Abstract
Protein ubiquitination plays an essential regulatory role within all eukaryotes. Large-scale analyses of ubiquitinated proteins are usually performed by combining affinity purification strategies with mass spectrometry. However, there is no reliable method to systematically differentiate ubiquitinated species from co-purified unmodified components. Here we report a simple strategy for the large-scale validation of ubiquitination by reconstructing virtual Western blots for proteins analyzed by gel electrophoresis and mass spectrometry. Because protein ubiquitination, especially polyubiquitination, causes dramatic shift of molecular weight, the difference between experimental and expected molecular weight was used to confirm the status of ubiquitination. Experimental molecular weight of putative yeast ubiquitin-conjugates was computed from the value and distribution of spectral counts in the gel using a Gaussian curve fitting approach. Unmodified proteins in yeast cell lysate were also analyzed as a control to assess the accuracy of the method. Multiple thresholds that incorporated the mass of ubiquitin and/or experimental variations were evaluated with respect to sensitivity and specificity. Ultimately, only ~30% of the candidate ubiquitin-conjugates were accepted based on the stringent filtering criteria, although they were purified under denaturing conditions. These accepted conjugates had an estimated false discovery rate of ~8% and primarily consisted of proteins larger than 100 kDa. Compared with another validation method (i.e. identification of ubiquitinated lysine sites), ~95% of the proteins with defined modification sites showed a convincing increase in molecular weight on the virtual Western blots. A second independent analysis indicated that the method can be simplified by excising less than ten gel bands. Therefore, this strategy establishes criteria necessary for the interpretation of ubiquitinated proteins.
INTRODUCTION
The ubiquitin (Ub) modification pathway is a highly regulated, transient and reversible event that is conserved amongst eukaryotes. The covalent modification of cellular substrates with Ub plays a principal regulatory role in many cellular processes, such as proteasome-mediated degradation1, 2, protein sorting3, inflammation4 and DNA-repair5. Ubiquitination occurs via the carboxyl terminus of the Ub glycine, which forms an isopeptide bond primarily with the ε-amino group of lysine residues on targeted substrates. This event is catalyzed by a cascade of enzymes that include Ub activating enzyme (E1), Ub-conjugating enzymes (E2s) and Ub-ligases (E3s)1, 6, 7. The substrates can either be mono-ubiquitinated (mono-Ub) or poly-ubiquitinated (poly-Ub) at a single or multiple Lys sites. Polyubiquitin chains are assembled when additional Ub molecules are conjugated to any of the seven lysine residues (K6, K11, K27, K29, K33, K48 and K63) or even N-terminal amine group of pre-existing Ub molecules8–10. Conversely, deubiquitination enzymes (DUBs) remove Ub from modified substrates to further contribute to dynamic ubiquitination process11, 12. Importantly, dysregulation of ubiquitination has profound impact on cellular functions and is involved in the pathogenesis of many diseases, including cancer and neurodegenerative disorders13, 14, and the inhibition of ubiquitin-proteasome system has been demonstrated to be a successful strategy to treat multiple myeloma. Thus, methodologies that assist in the global analysis of Ub-conjugates are essential for the characterization of pathways that are regulated by ubiquitination.
Recent advances in the development of mass spectrometry (MS)-based technologies have allowed for the detection and quantification of hundreds to thousands of proteins with accuracy and sensitivity16–18. Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) is typically used to analyze protein mixtures for large-scale proteomic applications and has become the preferred method for the analysis of ubiquitinated proteome19–22. However, because ubiquitinated conjugates are present at a low steady-state level, due in part to proteasome-mediated degradation and highly active deubiquitination enzymes in cells, it is difficult to identify Ub-conjugates without prior enrichment. To this end, various affinity approaches have been used to isolate Ub-conjugates, including Ub-antibodies23, 24, Ub-binding proteins25, 26, and epitope-tagged ubiquitin derivatives (e.g. FLAG, HA-tag, myc-tag, His-tag and biotin)8, 27–30. Co-purification of unmodified proteins is controlled using extensive wash and/or stringent denaturing conditions (e.g. 8 M urea), but many are still identified in enriched Ub samples. During nickel affinity chromatography, the contaminants are usually found to be endogenous His-rich or highly abundant proteins8. Additional efforts have also been made to reduce non-specific binding by introducing two-step affinity purification schemes28. The presence of protein contaminants is often exacerbated when employing non-tagging affinity strategies (e.g. Ub antibodies or Ub-binding proteins) under native conditions23. Thus, it is critical to distinguish Ub-conjugates from false-positive contaminants before subsequent functional studies31.
The common method for validating Ub-conjugates in large-scale proteomic analyses is the direct mapping of ubiquitination sites by MS/MS. Trypsin digestion of Ub-conjugates generates a di-glycine remnant (-GG, a monoisotopic mass of 114.043 Da) on modified lysine residues, producing unique MS/MS spectra that can be matched by database-searching algorithms8, 32, 33. One technical challenge is that complete mapping of modification sites requires almost 100% coverage of proteins/peptides “sequenced” by MS/MS. Thus, in large-scale analysis from yeast, only a small fraction of GG-sites can be mapped to peptides, matching to less than 10% of the proteins identified21. Therefore, secondary strategies are necessary to complement Ub site mapping to improve validation of large datasets.
Western blot analysis of immunoprecipitated Ub-conjugates is commonly used to confirm Ub-conjugates independently8. Two principles are utilized in the method: (i) ubiquitination causes dramatic increase in apparent molecular weight (MW) in Western blot, as Ub-conjugates display an increase of approximately 8 kDa after mono-ubiquitination and an even larger increase after poly-Ub events; (ii) ubiquitination often generates heterogeneous modified substrates that display as a ladder on the Western blot. However, this type of analysis becomes expensive and impractical for large-scale studies in which thousands of Ub-conjugate candidates are identified.
