Abstract
Genome-wide erasure of CpG methylation occurs along the paternal pronucleus in fertilized oocytes. This process involves an active, replication-independent enzymatic step, which has remained enigmatic. MBD3L1 and MBD3L2 are two mammalian homologues of the methyl-CpG-binding protein genes MBD2 and MBD3 that arose from recent gene duplication events. Expression of Mbd3l1 occurs specifically in haploid male germ cells. Mbd3l2 expression is restricted to metaphase II oocytes and zygotes making both proteins candidates for the zygotic demethylation process. Neither of these genes was able to promote reactivation of a methylation-silenced reporter gene. We created Mbd3l1 and Mbd3l2 knockout mice, which were viable and fertile. We show that demethylation of the paternal pronucleus in Mbd3l1−/− and Mbd3l2−/− mice is identical to that in wildtype controls. These data suggest that Mbd3l1 and Mbd3l2 are not involved in genome-wide demethylation of paternal genomes in mouse zygotes and are dispensable for normal development.
Keywords: Mbd3l1, Mbd3l2, demethylation, zygote
INTRODUCTION
DNA methylation is an important means of epigenetic gene regulation, and the de novo and maintenance methylation reactions carried out by DNA methyltransferases have been studied extensively (Chen and Li, 2006). Despite the stable and heritable features of DNA methylation patterns, genome-wide DNA demethylation occurs both in developing germ cells and in fertilized oocytes. In the primordial germ cells (PGCs), genome-wide demethylation occurs during a very short period of time (between E11.5 and E12.5) to allow reprogramming of sex-specific methylation patterns in both male and female germ cells (Reik et al., 2001; Hajkova et al., 2008; Latham et al., 2008). During preimplantation development, an asymmetric DNA demethylation pattern of parental genomes can be observed within the same oocyte cytoplasm beginning as early as 6 hours after fertilization when the paternal genome undergoes active DNA demethylation (Mayer et al., 2000; Oswald et al., 2000; Santos et al., 2002). The maternal genome is resistant to active demethylation, and undergoes passive demethylation that depends on the absence of maintenance methyltransferase DNMT1 during DNA replication in early development (Reik, 2007). This replication-independent DNA demethylation of the paternal genome would imply the existence of a mammalian DNA demethylase emzyme.
Despite intensive study, the DNA demethylase protein and its biochemical mechanism have not yet been identified convincingly. Several candidate pathways have been established including methyl-CpG binding domain (MBD) proteins, such as MBD2 and MBD4 that may either actively remove the methyl group from 5-methylcytosine (5meC) or can remove the entire methylated base or nucleotide. The functional role of MBD2 in transcriptional repression has been well characterized in mammalian somatic cells. However, the role of this protein in active DNA demethylation pathways is controversial (Bhattacharya et al., 1999; Hendrich et al., 1999; Ng et al., 1999; Wade et al., 1999). Immunostaining studies using an anti-5meC antibody with zygotes derived from Mbd2 knockout mice showed asymmetric methylation patterns that are consistent with control wildtype mice, leading to the conclusion that MBD2 does not have a DNA demethylating activity in zygotes (Santos et al., 2002). Although MBD3, a component of the NuRD complex, was reported to be unable to bind directly to methylated DNA, a growing body of evidence supports a crucial role for Mbd3 during embryonic development (Hendrich et al., 2001; Kantor et al., 2003; Kaji et al., 2007; Ruddock-D’Cruz et al., 2008). However, a function of MBD3 in DNA demethylation has not been reported. Recently, a role for the DNA damage response protein GADD45A in DNA demethylation has been reported for Xenopus GADD45A (Barreto et al., 2007) but could not be confirmed for human GADD45A (Jin et al., 2008). It is possible that the mammalian form has lost this activity and this gene is not specifically expressed in oocytes or zygotes.
Another mechanism of demethylation involving deamination of 5meC by cytidine deaminases, AID or APOBEC1, followed by DNA repair of the resulting T:G mismatch by base excision repair has also been suggested. To date no conclusive evidence has been published that these are the enzymes responsible for demethylation of the paternal genome in the zygote, although AID and APOBEC1 are expressed at this stage of development (Morgan et al., 2004). In plants, a demethylase pathway involving a DNA glycosylase activity has been identified (Agius et al., 2006; Gehring et al., 2006), but these proteins, ROS1 and DEMETER, do not appear to have mammalian homologues.
Suggestions for the identity of the DNA demethylase might come from the list of factors linked to epigenetic reprogramming during embryonic development. It seems reasonable that viable candidate genes could be those expressed in oocytes, zygotes, and 1- or 2- cell stage embryos where active DNA demethylation has been well documented (Oliveri et al., 2007; Jin et al., 2008). The cytoplasm of murine metaphase II (MII) oocytes is capable of remodeling the structure and epigenetic programming of the incoming paternal genome after fertilization, and of the nucleus of somatic cells following somatic cell nuclear transfer (SCNT) (Wilmut et al., 1997). This activity, however, is not present in pre-meiotic germinal vesicle (GV) oocytes. (Gao et al., 2002; Tesarik et al., 2003). Beaujean and co-workers have demonstrated that sheep sperm DNA can be demethylated in mouse oocytes by using interspecies intracytoplasmic sperm injection (Beaujean et al., 2004b), although in the zygotes of sheep no global demethylation of the paternal genome occurs during the first cycle of cleavage (Beaujean et al., 2004a). Interestingly, mouse sperm can also undergo demethylation to a limited extent when injected into sheep oocytes, suggesting a limited sperm-derived demethylation activity (Beaujean et al., 2004a; Beaujean et al., 2004b; Young and Beaujean, 2004). These findings may indicate that active demethylation of the paternal genome may involve both the sperm and the oocyte (Beaujean et al., 2004b).
