Abstract
The intergenic region (IGR) internal ribosome entry site (IRES) RNAs do not require any of the canonical translation initiation factors to recruit the ribosome to the viral RNA, they eliminate the need for initiator tRNA, and they begin translation from the A-site. The function of these IRESs depends on a specific three-dimensional folded RNA structure. Thus, a complete understanding of the mechanisms of action of these IRESs requires that we understand their structure in detail. Recently, the structures of both domains of the IGR IRES RNAs were solved by X-ray crystallography, providing the first glimpse into an entire IRES RNA structure. Here, I present an analysis of these structures, emphasizing how the structures explain many aspects of IGR IRES function, discussing how these structures have similarities to motifs found in other viral RNAs, and illustrating how these structures give rise to new mechanistic hypotheses.
Keywords: internal ribosome entry, RNA structure, X-ray crystallography, Dicistroviridae, translation initiation, pseudoknot, IRES
1. Introduction
Precisely regulated protein synthesis is essential for life as we know it; its importance is illustrated in the highly orchestrated, stepwise mechanism used by eukaryotic cells to initiate translation. The canonical process requires a modified nucleotide “cap” on the RNA’s 5′ end that is added during mRNA transcription and the coordinated action of well over 1 megadalton of eukaryotic initiation factor (eIF) proteins. The cap and eIFs recruit the ribosome to the mRNA, direct proper “scanning” to find the start codon, and direct the final stages of ribosome preparation as protein synthesis starts. Hence, canonical eukaryotic translation initiation is a cap- and protein-dependent process, a detailed description of which is beyond the scope of this chapter and therefore I refer the reader to an excellent recent review (Pestova et al., 2007).
Viruses do not have their own translation machinery, and must therefore use the host cell’s to produce the viral proteins from a virally encoded template RNA. One strategy to do this is to “cap” viral RNAs so that they will be recognized by translation initiation factors. Some viruses produce their own capping enzymes and some others use the host cell’s capping machinery. An example of the former is vaccinia virus (Broyles, 2003), an example of the latter is influenza virus (Engelhardt and Fodor, 2006). However, other viruses dispense with the need for the cap entirely, instead using specific sequences within the RNA template itself to drive ribosome recruitment and activation. These RNA sequences have come to be called internal ribosome entry sites (IRESs), and they are used by a variety of viruses of medical and economic importance, to include hepatitis C virus (HCV), foot-and-mouth-disease virus (FMDV), hepatitis A virus (HAV), poliovirus (PV), and human immunodeficiency-1 virus (HIV-1; note that HIV-1 genomic RNA is capped). IRESs also exist in cellular mRNAs, and they are now regarded as important players in regulating gene expression in both healthy and diseased cells as well as being critical for the successful infection of many viruses. Viral IRESs are the subject of several chapters in this special edition and also are the subject of two recent reviews (Doudna and Sarnow, 2007; Elroy-Stein and Merrick, 2007) and the reader is directed to there for a more general discussion of IRESs.