Herein we describe a robust method for large-scale validation of protein ubiquitination based on virtual Western blots reconstituted from MS data. MW information of every protein identified was extracted after 1D SDS gel and LC-MS/MS (1D geLC-MS/MS). To evaluate false discovery rate of the method, two geLC-MS/MS analyses were performed before and after Ub affinity purification. Multiple statistical analyses were implemented to improve the approach. Finally, we found that only ~30% of identified proteins in the Ub-conjugate samples survived the MW filtering, even though they were purified in the presence of 8M urea, suggesting that false discovery rate in previously published datasets of ubiquitinated proteome may be underestimated.
EXPERIMENTAL SECTION
Purification of ubiquitin conjugates from S. cerevisiae
The purification was performed similarly as described previously8, 31. Yeast strain SUB592 that expresses 6xHis-myc-ubiquitin as the only source of ubiquitin was grown at 30°C to log phase (OD600 0.7–1.5) and lysed in denaturing buffer (10 mM Tris-HCl, pH 8.0, 0.1 M NaH2PO4, 8 M urea, 10 mM β-mercaptoethanol). The total cell lysate was clarified by centrifugation at 70,000 g for 30 min and loaded twice onto a 0.5 ml Ni2+-NTA-agarose column (Qiagen). The column was extensively washed followed by elution with low-pH buffer (10 mM Tris, pH 4.5, 0.1 M NaH2PO4, 8 M urea).
Proteomic analyses by 1D geLC-MS/MS
Protein from yeast total cell lysate (TCL, ~100 μg) or affinity purified His-Ub-conjugates (UbC, ~100 μg) were reduced with 10 mM dithiothreitol (DTT) and alkylated with 50 mM iodoacetamide for 30 min in the dark. The samples were added with gel-loading buffer (10 mM Tris-HCl, pH 8.0, 2% SDS, 4% Ficoll, 0.02% bromophenol blue) and resolved on 6–12% olyacrylamide gradient SDS gels (0.75 mm thick, 14 mm wide, and 120 mm long). The gradient gel was selected to maximize the resolution. The gel was run at 200 V for ~4 hrs. After staining with Coomassie blue, the high and low retention factors (Rf values) for each gel band and MW marker (BenchMark™Protein Ladder, 10–220 kDa, Invitrogen) were measured. The gel lanes were cut into 54 and 40 gel bands for TCL and UbC samples, respectively, followed by in-gel trypsin digestion34. The peptide samples were analyzed by reverse phase nanoLC-MS/MS using a 75 μm i.d. self-packed fused silica C18 (5 μm beads) capillary column at a flow rate of ~0.3 μl/min32. Peptides were loaded and eluted for each analysis during a gradient in which the ions were detected, isolated and fragmented in a completely automated fashion on an ion trap mass spectrometer (Thermo Finnigan, San Jose, CA). MS/MS spectra were searched against a composite target/decoy yeast open reading frames (ORFs) database (downloaded in 2006 from the National Center for Biotechnology Information)35 using the SEQUEST algorithm (version 27)36. Searching parameters included the mass tolerance of precursor ions (±2 Da), no enzyme restriction, fixed modification of carboxyamidomethylated Cys (+57.0215 Da), dynamic mass shifts for oxidized Met (+15.9949 Da) and ubiquitinated Lys (114.0429)8. Only b and y ions were considered during the database match. Peptide matches were grouped by a combination of trypticity (fully, partial and non-tryptic) and precursor ion-charge state (1+, 2+, and 3+), and then filtered by XCorr and ΔCn values to reduce the false discovery rate to near zero35, namely, the filtering cutoffs were adjusted until no peptide matches from the decoy database could survive. Mass accuracy (15 ppm) was also used to filter incorrect peptides in samples analyzed by an LTQ-Orbitrap mass spectrometer. On occasion when Ub modified peptides had multiple lysine residues, the SEQUEST algorithm falsely assigned the ubiquitination site. Hence, all modified peptides were manually verified. All accepted proteins sharing peptides were grouped together, and the top protein with highest peptide matches was selected to represent the group. If other group members were identified by at least one unique peptide, they were also included in the group. In addition, the analysis of Ub conjugates was repeated on an LTQ-Orbitrap mass spectrometer except that eight gel bands were excised.
Experimental molecular weight derived from Gaussian curve fitting of protein spectral count distribution
(i) Assign average experimental MW to excised gel bands. Linear regression analysis was performed to establish a relationship between relative mobility (Rf) and the logarithmic (log) value of MW markers (10–220 kDa). Substitution of the average Rf value for each excised gel band into the equation for a straight line allowed for calculation of an average MW for each gel band. Linear extrapolation was used to estimate the MWs of gel bands excised outside the MW marker range (>220 kDa). (ii) Determine experimental MWs for identified proteins. Since many proteins were detected in multiple gel bands, we used spectral count (SC) as a semi-quantitative index in each gel band, and assumed that the scatter of protein spectral counts follows Gaussian distribution. Thus, the number of protein spectral counts (f) in each gel band was used to fit Gaussian distribution as a function of MW (x) (Eq. 1).
| (1) |
Where, “exp” is the exponential function, “a” is the amplitude, “μ” is the mean, representing the experimental MW of the protein, and “σ” is approximately equal to the sample standard deviation (SD), indicating the dispersion of a protein on the SDS gel. Because outliers have a significant influence on the curve fitting, data points that fell 2 SD away from the initial calculated MW (μ) were removed from further analysis. An iterative approach was used to define a, μ, and σ that best fit the experimentally determined variables (gel band MW and SC).
Statistical evaluation of the difference between experimental and predicted MWs (ΔMWs)
A central moving average algorithm37 was employed to smooth the curve of all ΔMWs versus predicted MWs. The “smoothed ΔMW” (ΔMWi)s for each data point (i) is the average of an odd number of consecutive 2n+1 points of experimentally assigned ΔMW (Eq. 2).
| (2) |
The odd number 2n+1 corresponds to the bin size (with minimum value of 31, in which the sample mean is almost identical to the mean of a normally distributed population). To reduce the effect of outliers, such as genuine ubiquitinated proteins, data points that fell outside two moving standard deviations (MSD) from the moving mean were discarded. In addition, proteins that had a MW difference greater or less than 50 kDa were treated as true outliers and, therefore, not considered for the moving average analysis. The smoothed ΔMW curve was then used to normalize all ΔMW values, and the associated moving standard deviation in each bin was recorded.