MBD3L1 and MBD3L2 are two mammalian homologues of the methyl-CpG domain proteins MBD2 and MBD3 that both lack the MBD, but can homo-and heterodimerize with MBD2 and MBD3 (Jiang et al., 2002; Jiang et al., 2004; Jin et al., 2005). Recently, we conducted an in silico study for the expression profile of Mbd3l2 in the mouse. We observed that the expression of mouse Mbd3l2 is elevated in oocytes and zygotes, but Mbd3l2 is not significantly expressed in the later stages of embryonic development and in somatic tissues. This pattern of expression is consistent with a role for Mbd3l2 in early embryonic development stages, similar to the NuRD complex or its components such as p66 and MBD3 (Hendrich et al., 2001; Kantor et al., 2003; Marino and Nusse, 2007; Oliveri et al., 2007; Ruddock-D’Cruz et al., 2008). The MBD3L1 protein is expressed predominantly in the round spermatids of the testis (Jiang et al., 2002). Thus, the unique expression patterns and the homology of MBD3L1 and MBD3L2 to methyl-CpG binding proteins prompted us to test the role of these genes in early development and in zygotic DNA demethylation.
RESULTS
Gene expression profiles of Mbd3l1 and Mbd3l2 in mouse developmental stages
Mbd3l1 is expressed predominantly in testis, specifically in round spermatids (Jiang et al., 2002). We have reported previously that MBD3L2 is expressed ubiquitously at low levels in human tissues using expression analysis by Northern blotting (Jin et al., 2005). However, increasing evidence of possible roles of the MBD proteins and the components of the NuRD complex in early development (Hendrich et al., 2001; Kantor et al., 2003; Marino and Nusse, 2007; Oliveri et al., 2007; Ruddock-D’Cruz et al., 2008) prompted us to investigate the roles of Mbd3l1 and Mbd3l2 in mouse development. We obtained the gene expression profiles of Mbd3l1 and Mbd3l2 in embryonic stages via the web interfaces of UniGene’s EST database (Table 1) and the Novartis Research Foundation’s SymAtlas (data not shown). Interestingly, we observed that Mbd3l2 was expressed predominantly in mouse oocytes, unfertilized ova, zygotes and early cleavage stages but Mbd3l2 mRNA was virtually absent at later embryonic stages and in adult tissues. We also noted limited expression of Mbd3l2 in 1-cell to 2-cell pre-implantation embryos according to the GEO database. Furthermore, Mbd3l2 transcripts were abundant in mature MII oocytes compared to immature germinal vesicle stage oocytes (GEO database), suggesting a role of Mbd3l2 during early development rather than during oogenesis. In contrast, Mbd3l1 mRNA was expressed specifically in testis tissue as expected (Table 1). Thus, the expression profiles of Mbd3l1 and Mbd3l2 suggested a possible role for these genes in gametogenesis, and early embryonic development. The specific expression of Mbd3l2 in oocytes and zygotes would be consistent with a role of Mbd3l2 in zygotic DNA demethylation. Expression of Mbd3l1 is specific to haploid male germ cells and, at least theoretically, this protein could be incorporated into sperm chromatin, and after fertilization could participate in demethylation of the paternal genome.
Table 1.
The expression profile of Mbd3l1 and Mbd3l2 in mouse developmental stagesa)
| Developmental Stages | Mbd3l1 (Mm.160088b)) | Mbd3l2 (Mm.17563) | ||
|---|---|---|---|---|
| TPMc) | Gene EST/Total EST | TPM | Gene EST/Total EST | |
| oocyte | 0 | 0/19488 | 102 | 2/19488 |
| unfertilized ovum | 0 | 0/20351 | 589 | 12/20351 |
| zygote | 0 | 0/28380 | 352 | 10/28380 |
| cleavage | 0 | 0/27724 | 468 | 13/27724 |
| morula | 0 | 0/38117 | 0 | 0/38117 |
| blastocyst | 0 | 0/69929 | 0 | 0/69929 |
| egg cylinder | 0 | 0/12254 | 0 | 0/12254 |
| gastrula | 0 | 0/28672 | 0 | 0/28672 |
| organogenesis | 0 | 0/128575 | 0 | 0/128575 |
| fetus | 0 | 0/670708 | 0 | 0/670708 |
| neonate | 0 | 0/107167 | 0 | 0/107167 |
| juvenile | 0 | 0/293392 | 0 | 0/293392 |
| adult | 72 | 74/1025953 | 0 | 0/1025953 |
| testis | 677 | 80/118155 | 0 | 0/118155 |
Expression profiles were determined by analysis of EST counts from Mus musculus UniGene database as of May 2008.
UniGene ID,
TPM indicates transripts per million.