Viral IRESs dispense with the need for a cap and all are driven by specific RNA sequences, but the mechanism by which they operate differs between viruses. Based on secondary structure similarities and their functional need for a subset of the eIFs or IRES trans-activating factors (ITAFs; cellular proteins not considered part of the translation machinery but co-opted by the IRES), the viral IRESs can be divided into several groups (Figure 1a). The most complex viral IRESs are unable to bind directly to the ribosome in binary complexes, and they functionally require several canonical eIFs and often ITAFs. These IRESs are found in picornaviruses (although some picornaviruses use a different sort of IRES, see below), and they are subdivided into three classes or types, based on secondary structures and mechanistic features. In this special edition, characteristics of these IRESs are discussed in a chapter by O. Fernandez-Miragall et al. on picornavirus structure (especially the FMDV IRES), and to some degree by G. Belsham (in the context of the divergent picornavirus IRESs). A somewhat less complex group of IRESs is able to bind directly to the small ribosomal subunit, and also requires fewer eIFs and no ITAFs (Figure 1a). The prototype of these IRES is the hepatitis C virus (HCV) IRES, which requires eIF3 and eIF2 to function, and which forms a specific structure that binds directly to the 40S subunit. This group also includes some pestiviruses as well as some picornaviruses, and they are discussed in this edition by P. Lukavsky in a chapter about HCV IRES structure and by G. Belsham who presents interesting features of the divergent picornavirus IRESs. The last group of IRESs use the most streamlined mechanism identified to date, in that they recruit both ribosomal subunits without using any eIFs. All IRESs of this group are found in the intergenic region (IGR) of a family of single-stranded positive-sense RNA viruses called the Dicistroviridae because they possess two open reading frames in a single viral RNA. The Dicistroviridae IGR IRES RNAs can be subdivided into two types, based primarily on sequence and secondary structure differences, which includes an extra stem-loop structure near the 3′ end of the type 2 IGR IRES RNAs. The IGR IRESs are discussed in this edition by N. Nakashima and T. Uchiumi, and they are also the subject of this chapter. In addition, it should be noted that there are other IRESs whose features are such that they do not seem to fit neatly into the above-defined groups. This includes atypical IRESs found in the 5′ UTR of Dicistroviridae, which are discussed in this edition by L.O. Roberts & E. Groppelli.
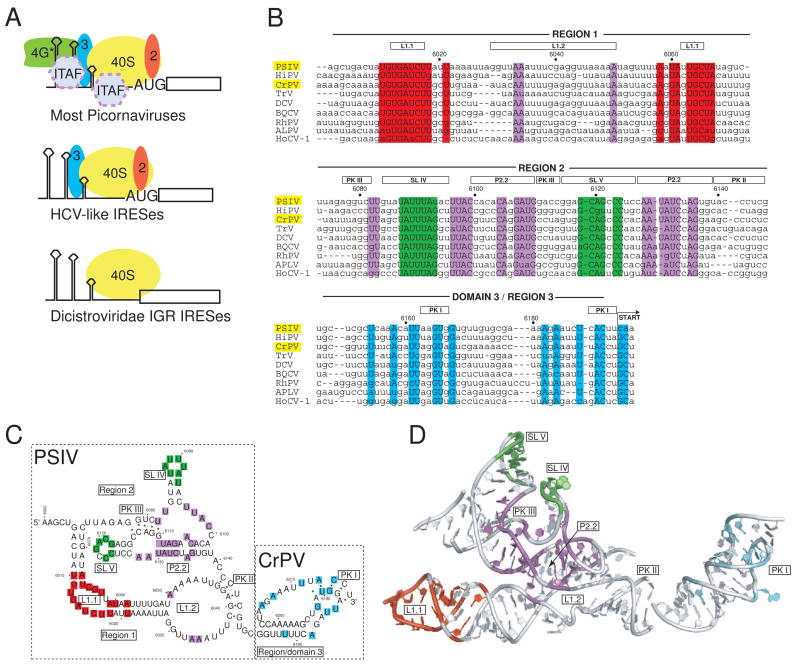
Fig. 1. IRES groups and the IGR IRES RNAs.