Reconstruction of virtual Western blot for identified proteins in silico based on spectral counts
The protein intensity in gel lanes was represented by two parameters, thickness and darkness, which were derived from spectral counts. As one protein may be identified in multiple gel bands, all spectral counts for the protein were first normalized by setting the maximal value to 50 and the remaining values proportionally. The SC values were equally divided into 7 grades for the thickness, as well as 50 levels for the darkness of protein bands, whereas the values less than 1 were considered to be too weak to detect. Moreover, the protein bands were plotted according to calculated relative mobility that was linearly correlated with the log of experimental MWs. All in-house programs were written in Perl and the GD.pm module was used for the drawing of virtual Western blots.
RESULTS AND DISCUSSION
A strategy to validate substrate ubiquitination by virtual Western blot based on 1D geLC-MS/MS
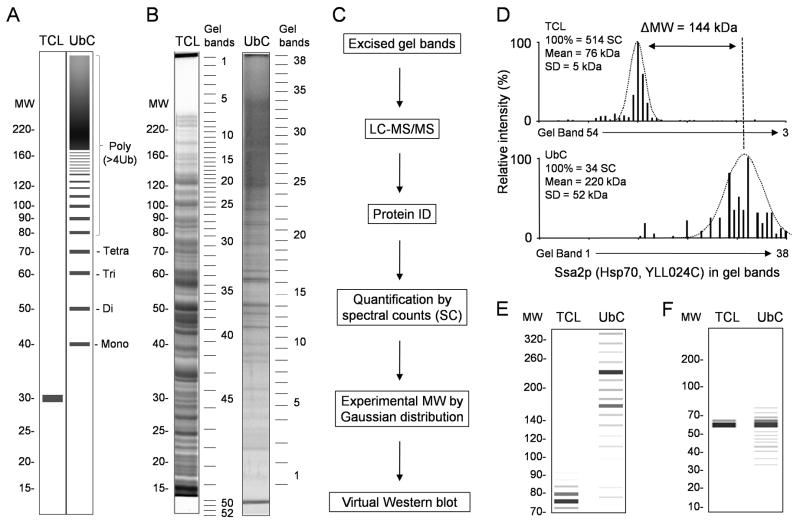
To examine if 1D geLC-MS/MS can be effectively used to reconstruct a virtual Western blot for every protein, we analyzed both yeast total cell lysate (TCL) and His-myc-tagged Ub-conjugates (UbC) that were enriched by a nickel affinity column under denaturing condition. Since His-myc-ubiquitin is expressed as a 98 amino acid protein, targeted substrates would display an increase of approximately 10 kDa for each Ub modification event as presented schematically (Fig. 1A). During the purification, the Ub-conjugates were substantially enriched to a similar level to that previously reported8, indicated by the characteristic high MW smearing (Fig. 1B).
Figure 1. A Strategy for validating ubiquitinated conjugates from 1D geLC-MS/MS analysis.
(A) A schematic Western blot image of a ubiquitinated protein in total cell lystate (TCL) and enriched His-tagged ubiquitin conjugates (UbC). Modification of the targeted substrate causes a dramatic shift in MW (an increase of ~10 kDa for mono-ubiquitination, ~20 kDa for Di-ubiquitination, etc.) compared with its precursor. Heterogeneous poly-ubiquitination is typically observed as high MW smears. (B) To experimentally estimate the degree of ubiquitination on targeted substrates, proteins from the TCL or UbC sample were analyzed by 1D SDS gel electrophoresis with protein MW marker (shown in kDa). The entire lanes were excised into gel bands as indicated, trypsin digested and peptides analyzed by LC-MS/MS. The stacking gel of the UbC was also cut into two fractions (#39 and #40, not shown) because it is well known that the stacking gel contains some Ub-conjugates that are very large or precipitated. (C) The experimental flow to assign approximate MWs from linear regression analysis (see Methods) and Gaussian curve fitting, and to reconstitute virtual Western blot images. (D) A representative Gaussian curve fitting to derive protein MW from the number and distribution of spectral counts. The difference in MW for a protein identified in both TCL and UbC provided an estimation of protein ubiquitination. (E) & (F) The in silico reconstructed Western blots of the proteins Ssa2p and Sro9p, respectively.
The proteins in the TCL or the UbC were well separated on a SDS gel, divided into 52 and 40 gel bands, respectively, trypsin digested and peptides analyzed by LC-MS/MS (Fig. 1B and 1C). Yeast database searches followed by stringent peptide filtering led to the acceptance of 1,976 and 1,246 proteins in the TCL and UbC samples, respectively. Of these, 749 proteins were identified in both samples. To reduce the interference of false identification of proteins on subsequent analysis, we applied the target/decoy strategy35 to evaluate false discoveries during database matching and used a number of parameters to filter the data until all decoy matches were removed, suggesting that the final false discovery rate of accepted peptides was minimized to near zero.