Testing a role for MBD3L1 and MBD3L2 in activation of methylation-silenced reporter plasmids
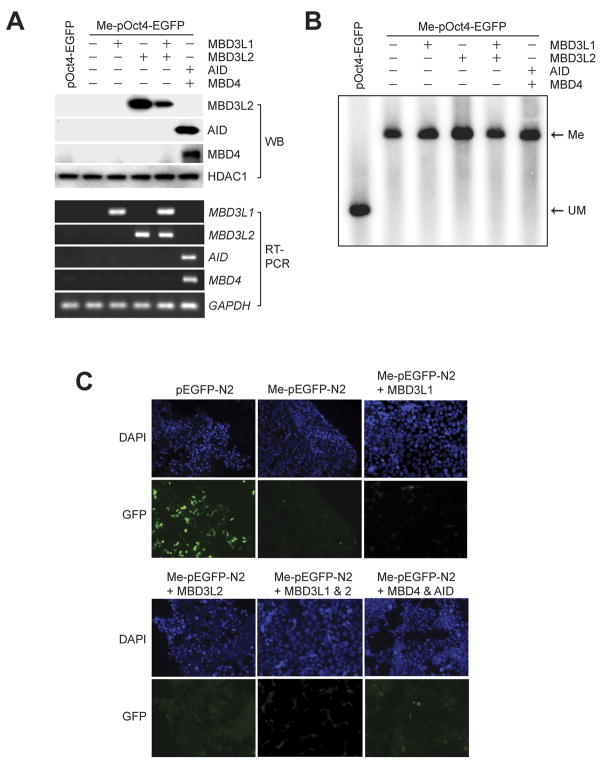
The unique developmental stage-specific expression of Mbd3l1 and Mbd3l2 in the mouse, and its homology to MBD proteins suggested that the two proteins could be involved in the process of global DNA demethylation. To elucidate this hypothesis, we first tested if overexpression of the two proteins has an effect on in vitro methylated reporter plasmids in somatic cells as a simple assay for detecting DNA demethylation activity. First, we confirmed that the transfected genes were expressed in HEK293 cells by Western blot (Figure 1A). Since no suitable antibody for MBD3L1 is available, we confirmed its expression by RT-PCR. Using a Southern blot assay, we tested the potential demethylation activity of MBD3L1 and MBD3L2 (Figure 1B). After cotransfection into HEK293 cells, the methylated and unmethylated pOct4-EGFP plasmids were recovered and digested with HincII and the methylation-sensitive restriction enzyme HpaII. No HpaII cleavage was observed indicating that no demethylation had occurred in presence of MBD3L1 and/or MBD3L2. The EGFP reporter plasmid (pEGFP-N2), which is controlled by the CMV promoter, was methylated in vitro at all CpG sites with SssI DNA methylase, and transiently transfected or co-transfected with MBD3L1 and/or MBD3L2 mammalian expression vectors into HEK293 cells. After 48 hours, expression of EGFP was determined by fluorescence microscopy (Figure 1C). The single transfected EGFP was expressed from the unmethylated pEGFP-N2 plasmid, but the methylated pEGFP-N2 was not expressed. The level of EGFP expression from methylated pEGFP-N2 in HEK293 cells expressing MBD3L1 or MBD3L2 was similar to that of the control (Figure 1C). These results suggest that MBD3L1 and MBD3L2 do not have an effect on the methylation-dependent silencing of the EGFP reporter plasmid and do not promote DNA demethylation. In order to test a potential role of the cytidine deaminase AID and the base excision repair protein MBD4 in 5-methylcytosine removal, we cotransfected these two genes in presence of methylated reporters. We did not obtain evidence for demethylation by AID and MBD4 in this system (Figure 1B, C).
Figure 1. Analysis of DNA demethylation with in vitro methylated plasmids.
A. Expression of all tested genes in transfected HEK293 cells was monitored by Western blotting (WB, upper panel) and RT-PCR (lower panel). B. A Southern blot assay was conducted to monitor DNA demethylation. The methylated and unmethylated pOct4-EGFP plasmids were recovered from HEK293 cells expressing the indicated genes and digested with HincII and the methylation-sensitive restriction enzyme HpaII. The digested fragments were subjected to Southern blot analysis with a 32P-labeled EGFP probe. HpaII-digested fragments were only detectable from the unmethylated pOct4-EGFP, indicating that demethylation did not occur in HEK293 cells that overexpress the tested genes. Me, methylated fragments; UM, unmethylated (HpaII-digested) fragments. C. HEK293 cells were transiently transfected with an unmethylated (pEGFP-N2) or CpG-methylated (Me-pEGFP-N2) plasmid for the controls. Cells were co-transfected with Me-pEGFP-N2 and pcDNA3.1-MBD3L1, pcDNA3.1-MBD3L2 or both, or with MBD4 and AID expression plasmids. Fluorescence microscopy observed at 48 hours after transfection indicates that the transfections have no effect on reactivation of EGFP. DAPI was used to visualize the nuclei.
Generation of Mbd3l1 and Mbd3l2 knockout mice
Although we can observe and evaluate the DNA methylation status of parental genomes in zygotes after visualization by immunostaining with anti-5-methylcytosine antibody, the process of DNA demethylation is not well understood. That is, in vitro experiments using over-expression of exogenous genes in somatic cells may not be a suitable means to identify the zygotic enzymatic DNA demethylation activity of candidate genes. To address these possible limitations and examine the function of Mbd3l1 and Mbd3l2 in mouse early developmental stages, we studied Mbd3l1 and Mbd3l2 knockout mice.