a) Cartoon of different strategies used by various IRES groups to recruit a 40S subunit to the IRES RNA. The most complex are those generally associated with the picornaviruses, which require several eIFs and often ITAFS in order to recruit the ribosome. These are subdivided into the entero-/rhinovirus class, the cardio-/apthoviruses class, and the hepatitis A virus class. The HCV-like IRESs (which include some picornaviruses) use a smaller set of eIFs and can bind directly to the ribosome. The most streamlined IRESs are shown at the bottom, which are the IGR IRESs from the Dicistroviridae. These IRESs bind directly to the ribosome, require no eIFs, and do not initiate from a canonical AUG start codon. More atypical IRESs, such as those from the 5′ UTR if the Dicistroviridae, are not shown. b) Sequence alignment of nine members of the Dicistroviridae IGR IRES group, all type 1. The approximate locations of the various regions and key secondary structure elements are indicated above the alignment, and the nucleotide numbers for the PSIV IRES are shown. In this review, I use the term “region” to refer to a logically connected group of secondary structure elements and reserve the use of the term “domain” for a part of the RNA shown to fold into an independent, stably folded three-dimensional structure. Locations in the structure that are highly conserved (sequence is the same in at least 7 of the 9 IRESs shown) are colored. The location of the non-AUG codon that is the start of translation is indicated, and the names of the two IRES RNAs for which crystal structures have been solved are highlighted in yellow. The alignment is based on previously published work (Jan, 2006; Kanamori and Nakashima, 2001). Abbreviations: Plautia stali intestine virus (PSIV), himetobi P virus (HiPV), cricket paralysis virus (CrPV), Triatoma virus (TRV), Drosophila C virus (DCV), black queen-cell virus (BQCV), Rhopalosiphum padi virus (RhPV), aphid lethal paralysis virus (APVL), Homalodisca coagulata virus-1 (HoCV-1). c) Secondary structure model of the ribosome-binding domain (regions 1+2) of the PSIV IGR IRES and the P-site domain (domain 3) of the CrPV IGR IRES, with secondary structure elements that are mentioned in the text labeled. The locations of conserved sequence are colored to match panel 1b. The secondary structure is drawn in a manner that allows it to be more directly compared to the structure shown in panel 1d. d) Combined crystal structures of the ribosome-binding domain of the PSIV IGR IRES (Pfingsten et al., 2006) and the P-site domain of the CrPV IGR IRES (Costantino et al., 2008), with structural elements labeled. Nucleotides that are highly conserved are colored to match panels 1b and 1c. The orientation of the two structures relative to one another matches their location when docked into cryo-EM reconstructions of CrPV IGR IRES-ribosome complexes (Costantino et al., 2008; Schuler et al., 2006; Spahn et al., 2004).
The Dicistroviridae IGR IRESs (hereafter referred to as simply the “IGR IRESs” for brevity) use the most streamlined mechanism yet identified to initiate translation, discussed in more detail in a recent review (Jan, 2006). The IGR IRES mechanism has several interesting features, which includes their ability to operate without any eIFs and to initiate from a non-AUG start codon and from the ribosome’s A-site (rather than P-site). They do not need initiator tRNAmet, ribosome subunit joining is GTP hydrolysis-independent, and the first translocation event occurs before a peptide bond is made (Jan et al., 2003; Pestova and Hellen, 2003; Sasaki and Nakashima, 1999; Sasaki and Nakashima, 2000; Wilson et al., 2000a; Wilson et al., 2000b; Yamamoto et al., 2007b). This single RNA sequence thus has the ability to bypass several key checkpoints in canonical translation initiation, a seemingly remarkable feat of manipulating the host cell’s machinery.
Discovery of the aforementioned features of IGR IRESs motivates efforts to understand this RNA-based mechanism. The IGR IRESs are divided into two types (designated type 1 and type 2) based primarily on secondary structure and sequence variation (N. Nakashima and T. Uchiumi, this edition), but all IGR IRES RNAs appear to comprise two independently and compactly folded domains that work together (Kanamori and Nakashima, 2001; Pfingsten et al., 2007). The larger “ribosome-binding domain” (regions 1 and region 2) is primarily responsible for recruiting both ribosomal subunits (Jan and Sarnow, 2002; Nishiyama et al., 2003), while the smaller “P-site domain” (region/domain 3) is placed into the ribosome’s P-site and helps direct initiation from the A-site (Jan et al., 2003; Jan et al., 2001). Although other IRESs have been shown to contain regions of higher-order structure or local areas of tightly folded RNA (Kieft et al., 1999), only the IGR IRESs possess a two domain-containing, overall compact fold (Costantino and Kieft, 2005). It is this compact, pre-folded RNA structure that drives the unusual mechanism of translation initiation used by the IGR IRESs and thus any effort to understand how this process works must include efforts to understand the IRES RNA’s three-dimensional structure. Recently, the structures of both independently folded IGR IRES RNA domains were solved by X-ray crystallography (Costantino et al., 2008; Pfingsten et al., 2006), and these structures allow interpretation of existing biochemical and functional data, help explain evolutionary conservation of sequences, and inform the development of new mechanistic models and the design of experiments to test those models. Several recent or forthcoming reviews discuss various aspects of what these structures tell us about IGR IRES function and the details of IGR IRES-ribosome interactions and hence those topics are not covered here (Kieft, 2008; Pfingsten and Kieft, 2008). Rather, I focus on specific features of these new crystal structures, how the structures help explain IGR IRES sequence conservation and some published biochemical observations, and how certain features are related to other viral RNA structures.