If a protein was only detected in a single gel band, it would be trivial to obtain its experimental molecular weight (MW). However, many proteins were identified in multiple gel bands. To achieve the best approximation of experimental MW for every protein, we assumed that the distribution of protein abundance along 1D SDS gel follows Gaussian distribution. As the protein abundance can be reflected by its spectral count that is the number of tandem spectra to sequence the protein38, 39, an iterative Gaussian curve fitting approach was employed to compute the mean and standard deviation (SD) from its spectral count distribution (Fig. 1C and 1D). The mean represented an experimental MW of the protein and SD indicated its dispersion on the gel. Virtual Western blots in silico were used to facilitate the visualization of MW differences for those proteins identified in both UbC and TCL samples (Fig. 1E and 1F). For example, after Gaussian curve fitting, the yeast protein Ssa2p (Hsp70, YLL024C) was assigned a MW of 76 and 220 kDa in the TCL and UbC samples, respectively. The increase in MW (ΔMW = 144 kDa) observed for Ssa2p in UbC compounded with its dispersed spectral counts (SD = 52 kDa) suggested that this protein was truly modified by ubiquitin. In contrast, the protein Sro9p (YCL037C) showed no obvious MW shift, suggesting that it was a co-purified protein without ubiquitin modification.
The accuracy of experimental MW derived from the Gaussian curve fitting approach
The accuracy of experimental MWs relies on precision of protein relative mobility and gel band excision. Although the relative mobility under the SDS denaturing condition is primarily dependent on the size, protein migration on a SDS gel may be affected by remaining secondary and higher order structures. Moreover, whereas the relative mobility of a protein is inversely correlated with the logarithmic (log) value of its MW, in this study the linear relationship between them was not perfect, as indicated by the working curve from the MW marker (supplemental Fig. 1). The result is consistent with the previous report that proteins of extremely large or small size do not follow the anticipated linearity during electrophoresis40. Nevertheless, during the fitting of a linear line, the MW marker (15 kDa to 220 kDa) displayed a R2 value of 0.993. In addition, other experimental conditions, such as amount of protein loaded, also influence the MW resolution. Therefore, the loading capacity was examined and it was shown that ~60 μg of protein could be well resolved in a gel lane (0.75 mm thick, 7 mm wide, and 120 mm long, supplemental Fig. 2). We thus loaded ~100 μg of protein in the gel wells twice this size (see Experimental Section).
To minimize systematic experimental errors in the calculation of experimental MWs, we presumed that the difference between the experimental and predicted MWs (ΔMW = experimental – predicted MW) of proteins was centered at zero based on the null hypothesis. A central moving average algorithm37 was applied to normalize the ΔMWs after removing outliers. Indeed, in the sample of total cell lysate, the vast majority of proteins had experimental MWs in agreement with their predicted MWs, and the ΔMWs were generally small and showed a reasonable Gaussian distribution, consistent with the null hypothesis (Fig. 2A, 2C, 2E, and supplementary table S1). In contrast, in the Ub-conjugate sample, a significant portion of proteins had a clear shift towards higher MW regions, indicating the presence of ubiquitinated species (Fig. 2B, 2D, 2F, and supplementary table S2). It should be noted that the normalization step did not dramatically change the distribution pattern of ΔMWs, as indicated by the comparison of either the TCL or the UbC proteins before and after normalization (Fig. 2A versus Fig. 2C; Fig. 2B versus Fig. 2D), because the proteins with large ΔMWs were treated as outliers and thus did not influence the application of the moving average algorithm.
Figure 2. Evaluation of MW differences between predicted and experimental protein MWs.
(A) & (B) The distribution of ΔMW values (experimental – predicted MWs) along the predicted MW in the TCL (n =1,976) and UbC (n =1,246) before normalization by moving average algorithm. Each grey point represents a single protein, and the dashed line represents ΔMW equal to zero. (C) & (D) The distribution after normalization in the TCL and UbC, respectively. (E) & (F) The ΔMW histogram of the proteins identified in the TCL and UbC.
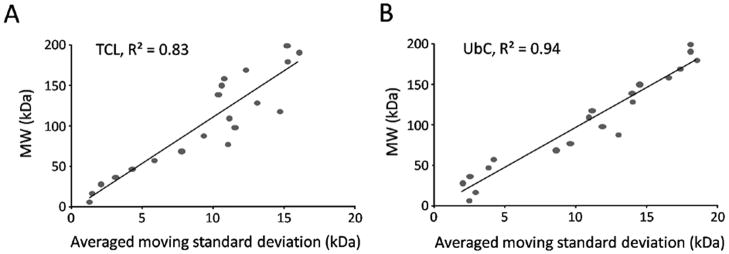
Importantly, the algorithm computed for every data point (i.e. one protein) a moving standard deviation (MSD) as an indicator of the precision of the derived ΔMW (Fig. 3). It should be emphasized that the MSD is not related to the SD fitted from Gaussian curve analysis (Fig. 1C), which was a measurement of protein dispersion based on spectral counts. As expected, in both samples, the MSD of identified proteins increased almost linearly with the experimental MWs, essentially reflecting limited resolving power of gel electrophoresis. Therefore, the precision in the MW measurement by electrophoresis generally decreased as the mass of the proteins increased.
Figure 3. Linear correlation of moving standard deviation (MSD) with measured MWs.
The central moving average approach was used for computational correction of protein ΔMW values and for the calculation of MSD that indicated the precision of the measurement of ΔMWs. To evaluate the relationship between MSD and experimental MWs, proteins in the range of 0–200 kDa were divided into 20 bins, in which MSDs associated with the proteins were averaged. The averaged MSDs were then plotted against the experimental MWs. (A) and (B) represent TCL and UbC, respectively.