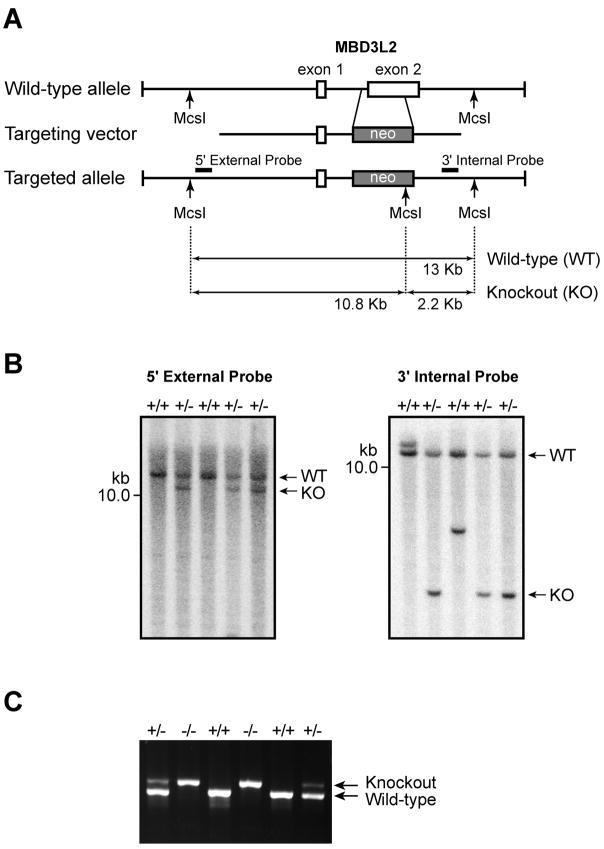
The generation of Mbd3l1 knockout mice has been described previously (Jiang et al., 2004). The Mbd3l1-deficient mice are viable and fertile, but zygotic demethylation in these mice has not yet been examined. The Mbd3l2 targeting vector was designed to replace 0.95-kb of exon 2, which represents 70% of the Mbd3l2 coding sequence, with the 1.8-kb loxP-neomycin resistance (PGKneo) cassette (Figure 2A). Correctly targeted ES cell clones were screened by PCR, and genotypes were confirmed by Southern blotting using external and internal probes after digestion with MscI restriction enzyme (Figure 2B). Blastocyst embryos were injected with the recombinant ES cell clones, and then transferred to the uteri of recipient females. Heterozgyous Mbd3l2 knockout mice were produced by breeding the male chimeras with 129SV and C57BL/6J wild-type female mice. The heterozygous mice were inter-bred, and homozygous mice were obtained at the expected Mendelian 1:2:1 ratio. In Figure 2C, the genotype of Mbd3l2 homozygous mutant mice was confirmed by PCR-based genotyping using the genomic DNA of Mbd3l2+/+ and Mbd3l2+/− littermates as a control. Homozygous Mbd3l2 knockout mice were viable and fertile, and have shown no adverse health effects after ten months of observation.
Figure 2. Generation of Mbd3l2 knockout mice.
A. The Mbd3l2 targeting vector was designed to replace 0.95-kb of exon 2, which presents 70% of the Mbd3l2 coding sequence, with the loxP-neomycin resistance (PGKneo) cassette. The resultant construct replaces exon 2 of Mbd3l2 with the 1.8-kb PGKneo cassette. B. Correctly targeted ES cell clones were confirmed by Southern blotting using digestion with MscI restriction enzyme and external and internal probes. The example shows MscI-digested DNA hybridized with the 5′-external and 3′-internal probes, and three clones (+/−) show the correct size band for a homologous recombination event. C. PCR based genotyping of Mbd3l2 knockout mice using the genomic DNA derived from littermate mice born from Mbd3l2+/− mice. The gel shows data for two wildtype (+/+), two heterozygous (+/−), and two homozygous knockout (−/−) mice.
Immunofluorescence staining with anti-5meC antibody in Mbd3l1−/− and Mbd3l2−/− zygotes
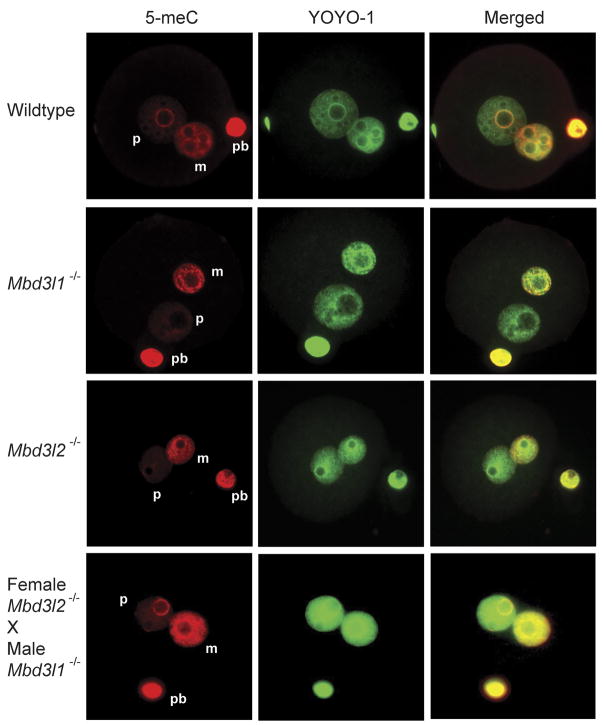
To test if the global demethylation of the paternal genome is affected by the deletion of Mbd3l1 or Mbd3l2 in one cell-stage zygotes, we immunostained zygotes with an anti-5-meC antibody. The zygotes were incubated with anti-5-meC antibody, and then with an Alexa 568-conjugated secondary antibody (red). The genomic DNA of the parental pronuclei was counterstained with YOYO-1 (green). In Figure 3, positive signals with 5-meC antibody staining were detectable only over the maternal (smaller) pronuclei in zygotes derived from Mbd3l1−/− or Mbd3l2−/− null mice. These asymmetric DNA methylation patterns of parental genomes in the zygotes are consistent with the methylation patterns seen in wildtype mice, suggesting that Mbd3l1 and Mbd3l2 do not affect global DNA demethylation in mouse early developmental stages. Since Mbd3l2 may be a maternal effect gene, we used F3 generation zygotes derived by breeding Mbd3l2−/− males with Mbd3l2−/− females.