2. IGR IRES structure was solved in two parts
High resolution structural determination of biological macromolecules requires a strategy based on the target’s architectural features. In the case of the IGR IRES RNAs, the fact that the structure comprises two independently folded domains suggested a strategy of independently solving each domain’s structure. In addition, the various IGR IRESs share a similar three-dimensional fold and seem to use the same mechanism to initiate translation despite considerable sequence variation between viral species, suggesting that structures of these domains from any member would yield information about the group as a whole (Cevallos and Sarnow, 2005; Hatakeyama et al., 2004; Pfingsten et al., 2007). The strategy of solving the structures of each domain separately was successful as the larger ribosome-binding domain of the Plautia stali intestine virus (PSIV) IRES and the smaller P-site domain of the cricket paralysis virus (CrPV) IRES were solved to 3.1 and 2.4 Å resolution (Costantino et al., 2008; Pfingsten et al., 2006), respectively. These two structures can now be combined to yield the first complete structural description of an IRES RNA. A chimera RNA consisting of these two structures appears to be functional (unpublished data), which suggests that there is modularity in the overall architecture of the IGR IRESs and further validating the overall structural model.
It is interesting to put the strategy used to pursue the IGR IRES structure in perspective with that used to study the HCV IRES (and related IRESs). In the case of the HCV IRES, a more extended structure has mandated that the RNA be solved in smaller pieces in a “divide and conquer” effort (Collier et al., 2002; Kieft et al., 2002; Klinck et al., 2000; Locker et al., 2007; Lukavsky et al., 2003; Lukavsky et al., 2000; Rijnbrand et al., 2004). The resultant structures can then be combined with cryo-EM representations to give a model of the RNA as a whole (Boehringer et al., 2005).
3. Three-dimensional structure, sequence conservation, and evolution
The crystal structures of both IGR IRES RNA domains provide a way to interpret observed sequence conservation and understand how those conserved sequences combine to produce a functional IRES. To date, 14 Dicistroviridae family members have been identified and sequenced; all contain an IGR IRES, and as previously mentioned, these IRESs are divided into two types based mostly on their secondary structures (Jan, 2006); (N. Nakashima and T. Uchiumi, this edition). Both crystal structures are of domains from type 1 IGR IRESs. Sequence alignment of nine class 1 IGR IRESs show that although there is a substantial amount of sequence diversity, there are also parts that are well conserved and these are distributed throughout the sequence (Figure 1b). In some cases, stretches of several nucleotides in a row are conserved, whereas in other cases the conserved nucleotides are more isolated. A phylogenetic analysis of sequence conservation combined with mutagenesis was used in an elegant set of experiments to identify the secondary and tertiary structures of the IGR IRESs (Kanamori and Nakashima, 2001). These two-dimensional diagrams show that many conserved bases cluster in specific secondary structure elements important for functions such as 40S subunit binding (Figure 1c), but could not explain the conservation of all bases or how they relate in three-dimensional space. Since probing experiments subsequently showed a common three-dimensional fold shared among diverse IGR IRESs (Pfingsten et al., 2007), it seemed likely that many conserved bases would have a role in stabilizing this fold.