Criteria of MW shift for validating ubiquitinated proteins
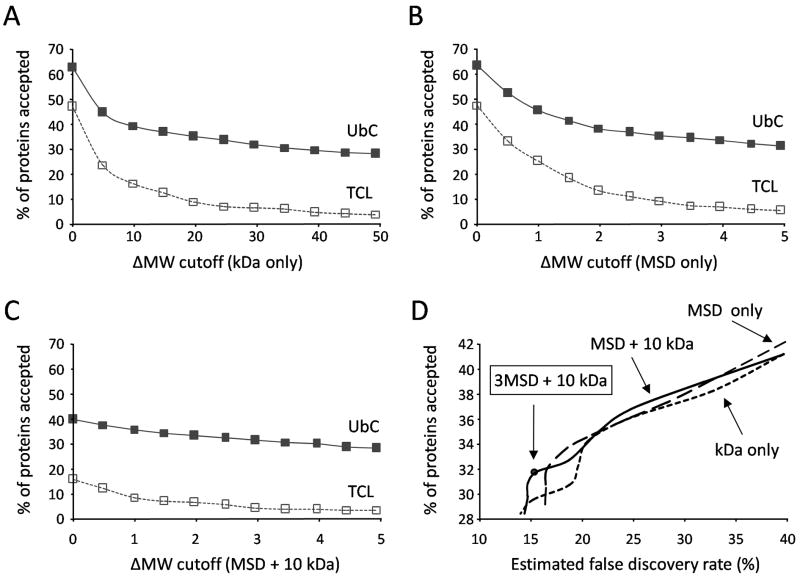
To validate the candidate Ub-conjugates based on an increased experimental MW, it was necessary to optimize ΔMW thresholds that would effectively separate real Ub-conjugates from other protein contaminants. Ostensibly, it would be reasonable to accept all proteins in UbC above a ΔMW threshold of 10 kDa as being ubiquitinated (i.e. true positives) because this is the minimum MW increase caused by a single His-myc-ubiquitin modification. However, without regard of experimental errors, a 10 kDa increase might not satisfy the requirement for ubiquitination of all proteins. To test this hypothesis the percentage of proteins accepted as true positives were monitored with increasing ΔMW thresholds based on the magnitude (kDa) and/or associated dynamic MSD (Fig. 4). In parallel, we analyzed the TCL proteins in the same manner, and considered any proteins accepted in the TCL as false discoveries. Under the identical threshold, the percentage of false discoveries in the UbC was assumed to be the same as that in the TCL dataset. To reduce the effect of protein properties on this assumption, in this analysis the population of TCL was limited to only those proteins also identified in the UbC samples (n = 749), since they essentially represent precursor proteins void of Ub modification.
Figure 4. ΔMW criteria to validate protein ubiquitination.
(A–C) The percentage of proteins accepted in the Ub-conjugates (UbC, solid line) and the total cell lyste (TCL, dashed line) were monitored with increasing ΔMW thresholds based on (A) kDa alone, (B) moving standard deviation (MSD) only, or (C) MSD + 10 kDa. (D) The change of cutoffs in any method led to varied sensitivity (indicated by % of proteins accepted in UbC) and specificity (reflected by false discovery rate), exhibited in a ROC curve. The false discovery rate was derived by the formula (% of proteins accepted in the TCL/% of proteins accepted in the UbC).
Three different approaches for setting up ΔMW cutoff were compared (Fig. 4). Proteins that had higher experimental MWs than predicted were of interest, which corresponded to 63% and 47% of proteins in the UbC and TCL, respectively. When the threshold of ΔMW moved from 0 to 10 kDa, the proteins accepted in both samples was markedly reduced (Fig. 4A). Although a more precipitous drop in acceptance was observed for TCL, 16% were still above the 10 kDa threshold, suggesting that a simple increase in the MW of His-myc-ubiquitin alone was not a reliable indicator of protein ubiquitination for most proteins analyzed. We then evaluated the thresholds using MSD alone. Here a more modest reduction in proteins accepted was observed as the MSD threshold increased (Fig. 4B). Interestingly, when the static 10 kDa cutoff was used in combination with the dynamic MSD, the false discoveries appeared to be controlled at much lower level without obvious reduction of the yield of Ub-conjugates (Fig. 4C). The three methods were further compared in a receiver operating characteristic (ROC) plot, which displayed the percentage of proteins accepted (i.e. sensitivity) to the estimated false discovery rate (Fig. 4D). A reasonable compromise between sensitivity and specificity was achieved using a threshold of 10 kDa + 3 MSD. This condition resulted in the acceptance of 32% (396/1246) of UbC proteins, but only 5% of TCL proteins. If the same fraction (5%) of the UbC proteins were considered as false discoveries, the false discovery rate was estimated to be 16% (5%/32%), namely, ~63 false discoveries in 396 UbC proteins.
False discoveries in the Ub-conjugates resulted from, at least partially, the inaccuracy of predicted MWs of unmodified proteins. Although 84% of proteins identified in the TCL had computed MWs within ±3 MSD of their predicted MW (Fig. 2), consistent with previous studies that used MW location from 1D geLC-MS/MS to aid in protein identification41, 42, there were still 16% of proteins that showed inconsistency. This is probably caused by posttranslational modifications (e.g., glycosylation and lipidation) or alternative splicing and proteolysis events.
To further reduce the false discoveries, we corrected the calculation of MW difference using experimental MW detected in the TCL as reference (ΔMWc = experimental MW in the UbC – that in the TCL) and applied the threshold of 10 kDa with the addition of 3 MSD. As a consequence, 33 more proteins were removed from the list, resulting in a final dataset of 363 Ub-conjugates (29% of 1246 proteins, supplementary table S3) with an estimated false discovery rate of 8% [(63–33)/363]. The virtual Western blots of those proteins were also generated and shown with web links (supplementary table S4). These proteins best represented protein ubiquitination based on their increased MW after 1D geLC-MS/MS analysis.
Comparison of the virtual Western blot strategy with other validation methods
During the affinity enrichment of His-tagged Ub-conjugates by Nickel chromatography, a negative control experiment was previously performed using a wild-type yeast strain, which do not express His-tag Ub, in order to identify potential false-positive contaminants8. In that report, a total of 50 proteins were identified in the wild-type dataset. They were either endogenous His-rich or highly abundant proteins in cells, and were simply removed from the list of candidate ubiquitinated proteins. In the UbC sample, 36 of these contaminants were also detected, of which 94% (34/36) were discarded using this newly developed virtual Western blot strategy (Fig. 5A, Supplementary Table S5). The result strongly supported the accuracy of the ΔMW criteria as an approach to predict ubiquitination. Only 2 out of 36 potential contaminated proteins were found in the final dataset. However, it could not be ruled out that the two accepted proteins were ‘genuinely’ modified since His-rich or highly abundant proteins “contaminants” could be ubiquitinated. Indeed, one of the two proteins (Hbt1p, YDL223C) was detected in a series of gel bands in the SDS gel, reminiscent of heterogeneous polyubiquitinated protein pattern.