Figure 3. Asymmetric methylation patterns of zygotes derived from wildtype, Mbd3l1 and Mbd3l2 null mice.
The zygotes were immunostained with 5-meC antibody and Alexa 568-conjugated secondary antibody (red), and counterstained with YOYO-1 (green). The positive signals with 5-meC antibody staining were detectable only over the maternal (smaller) pronuclei in zygotes derived from wildtype, Mbd3l1−/− and Mbd3l2−/− mice. The bottom panels show zygote staining from a cross of Mbd3l1−/− male mice and Mbd3l2−/− female mice. Counterstain with YOYO-1 shows the presence of the paternal and maternal pronuclei. P, paternal pronulceus; m, maternal pronucleus; pb, polar body.
We next examined the potential for redundancy of Mbd3l1 and Mbd3l2 so that a single knockout would not lead to a demethylation phenotype. In the mouse genome, the two genes are only 40 kb apart and it is difficult to create a conventional double knockout mouse. Because of the specific expression patterns of the two genes, we reasoned that it was possible to create a double knockout phenotype by mating Mbd3l1−/− male mice with Mbd3l2−/− female mice, because the sperm that would potentially carry Mbd3l1 into the zygote will be null for Mbd3l1 and the oocyte that would express Mbd3l2 will be null for Mbd3l2. However, paternal genome DNA demethylation was normal in this cross (Figure 3, bottom panels).
DISCUSSION
Despite extensive studies, the identity and composition of the mammalian DNA demethylase machinery remains unknown. Interestingly, in silico studies showed that mouse Mbd3l2, a gene coding for a protein homologous to the methyl-CpG binding domain containing proteins Mbd2 and Mbd3, is predominantly expressed in oocytes and in 1 cell to 2 cell stages of embryos. Furthermore, Mbd3l2 transcripts were highly accumulated in mature MII oocytes (GEO database; data not shown) that have the specialized ability to epigenetically reprogram the sperm genome (Gao et al., 2002; Tesarik et al., 2003). Although human MBD3L2 acts as a transcriptional repressor, is a component of the NuRD complex, and has the potential to recruit the MeCP1 complex away from methylated DNA in human cell lines (Jin et al., 2005), the function of MBD3L2 in vivo is still unknown. Thus, the expression profile of mouse Mbd3l2 strongly suggests a role of this protein in mouse early developmental stages. The in vitro preliminary data and its homology to methyl-CpG binding (MBD) proteins MBD2 and MBD3 prompted us to test the role of MBD3L2 in genome-wide DNA demethylation. Overexpressed human MBD3L2 did not affect the methylation status of in vitro methylated GFP plasmids in human HEK293 cells (Figure 1). Also, immunostaining of zygotes using anti-5meC antibody showed asymmetrical DNA methylation of the paternal pronucleus equivalent to wildtype controls (Figure 3). The antibody staining technique has of course its detection limits. Thus, we cannot exclude the possibility that specific sequences of the mouse genome may be demethylated by Mbd3l2. However, the results suggest that Mbd3l2 is not involved in global DNA demethylation in mouse early developmental stages.
The testis-specific gene Mbd3l1 has been initially characterized in our lab, but its in vivo function is still unclear (Jiang et al., 2004). The possible involvement of Mbd3l1 in a sperm-derived demethylation pathway in zygotes was predicted by the unique expression pattern and its homology to methyl-CpG binding (MBD) proteins MBD2 and MBD3. Over-expression of Mbd3l1 had no effect on de-repression of CMV-EGFP reporter plasmids silenced by in vitro methylation, and the methylation-specific immunostaining assays with zygotes derived from Mbd3l1 knockout mice appeared normal (Figures 1 and 3).
It was reported that base-excision repair has an important role in the erasure of DNA methylation from imprinted plant genes (Agius et al., 2006; Gehring et al., 2006). Although these proteins, ROS1 and DEMETER, do no have recognizable mammalian homologues, it is possible that DNA glycosylase activity might be involved in mammalian demethylase pathways. It was proposed that the mammalian cytidine deaminases AID and APOBEC1 in vitro have 5meC deaminase activity (Morgan et al., 2004), and that the resulting thymine may be removed by base excision repair pathways. Interestingly, AID and APOBEC1 are expressed in ovaries and oocytes (Morgan et al., 2004), and the genes are located in a cluster of genes with Stella, Gdf3 and Nanog, genes which have a critical role in mouse embryonic development. Therefore, we tested if an efficient coupling of the deamination by AID and the base excision by MBD4 might have a role in DNA demethylation. However, we failed to observe DNA demethylation by the deamination-coupled DNA glycosylase enzymatic activity using transfection assays (Figure 1). Above all, oocytes lacking Mbd4 still have DNA demethylation activity (Santos and Dean, 2004). Thymidine DNA glycosylase (TDG) and MBD4 have similar T/G mismatch cleavage activity, and, could in theory, participate in a deamination-coupled demethylation process. Although the TDG protein has recently been implicated in cyclical DNA demethylation occurring at promoters in somatic cells (Kangaspeska et al., 2008; Metivier et al., 2008), its role in paternal genome demethylation in zygotes remains to be tested.