When conserved bases are mapped onto the IGR IRES crystal structures, they cluster into four locations that correspond to their role in IRES function (Figure 1d). First, both apical loops of stem-loops IV and V (SL IV and SL V) contain many conserved bases. This cluster is expected from the secondary structure, and footprinting and mutation analysis revealed that the role of these stem-loops is to bind the 40S subunit (Costantino and Kieft, 2005; Jan and Sarnow, 2002; Nishiyama et al., 2003). A second cluster is found in internal loop L1.1, which footprinting suggested makes contact to the 60S subunit with the assembled 80S ribosome (Nishiyama et al., 2003). Docking of the crystal structure into cryo-EM reconstructions and functional analysis of mutants confirm that L1.1 is necessary for stable 60S association with the IRES (Pfingsten et al., 2006; Schuler et al., 2006; Spahn et al., 2004), and hence the role of this cluster of conserved sequence within the three-dimensional structure is clear. A third cluster of conservation consists of bases that are more widely distributed in the sequence than those previously mentioned (Figure 1b, c). These bases are found in the molecule’s tightly packed “core”, largely centered on helix P2.2 and many of them are involved in non-canonical intramolecular interactions that likely serve to stabilize the folded structure (Figure 1d). The role of these bases could not be predicted from secondary or tertiary structure diagrams, but when they are mapped onto the three-dimensional structure their likely role is apparent (discussed in more detail in a later section). Finally, a fourth cluster is found in domain 3, the P-site domain. The location of these conserved bases suggest that they must be important for stabilizing the domain structure or interacting with the ribosome. The crystal structure of this domain shows it is a precise mimic of a tRNA anticodon-mRNA codon interaction and several of these bases are involved in that structural mimicry (discussed in more detail in a later section).
Sequence conservation is a result of evolution, and thus the distribution of conserved sequences within the IGR IRES three-dimensional structure provides insight into the evolution of this RNA structure. If modern IGR IRESs share a common ancestor RNA with a structure similar to that observed today, it can be proposed that natural selection has maintained sequences that specifically bind the 40S subunit, specifically interact with the 60S subunit, and form a precise tRNA-mRNA structural mimic within a specific three-dimensional architecture stabilized by yet other conserved bases. Genetic drift has allowed other sequences to change and likely allowed some variation in the details of the intramolecular interactions that stabilize the fold, but the necessary architecture is maintained. Since there is evidence of many more Dicistroviridae-like viruses that have yet to be described, it will be interesting to see how their sequence conservation matches those already discovered.
4. Specific IGR IRESs structural features
The IGR IRES mechanism starts with 40S subunit recruitment to the folded IRES, followed by 60S subunit recruitment to form the 80S ribosome (Jan, 2006) (Figure 2). In addition, direct 80S ribosome recruitment has also been reported (Pestova et al., 2004). In the ribosome, region 2 of the IGR IRES RNA interacts with the 40S subunit and region 1 with the 60S subunit. Hence, there is a “division of labor” in terms of ribosomal subunit binding, but the two regions that perform these tasks pack against one another in the structure. Binding of the 60S subunit to the IRES has not been reported in the absence of 40S subunit binding, and this suggests the two regions act together within the folded ribosome-binding domain to assemble the 80S ribosome. Once a tRNA is delivered to the assembled 80S ribosome’s A-site, translocation occurs, followed by protein synthesis. In the following sections, I discuss structural IGR IRES elements roughly in the order in which they act, realizing that some elements work simultaneously within a highly coordinated process. I will present a few key features of the IGR IRES RNAs, describe what the crystal structures reveal in regard to how three-dimensional structure drives function, and briefly put these three-dimensional structures into context with other RNA structures, as many motifs in the IGR IRESs are similar to previously solved structures of very different function.