Figure 5. The overlap between validated Ub-conjugate dataset with other negative or positive controls.
(A) A total of 363 proteins were accepted from the UbC based on their increase in MW for 1D geLC/MS/MS. Proteins having identified GG-sites (positive controls, circle of solid line) and previously defined contaminants (negative controls, circle of dashed line) were used to assess the dataset. The ubiquitinated proteins that were filtering by the virtual Western blot method contained 25 out of 26 positive controls, but only 2 out of 36 negative controls. (B) Comparison of this study with previously published datasets. The number of overlapped proteins between any two datasets was indicated. See more details in supplemental Table S7.
Another approach to confirm Ub candidate substrates is to directly identify ubiquitinated sites with Gly-Gly tag (GG-sites) on modified peptides by tandem mass spectrometry. In this study proteins identified with GG-sites served as positive controls since they were thought to best represent bona fide Ub-conjugates. In the UbC sample, 44 GG-sites were mapped by MS/MS spectra to 26 candidate ubiquitin conjugates including Ub itself (Fig. 5A, Supplementary Table S6). Of these 96% (25/26) passed the optimal threshold (10 kDa + 3 MSD) based on experimental increase in MW, indicating a high degree of consistency of the two independent validation methods.
Interestingly, the single protein (Rps3p, YNL178W) that did not meet the MW criteria still displayed an increased MW, maybe resulting from mono-ubiquitination. Further development of the method is required to confirm the proteins conjugated by a single ubiquitin molecule. However, mono-ubiquitinated species may not represent a large portion of discarded proteins because of low recovery during purification30. Mono-ubiquitination of proteins can also induce large MW shift by modify multiple sites (i.e. poly-mono-ubiquitination). Alternatively, it is possible that the GG site in Rps3p was falsely assigned during database matching.
To this end, we have encountered several pitfalls during the identification of GG-sites by matching MS/MS spectra with database. (i) Peptides can be falsely assigned as ubiquitinated if they contain internal Lys residue(s) and adjacent N- or C-terminal amino acid residues that have similar mass to GG-modification, such as GG themselves (average mass of 114.1 Da), L (113.2 Da), I (113.2 Da), N (114.1 Da), or D (115.1 Da). For instance, when mass tolerance of precursor ions is not strict, a peptide LK(X)nR (X represents any residue) displays almost identical MS/MS patterns to L.K*(X)nR (the asterisk indicates GG-tag, and the dot indicates the cleavage site). These mismatches can be removed by examining the tryptic state of the peptides, as the peptide of L.K*(X)nR is not fully tryptic. High mass accuracy filtering is also effective to correct for these types of errors. (ii) Although it is possible to generate LR74GG tag on modified Lys due to incomplete tryptic cleavage, we have rarely found ubiquitinated peptides carrying this large tag in numerous large-scale analyses. This is supported by the report that the R74 residue is the most accessible tryptic site in ubiquitin under native condition43 and can be excised at high efficiency under denaturing conditions44. Therefore, more caution should be taken to examine LRGG modified peptide assignments. (iii) Occasionally a mono-ubiquitinated peptide has several lysine residues, and the SEQUEST algorithm36 ambiguously assign the GG modification to a Lys residue, such as the C-terminal lysine (Supplementary Fig. S3). Recently, it has been reported that trypsin is able to cleave at the C-terminal GG-modified Lys sites45. To test this hypothesis, we carried out a digestion experiment using a synthetic ubiquitin GG-peptide that was shown to be cleaved. During the time course of incubation, the signal of the GG-peptide was stable in 18 hours. In contrast, a similar amount of neurotensin peptide was completely digested in only 10 minutes, strongly arguing that the GG-modified Lys residue is not susceptible to trypsin digestion under the condition (Supplementary Fig. S4). (iv) Ubiquitinated peptides could be simply generated by random mismatching, especially when a large number of MS/MS spectra are used to search again a small database. Therefore, additional MW information of proteins presented here can assist the interpretation of these modified peptides.
Comparison with previously published datasets of ubiquitinated proteins
We carried out the comparison between this study and other three datasets reported by Tagwerker et al.28 and Mayor et al.22, 27 (Fig. 5B, Supplementary Table S7). The overlapped putative Ub-conjugate between any two datasets was lower than 30%, even from the same research group, implying that the purification was highly variable and possibly contained contaminants. Alternatively, the variations could be partially explained by different strains, cell culture conditions, purification methods, mass spectrometry platforms, and most importantly, filtering criteria. More interestingly, out of 253 proteins identified by Tagwerker and coworkers, 166 proteins were shared with our list of Ub-conjugate candidates before MW filtering, and 58 proteins were overlapped with the list after filtering. Apparently, ~35% (58/166) of proteins passed the MW threshold. Similar results (32/92 = 35%, 47/129 = 36%) were obtained by comparison with the other datasets, supporting the idea that a similar percentage of false positives might be present in those analyses employing denaturing conditions.
Simplification of the virtual Western blot strategy
Typically MW resolution is increased if more gel bands are excised, however, it was laborious to cut one sample lane into as many as 52 gel bands. To reduce the workload, we tested if some gel bands contained more ubiquitinated species than others. Indeed, no proteins in the final accepted ubiquitinated dataset had an experimental MW below 40 kDa and 84% of the proteins had experimental MWs greater than 100 kDa (Fig. 6A). Although the accepted Ub-conjugate list did not have proteins with a computed MW below 40 kDa, some of these proteins spread into a number of gel bands and were still detected in lower MW range possibly owing to partial degradation (Fig. 6B). However, at least 80% of proteins identified in the gel bands below 40 kDa were filtered out by the stringent criteria. In contrast, the filtered percentage substantially reduced to <40% in the gel bands above 100 kDa (Fig. 6B). Together these data indicated that majority of Ub-conjugates could be identified from gel bands larger than 100 kDa, in agreement with the observation that UbC proteins run as high MW smears above 100 kDa (Fig. 1B). More importantly, the results suggest that the analysis of Ub-conjugates could be simplified by focusing on gel bands with high MWs.