The biological function of Mbd3l1 and Mbd3l2 remains unknown. MBD3L-like genes have only been found in placental mammals. The two genes have arisen by a relatively recent gene duplication event. They are embedded in a gene-rich, rearrangement-prone genomic region on chromosome 19 of the human and chromosome 9 of the mouse genomes. A comparison between chicken and mammalian genomes has shown that synteny between the loci MGC33407 and MUC16 is disrupted by lineage-specific gene expansion of OR2Z1, ZNF558, and MBD3L1 in humans and is associated with a duplication site (Gordon et al., 2007). This expansion is conserved in all other mammalian species examined. In humans, the species-specific expansion at this site includes an expansion producing five presumably functional MBD3L2-like loci that are 97% identical at the DNA level and over 98% identical at the protein level suggesting very recent primate-specific duplications in this region (Gordon et al., 2007). However, in mouse there are single functional Mbd3l1 and Mbd3l2 genes on chromosome 9 and one Mbd3l2 pseudogene is found on the X chromosome. Also of note, the homology between mouse Mbd3l1 and human MBD3L1 proteins is quite low (62% identical, 73% similar) and the homology between mouse Mbd3l2 and human MBD3L2 proteins is even lower (37% identical, 54% similar). This suggests that the two genes are very recent additions to the mammalian gene repertoire and are still continuing to evolve. This may, at least in part, explain why Mbd3l1 and Mbd3l2 are nonessential genes in the mouse.
EXPERIMENTAL PROCEDURES
Cell culture and transfection
HEK293 and MCF7 cells were maintained as a monolayer in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 standard incubator. DNA transfections were carried out in Opti-MEM-I medium using Lipofectamine 2000 (Invitrogen; Carlsbad, CA) according to the manufacturer’s instructions.
Plasmids and in vitro methylation
The ORF human cDNAs of MBD3L1 and MBD3L2 were subcloned in frame into the BamHI-NotI and BamHI-XhoI sites of pcDNA3.1 (+) (Invitrogen; Carlsbad, CA), respectively. The FLAG-tagged MBD4 construct was made by insertion of the human cDNA of MBD4 into the EcoRI-XhoI sites of pcDNA3.0-FLAG, which was modified from pcDNA3.0 (Invitrogen; Carlsbad, CA). The vector encoding human activation induced cytidine deaminase (AID) was a kind gift from Tim O’Connor (City of Hope, Duarte, CA). All recombinant clones were verified by DNA sequencing. For the pOct4-EGFP construct, the cytomegalovirus (CMV) promoter of the pEGFP-N2 vector (Clontech; Palo Alto, CA) was replaced with the 2.4-kb mouse Oct4 gene promoter. For this purpose, the AseI and EcoRI fragment of the pEGFP-N2 plasmid encoding the CMV promoter was excised by restriction nuclease cleavage. Next, the 2.4-kb Oct4 promoter region upstream of the translational start site was amplified by PCR followed by AseI and EcoRI digestion of the PCR product and insertion into the AseI and EcoRI sites of the pEGFP-N2 plasmid. The constructed pOct4-EGFP plasmid was verified by DNA sequencing. For in vitro methylation of plasmids, 10 μg of pEGFP-N2 or pOct4-EGFP were methylated in vitro with 20 units of SssI (pEGFP-N2), or with HpaII and HhaI DNA methylases (pOct4-EGFP) (New England Biolabs), respectively. These methylated plasmids were then phenol/chloroform extracted, ethanol-precipitated and resuspended in TE buffer. The extent of methylation was confirmed by methylation-sensitive restriction enzyme, HpaII, digestion.
Western blot and RT-PCR
For Western blot analysis, HEK293 nuclear extracts were prepared by using NE-PER Reagents (Pierce; Rockford, IL) and were separated by SDS/PAGE and blotted onto PVDF membranes. The membranes were blocked with 5% nonfat milk at 4°C overnight. After washing, the overexpressed proteins were detected using anti-MBD3L2 antiserum (1:7000), anti-AID (Cell Signaling Technology Inc., 30F12; 1:1000), anti-MBD4 (Santa Cruz, E-19; 1:2000) or anti-HDAC1 (Santa Cruz, H-51; 1:3000) antibodies, followed by peroxidase-conjugated anti-rabbit, anti-mouse or anti-goat IgG (Jackson Laboratory; Bar Harbor, ME) secondary antibodies. The signal was visualized by using ECL-Plus (Amersham Pharmacia Biotech). For RT-PCR, total RNAs were isolated from HEK293 cells using the RNeasy Mini Kit (Qiagen; Valencia, CA). cDNAs were produced with SuperScriptIII reverse transcriptase (Invitrogen; Carlsbad, CA) and amplified by 25 cycles of PCR with Platinum Taq DNA polymerase High Fidelity (Invitrogen; Carlsbad, CA) according to the manufacturer’s instructions. The following primers were used: for MBD3L1, 5′-AGGTCTGCTGGCAGAGGAGACT-3′ (forward) and 5′-GGTCTCTCACTTTCTCTGCCTCA-3′ (reverse); for MBD3L2, 5′-CTGTTCTGGGGAAGCTCAAAAGGAAC-3′ (forward) and 5′-TGACCTGGTTGTCAGGATGAGACCTG-3′ (reverse); for MBD4, 5′-GGATGTAGGAAGAGCTGTTCA-3′ (forward) and 5′-CACTGAGGGTCTCACCACAT-3′ (reverse); for AID, 5′-AAATGTCCGCTGGGCTAAGG-3′ (forward) and 5′-GGAGGAAGAGCAATTCCACGT-3′ (reverse); for GAPDH, 5′-CCTGTTCGACAGTCAGCCG-3′ (forward) and 5′-CGACCAAATCCGTTGACTCC-3′ (reverse).