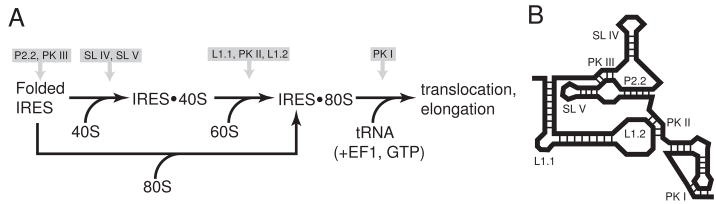
Fig. 2. Mechanism of IGR IRES translation initiation.
a) Simplified pathway of ribosome recruitment by the IGR IRES. The IRES first bind the 40S subunit, then the 60S subunit to form the 80S ribosome, or alternately has been shown in vitro to be able to bind a preformed 80S ribosome (Pestova et al., 2004). Once in the 80S ribosome, the delivery of an aminoacylated tRNA to the A-site drives a translocation event. The approximate steps in the scheme where various IRES secondary or tertiary structural elements act are indicated above the diagram. b) Simple cartoon representation of a class 1 IGR IRES secondary structure with the structural elements shown in panel 2a labeled.
4.1. Positioning stem-loops for 40S binding: Pseudoknot III and P2.2
The first step in the simple scheme in figure 2a is binding of the 40S subunit, and this requires interactions with SL IV and SL V of the IRES, both of which are positioned in space by highly structured elements within the “core” of the IRES. The structures of the apical loops of SL IV and SL V that make direct contact with the 40S subunit are still unsolved, as these sequences were changed to obtain diffracting crystals of the ribosome-binding domain (Pfingsten et al., 2006). However, the structures of pseudoknot III (PK III) and helix P2.2 elements that correctly position these stem-loops are known, and their disruption through mutation results in IRESs unable to bind the 40S subunit (Costantino and Kieft, 2005; Jan and Sarnow, 2002; Nishiyama et al., 2003). These structures and the positioned stem-loops together comprise region 2, which has 40S subunit binding activity even when isolated from the rest of the IRES RNA (Nishiyama et al., 2003).
The PK III interaction connects the base of SL V with SL IV through long-range base-pairing, and together with helix P2.2 forms a three-dimensional structure that results in the two stem-loops positioned adjacent to one another in a nearly parallel arrangement. This is achieved because region 2 forms a classic H-type pseudoknot architecture (Figure 3a). In general, pseudoknots contain two helical stems and three loops, but several different architectures are possible (Brierley et al., 2007; Hilbers et al., 1998). In most H-type pseudoknots, the two helical stems stack coaxially, loop 1 lies in the major groove of helix 2, loop 2 is short or nonexistent, and loop 3 lies along the minor groove of helix 1. In region 2 of the IGR IRES, the classic H-type pseudoknot architecture is maintained. Helix P2.2 and the helix formed by the base pairs of PK III stack coaxially and each of the two loops contains an entire stem-loop. SL IV comprises loop 1 and lies in the major groove of P2.2, and SL V is contained in loop 3, where it emerges from the minor groove of PK III (Figure 3a). As SL IV emerges from the minor groove, it forms a fairly sharp bend or kink that places it nearly perpendicular to the stack. Figure 3a shows a frameshifting pseudoknot from another virus, the simian retrovirus-1 (SRV-1) (Michiels et al., 2001). Despite performing a very different task, this frameshifting pseudoknot bears remarkable architectural similarity to the fold in region 2 of the PSIV IRES in terms of how the stems relate to one another, and how the loops pack into the major and minor grooves. Thus, although the details and apparent complexity of the structures are different, the same essential architecture underlies these two functionally different RNAs. A more in-depth analysis of the diversity of pseudoknot structures found in viral RNAs is available in a recent review (Brierley et al., 2007) and pseudoknots are also discussed by D. Giedroc and P. Cornish in a chapter in this edition.