Figure 6. The distribution of filtered Ub-conjugates in gel bands during 1D geLC-MS/MS. (.
A) The accepted ubiquitinated proteins accumulated with the increase of experimental MWs. After Gaussian curve fitting analysis no protein with a computed MW below 40 kDa was considered ubiquitinated. (B) The percentage of proteins that were filtered out by the virtual Western blot strategy in every gel band.
To test this approach, we performed a small scale analysis of yeast purified Ub-conjugates. In this case, only eight gel bands above 50 kDa were analyzed (Supplementary Fig. S5). A total of 353 proteins were identified and their ΔMW distribution was similar to that in the experiment with 40 gel bands. After applying the same criteria to filter the Ub-conjugate candidates, the percentage of acceptance was 31% (108/353 proteins) that was slightly higher than 29% in the dataset above, which could be explained by the fact that there are less false positives in the gel bands with high MWs. This independent analysis demonstrated the feasibility of the simplified strategy for validating the ubiquitinated proteome.
CONCLUSION
Although recent large-scale proteomics studies have implemented LC-MS/MS for the identification of affinity purified Ub-conjugates8, 23, 27, 28, 30, it remains challenging to differentiate genuine Ub-conjugates from co-isolated unmodified species, since GG-peptides alone can only validate a very small fraction of candidate Ub-conjugates, and immunoprecipitation of individual candidate Ub-conjugates becomes impractical to confirm all identified proteins. In this study we describe a method for the large-scale validation of protein ubiquitination from 1D geLC-MS/MS analysis. The estimated MW for all proteins was computed from the value and distribution of spectral counts in 1D SDS gels using a Gaussian curve fitting approach. This data was used to recapitulate virtual western blots for each protein analyzed. Therefore, relative to the MW of its unmodified precursor, the approximate degree and diversity of ubiquitination for each candidate Ub-conjugate was obtained.
In spite of the presence of some false discoveries (~8%) and false negatives (due to mono-ubiquitination), the virtual Western blot strategy proved to be an effective tool to assess differences in MW caused by ubiquitination. This type of analysis provides much needed validation of ubiquitinated proteins identified after affinity enrichment and complements previous small-scale validation schemes.
Supplementary Material
Figure S1. The linear correlation between log10(MW) of protein markers with their relative mobility in the total cell lysate (TCL). Similar data was obtained in the Ub-conjugates (UbC, data not shown).
Figure S2. Examination of loading capacity for 1D SDS gel. Yeast protein (20–160 μg) was titrated on a thin SDS gel (0.75 mm thick, 7 mm wide). After running, the gel was stained with Coomassie blue G-250 to evaluate the resolution.
Figure S3. False assignment of GG-modification to the C-terminal Lys residue during database search. (A) The MS/MS spectra with the b and y series of ions indicated. The labeled ions in parentheses were derived from the peptide with internal GG-modification. (B) The mass of b and y series of ions of both peptides with C-terminal or internal GG-modification, respectively. Underlined are matched ions. Although the internal modified peptide had more continuously matched ions than the C-terminal modified peptide, it was assigned with a slightly lower Xcorr score.
Figure S4. Internal GG-modified Lys residue cannot serve as tryptic site. Similar amount (~1picomole) of ubiquitin GG-peptide at K27 (TITLEVESSDTIDNVK*SK, the asterisk indicates the modification) was mixed with neurotensin (NT, pyroELYENKPRRPYIL with one internal tryptic site as a positive control) and bradykinin (RPPGFSPFR as a negative control). The mixture was treated with trypsin at 10 ng/μl. a series of time points were taken and analyzed by LC-MS/MS using LTQ-Orbitrap: 0 min without trypsin addition (A), 5 min on ice (B), and 10 min (C), 1 hr and 18 hr at 37°C. (D) The extracted ion currents of the three peptides, the digested product of neurotensin, and the autocleavage peptide of trypsin. Clearly, the trypsin worked highly effectively to digest neurotensin but not the GG peptide.
Figure S5. Independent assessment of simplified virtual Western blot strategy. (A) 1D gel LC-MS/MS was used to assess a second batch of candidate ubiquitin conjugates in which only eight gel bands were analyzed. (B) To assess the presence of protein ubiquitination, the experimental and predicted MW for all 353 proteins were compared after Gaussian curve fitting of spectral count intensity. The distribution of experimental and expected MW difference (ΔMW) was shown in the figure. Moreover, the accepted proteins were further filtered by adjusting ΔMW values using the TCL as the reference, leading to the final list of 108 Ub-conjugates.
Acknowledgments
We thank Dr. Daniel Finley for providing the yeast strains. We also thank the members in the lab for critical reading of the manuscript. This work was partially supported by the NIH grants (DK069580, CA126222 and the Emory Alzheimer’s Disease Center AG025688).