DNA methylation analysis using Southern blot
Transfected plasmids were recovered by using QIAquick PCR-purification kits (Qiagen). The recovered plasmids were digested with HincII and HpaII and analyzed by Southern blotting using an EGFP gene probe.
GFP expression assay
HEK293 and MCF7 cells were grown on poly-L-lysine-coated coverslips in a six-well tissue culture dish and then co-transfected with a total of 2 μg of pEGFP-N2 or methylated pEGFP-N2 and pCDNA3.1-MBD3L1, pCDNA3.1-MBD3L2, MBD4 and/or AID. The cells were incubated for 48 hours and were then washed twice with cold PBS. The cells were fixed in 3.7% (v/v) formaldehyde in PBS for 10 min at room temperature and were then washed with PBS. The cells were stained with 0.25 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma), washed three times with PBS and mounted with Fluoromount G (Southern Biotechnology Associates; Birmingham, AL). Images were captured using a fluorescence microscope (Olympus IX81) and processed by ImageProPlus high-end image analysis software (Media Cybernetics, Silver Spring, USA) and Adobe Photoshop software.
Generation of Mbd3l2 knockout mice
The Mbd3l2 targeting vector was designed to replace 0.95-kb of exon 2, which presents 70% of the Mbd3l2 coding sequence, with the loxP-neomycin resistance (PGKneo) cassette. The targeting vector was made by high fidelity PCR with the proof-reading capable PfuUltra II Fusion HS DNA polymerase (Stratagene) and 129S1 mouse ES cell genomic DNA as the template. A 5-kb fragment encompassing the promoter region and exon 1, and a 1.65-kb fragment containing sequence downstream of exon 2 were used as long and short arms, respectively. The resultant construct replaced exon 2 of Mbd3l2 with the 1.8-kb PGKneo cassette. The constructed targeting vector was verified by DNA sequencing. Correctly targeted ES cell clones were screened by PCR, and confirmed by Southern blotting using external and internal probes after digestion with MscI restriction enzyme. The City of Hope Transgenic Mouse Core Facility was used to generate the targeted ES cell lines and for the production of mouse chimeras. Germline transmission was achieved by breeding the male chimeras with C57BL/6J females, and then the heterozygous Mbd3l2 knockout mice were obtained and confirmed by using Southern blot assays with external and internal probes.
Genotyping
A small piece of mouse tails was digested in 200 μl 50 mM TrisCl, pH 8.0, 20 mM EDTA, 0.45% Tween-80, 0.45% NP-40 and 0.5 mg/ml proteinase K overnight at 56°C. The digested samples were diluted in sterile water and denatured in boiling water for 5 min; then 2 μl was used for the multiplex PCR reaction using the following multiplex PCR primers: the forward primer (L2KO3U2) 5′-CAAAAGCTTAACTCCGGCTAAGCAGGTCTT-3′, the reverse primer (L2KO3L7) 5′-TCCTGACATTGAAAGACCTCCAAAGT-3′ and BPAU1 5′-GATTGGGAAGACAATAGCAGGCA-3′. L2KO3U2 and L2KO3L7 produce a 2-kb fragment, which is the wild type allele, and BPAU1 and L2KO3L7 produce a 1.8-kb fragment, which is the targeted locus. PCR cycling conditions in 25 μl of reaction volume were as follows: 95°C for 3 min and then 36 cycles of PCR at 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec, followed by a final extension step at 72°C for 2 min.
Preparation of mouse zygotes
Mouse zygotes were collected from superovulated females. 6 to 8-week-old female Mbd3l1−/− (129S1) mice or 6-week-old female Mbd3l2−/− (129S1 × C57BL/6J) mice were superovulated by intraperitoneal injection of 5 IU pregnant mares serum gonadotropin (PMSG), and 47 h later, by injection of 5 IU human chorionic gonadotropin (hCG). The females were mated with 14-week-old males of the same strains. Zygotes were collected from oviducts of the females at 25 hr post-hCG injection. The cumulus cells were removed by digestion with hyaluronidase at room temperature, and the zygotes were washed two times with M2 medium. The zonae pellucidae were removed with acid thyrode solution, and the zygotes were washed with Ca++-Mg++-protein-free M2 on agarose coated Petri dishes, and then attached to poly-L-lysine-coated coverslips in pre-warmed Ca-Mg-protein-free M2 medium.
