription factor plays a key role in regulating the growth of cervical cancer cells in vivo. Oncogene 20:4899–4903 [DOI] [PubMed]
- Obasanjo-Blackshire K, Mesquita R, Jabr RI, Molkentin JD, Hart SL, Marber MS, Xia Y, Heads RJ (2006) Calcineurin regulates NFAT-dependent iNOS expression and protection of cardiomyocytes: co-operation with Src tyrosine kinase. Cardiovasc Res 71:672–683 [DOI] [PubMed]
- Punn A, Mockridge JW, Farooqui S, Marber MS, Heads RJ (2000) Sustained activation of p42/p44 mitogen-activated protein kinase during recovery from simulated ischaemia mediates adaptive cytoprotection in cardiomyocytes. Biochem J 350 Pt 3:891–899 [PMC free article] [PubMed]
- Sakamoto K, Urushidani T, Nagao T (2000) Translocation of HSP27 to sarcomere induced by ischemic preconditioning in isolated rat hearts. Biochem Biophys Res Commun 269:137–142 [DOI] [PubMed]
- Sugars KL, Budhram-Mahadeo V, Packham G, Latchman DS (2001) A minimal Bcl-x promoter is activated by Brn-3a and repressed by p53. Nucleic Acids Res 29:4530–4540 [DOI] [PMC free article] [PubMed]
- Sun Y, MacRae TH (2005) Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci 62:2460–2476 [DOI] [PMC free article] [PubMed]
- Tallot P, Grongnet JF, David JC (2003) Dual perinatal and developmental expression of the small heat shock proteins [FC12]aB-crystallin and Hsp27 in different tissues of the developing piglet. Biol Neonate 83:281–288 [DOI] [PubMed]
- Theil T, Rodel B, Spiegelhalter F, Moroy T (1995) Short isoform of POU factor Brn-3b can form a heterodimer with Brn-3a that is inactive for octamer motif binding. J Biol Chem 270:30958–30964 [DOI] [PubMed]
- Turner EE, Jenne KJ, Rosenfeld MG (1994) Brn-3.2: a Brn-3-related transcription factor with distinctive central nervous system expression and regulation by retinoic acid. Neuron 12:205–218 [DOI] [PubMed]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, Shows TB, Nathans J (1995) The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci 15:4762–4785 [DOI] [PMC free article] [PubMed]
- Xiang M, Gan L, Zhou L, Klein WH, Nathans J (1996) Targeted deletion of the mouse POU domain gene Brn-3a causes a selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc Natl Acad Sci USA 93:11950–11955 [DOI] [PMC free article] [PubMed]