Abbreviations
- Ub
ubiquitin
- MS
mass spectrometry
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- TCL
total cell lysate
- and UbC
Ub-conjugates
References
- 1.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. Trends Biochem Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Hicke L, Dunn R. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 4.Chen ZJ. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickart CM. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 6.Weissman AM. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 7.Hochstrasser M. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 8.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 9.Pickart CM, Fushman D. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkinson KD. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 12.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Taylor JP, Hardy J, Fischbeck KH. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg AL. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 15.Voorhees PM, Orlowski RZ. Annu Rev Pharmacol Toxicol. 2006;46:189–213. doi: 10.1146/annurev.pharmtox.46.120604.141300. [DOI] [PubMed] [Google Scholar]
- 16.Yates JR, 3rd, Gilchrist A, Howell KE, Bergeron JJ. Nat Rev Mol Cell Biol. 2005;6:702–714. doi: 10.1038/nrm1711. [DOI] [PubMed] [Google Scholar]
- 17.Domon B, Aebersold R. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 18.Mann M. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 19.Kirkpatrick DS, Denison C, Gygi SP. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser P, Huang L. Genome Biol. 2005;6:233. doi: 10.1186/gb-2005-6-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu P, Peng J. Biochim Biophys Acta. 2006;1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Mol Cell Proteomics. 2007;6:1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Proteomics. 2005;5:4145–4151. doi: 10.1002/pmic.200401280. [DOI] [PubMed] [Google Scholar]
- 24.Vasilescu J, Smith JC, Ethier M, Figeys D. J Proteome Res. 2005;4:2192–2200. doi: 10.1021/pr050265i. [DOI] [PubMed] [Google Scholar]
- 25.Maor R, Jones A, Nuhse TS, Studholme DJ, Peck SC, Shirasu K. Mol Cell Proteomics. 2007;6:601–610. doi: 10.1074/mcp.M600408-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Tomlinson E, Palaniyappan N, Tooth D, Layfield R. Proteomics. 2007;7:1016–1022. doi: 10.1002/pmic.200601008. [DOI] [PubMed] [Google Scholar]
- 27.Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Mol Cell Proteomics. 2005;4:741–751. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. Mol Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Kirkpatrick DS, Weldon SF, Tsaprailis G, Liebler DC, Gandolfi AJ. Proteomics. 2005;5:2104–2111. doi: 10.1002/pmic.200401089. [DOI] [PubMed] [Google Scholar]
- 30.Jeon HB, Choi ES, Yoon JH, Hwang JH, Chang JW, Lee EK, Choi HW, Park ZY, Yoo YJ. Biochem Biophys Res Commun. 2007;357:731–736. doi: 10.1016/j.bbrc.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Peng J, Cheng D. Methods Enzymol. 2005;399:367–381. doi: 10.1016/S0076-6879(05)99025-3. [DOI] [PubMed] [Google Scholar]
- 32.Peng J, Gygi SP. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 33.Marotti LA, Jr, Newitt R, Wang Y, Aebersold R, Dohlman HG. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 35.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 36.Eng J, McCormack AL, Yates JR., 3rd J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 37.Kenney JF, Keeping ES. Princeton, NJ: Van Nostrand; 1962. p. 223. [Google Scholar]
- 38.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 39.Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, Paiement J, Kearney RE, Nilsson T, Bergeron JJ. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 40.Neville DM., Jr J Biol Chem. 1971;246:6328–6334. [PubMed] [Google Scholar]
- 41.Ahmad QR, Nguyen DH, Wingerd MA, Church GM, Steffen MA. Proteome Sci. 2005;3:6. doi: 10.1186/1477-5956-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park GW, Kwon KH, Kim JY, Lee JH, Yun SH, Kim SI, Park YM, Cho SY, Paik YK, Yoo JS. Proteomics. 2006;6:1121–1132. doi: 10.1002/pmic.200500318. [DOI] [PubMed] [Google Scholar]
- 43.Wang M, Cheng D, Peng J, Pickart CM. EMBO J. 2006;25:1710–1719. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu P, Cheng D, Duong DM, Rush J, Roelofs J, Finley D, Peng J. Israel J Chem. 2006;46:171–182. [Google Scholar]
- 45.Denis NJ, Vasilescu J, Lambert JP, Smith JC, Figeys D. Proteomics. 2007;7:868–874. doi: 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The linear correlation between log10(MW) of protein markers with their relative mobility in the total cell lysate (TCL). Similar data was obtained in the Ub-conjugates (UbC, data not shown).
Figure S2. Examination of loading capacity for 1D SDS gel. Yeast protein (20–160 μg) was titrated on a thin SDS gel (0.75 mm thick, 7 mm wide). After running, the gel was stained with Coomassie blue G-250 to evaluate the resolution.
Figure S3. False assignment of GG-modification to the C-terminal Lys residue during database search. (A) The MS/MS spectra with the b and y series of ions indicated. The labeled ions in parentheses were derived from the peptide with internal GG-modification. (B) The mass of b and y series of ions of both peptides with C-terminal or internal GG-modification, respectively. Underlined are matched ions. Although the internal modified peptide had more continuously matched ions than the C-terminal modified peptide, it was assigned with a slightly lower Xcorr score.
Figure S4. Internal GG-modified Lys residue cannot serve as tryptic site. Similar amount (~1picomole) of ubiquitin GG-peptide at K27 (TITLEVESSDTIDNVK*SK, the asterisk indicates the modification) was mixed with neurotensin (NT, pyroELYENKPRRPYIL with one internal tryptic site as a positive control) and bradykinin (RPPGFSPFR as a negative control). The mixture was treated with trypsin at 10 ng/μl. a series of time points were taken and analyzed by LC-MS/MS using LTQ-Orbitrap: 0 min without trypsin addition (A), 5 min on ice (B), and 10 min (C), 1 hr and 18 hr at 37°C. (D) The extracted ion currents of the three peptides, the digested product of neurotensin, and the autocleavage peptide of trypsin. Clearly, the trypsin worked highly effectively to digest neurotensin but not the GG peptide.
Figure S5. Independent assessment of simplified virtual Western blot strategy. (A) 1D gel LC-MS/MS was used to assess a second batch of candidate ubiquitin conjugates in which only eight gel bands were analyzed. (B) To assess the presence of protein ubiquitination, the experimental and predicted MW for all 353 proteins were compared after Gaussian curve fitting of spectral count intensity. The distribution of experimental and expected MW difference (ΔMW) was shown in the figure. Moreover, the accepted proteins were further filtered by adjusting ΔMW values using the TCL as the reference, leading to the final list of 108 Ub-conjugates.