Zygote immunostaining
The zygotes were fixed in 3.7% formaldehyde for 15 min at room temperature and were then washed with PBS twice for 10 min. Zygotes were permeabilized with 0.2% Triton-X100 for 30 min at room temperature and were then washed with PBS twice. The zygotes were treated with 100 μg/ml RNase A in PBS for 30 min at 37°C and with 4 N HCl in 0.1% Triton-X100 for 10 min at room temperature, and then neutralized with 0.1 M Tris-HCl (pH 8.0) for 30 min at room temperature. After washing twice with PBS, the zygotes were blocked with 1% BSA (Sigma), 0.05% Tween-20 in PBS for 30 min at room temperature, followed by several washes with 0.05% Tween-20 in PBS. For the visualization of DNA methylation, the zygotes were incubated with a mouse monoclonal anti-5-MeC antibody (1:2000 dilution in 0.05% Tween-20 in PBS; Eurogentec, Seraing, Belgium) for 1 h at room temperature, then washed several times in 0.05% Tween-20 in PBS and incubated with Alexa 568-conjugated anti-mouse secondary antibody (1:1000 dilution in 0.05% Tween-20 in PBS; Molecular Probes, Inc., Eugene, OR) for 1 h at room temperature, followed by several washes with 0.05% Tween-20 in PBS. Before observation, for DNA staining, the zygotes were incubated in 50 nM of YOYO-1 iodide (Molecular Probes, Inc., Eugene, OR) for 10 min at room temperature. After washing three times with PBS and mounting with Fluoromount G (Southern Biotechnology Associates; Birmingham, AL), images were captured using a fluorescence microscope (Olympus IX81) and processed by ImageProPlus high-end image analysis software (Media Cybernetics, Silver Spring, USA) and Adobe Photoshop.
Acknowledgments
This work has been supported by NIH grant CA104967 to G.P.P.
References
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci U S A. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Beaujean N, Taylor J, Gardner J, Wilmut I, Meehan R, Young L. Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer. Biol Reprod. 2004a;71:185–193. doi: 10.1095/biolreprod.103.026559. [DOI] [PubMed] [Google Scholar]
- Beaujean N, Taylor JE, McGarry M, Gardner JO, Wilmut I, Loi P, Ptak G, Galli C, Lazzari G, Bird A, Young LE, Meehan RR. The effect of interspecific oocytes on demethylation of sperm DNA. Proc Natl Acad Sci U S A. 2004b;101:7636–7640. doi: 10.1073/pnas.0400730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol. 2006;301:179–201. doi: 10.1007/3-540-31390-7_6. [DOI] [PubMed] [Google Scholar]
- Gao S, Gasparrini B, McGarry M, Ferrier T, Fletcher J, Harkness L, De Sousa P, Wilmut I. Germinal vesicle material is essential for nucleus remodeling after nuclear transfer. Biol Reprod. 2002;67:928–934. doi: 10.1095/biolreprod.102.004606. [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L, Yang S, Tran-Gyamfi M, Baggott D, Christensen M, Hamilton A, Crooijmans R, Groenen M, Lucas S, Ovcharenko I, Stubbs L. Comparative analysis of chicken chromosome 28 provides new clues to the evolutionary fragility of gene-rich vertebrate regions. Genome Res. 2007;17:1603–1613. doi: 10.1101/gr.6775107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Jiang CL, Jin SG, Lee DH, Lan ZJ, Xu X, O’Connor TR, Szabo PE, Mann JR, Cooney AJ, Pfeifer GP. MBD3L1 and MBD3L2, two new proteins homologous to the methyl-CpG-binding proteins MBD2 and MBD3: characterization of MBD3L1 as a testis-specific transcriptional repressor. Genomics. 2002;80:621–629. doi: 10.1006/geno.2002.7001. [DOI] [PubMed] [Google Scholar]
- Jiang CL, Jin SG, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with methyl-CpG-binding protein 2 (MBD2) and components of the NuRD complex. J Biol Chem. 2004;279:52456–52464. doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Jiang CL, Rauch T, Li H, Pfeifer GP. MBD3L2 interacts with MBD3 and components of the NuRD complex and can oppose MBD2-MeCP1-mediated methylation silencing. J Biol Chem. 2005;280:12700–12709. doi: 10.1074/jbc.M413492200. [DOI] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Kantor B, Makedonski K, Shemer R, Razin A. Expression and localization of components of the histone deacetylases multiprotein repressory complexes in the mouse preimplantation embryo. Gene Expr Patterns. 2003;3:697–702. doi: 10.1016/j.modgep.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Latham T, Gilbert N, Ramsahoye B. DNA methylation in mouse embryonic stem cells and development. Cell Tissue Res. 2008;331:31–55. doi: 10.1007/s00441-007-0537-9. [DOI] [PubMed] [Google Scholar]
- Marino S, Nusse R. Mutants in the mouse NuRD/Mi2 component P66alpha are embryonic lethal. PLoS ONE. 2007;2:e519. doi: 10.1371/journal.pone.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Oliveri RS, Kalisz M, Schjerling CK, Andersen CY, Borup R, Byskov AG. Evaluation in mammalian oocytes of gene transcripts linked to epigenetic reprogramming. Reproduction. 2007;134:549–558. doi: 10.1530/REP-06-0315. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Ruddock-D’Cruz NT, Xue J, Wilson KJ, Heffernan C, Prashadkumar S, Cooney MA, Sanchez-Partida LG, French AJ, Holland MK. Dynamic changes in the localization of five members of the methyl binding domain (MBD) gene family during murine and bovine preimplantation embryo development. Mol Reprod Dev. 2008;75:48–59. doi: 10.1002/mrd.20712. [DOI] [PubMed] [Google Scholar]
- Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127:643–651. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Tesarik J, Martinez F, Rienzi L, Ubaldi F, Iacobelli M, Mendoza C, Greco E. Microfilament disruption is required for enucleation and nuclear transfer in germinal vesicle but not metaphase II human oocytes. Fertil Steril. 2003;79(Suppl 1):677–681. doi: 10.1016/s0015-0282(02)04816-1. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Young LE, Beaujean N. DNA methylation in the preimplantation embryo: the differing stories of the mouse and sheep. Anim Reprod Sci. 2004;82–83:61–78. doi: 10.1016/j.anireprosci.2004.05.020. [DOI] [PubMed] [Google Scholar]