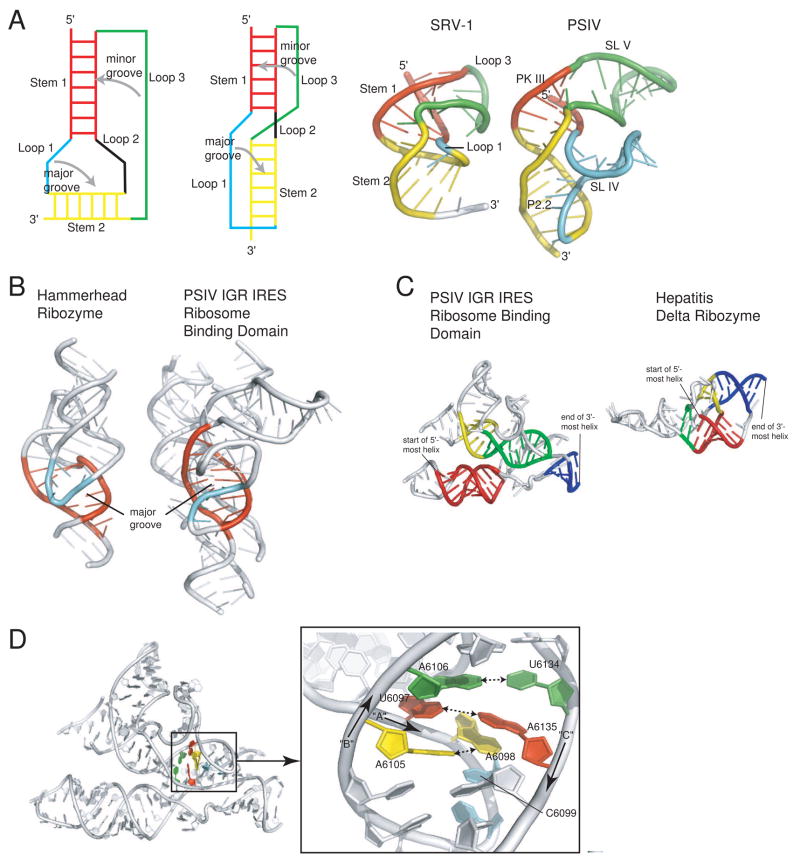
Fig. 3. Structural features that stabilize the core of the PSIV IGR IRES.
a) At left are two diagrams of a classical H-type pseudoknot architecture, with the various stems and loops labeled and the placement of loops into major and minor grooves indicated. In classic pseudoknot packing the two stems stack coaxially (red and yellow), loop 1 is in the major groove (cyan), and loop 3 is in the minor groove (green). At right is a comparison of the NMR structure of a frameshifting pseudoknot from SRV-1 (left) with the structure of only region 2 of a the PSIV IGR IRES (right). The two structures are colored to match the diagrams on the left and to show the location of analogous elements in each. The classic architecture is observed in both of these structures. In the case of the PSIV IGR IRES, both loops are expanded to include stem-loop structures, and one of the stems (P2.2) is underwound to open the major groove and allow docking of SL IV. b) Comparison of the structures of the full hammerhead ribozyme (left) with the PSIV IGR IRES ribosome-binding domain (right). In each, an underwound stem element is colored red. The molecules are oriented such that the view is directly into the widened/opened major groove of these helical elements. In both cases, another structure element docks into this groove. In the case of the hammerhead ribozyme, this is a loop structure, while in the PSIV IGR IRES it is a single stranded region at the base of a stem-loop. c) Comparison of the nested pseudoknot structures of the PSIV IGR IRES ribosome binding domain (left) with the nested pseudoknot structure of the hepatitis delta ribozyme (right). Each structure has four helices that define the nested architecture, and in each structure the analogous helices are colored to match. Both are nested pseudoknots, but different topologies create different folds. d) At left is the full structure of the PSIV IGR IRES ribosome-binding domain; a site of interesting intramolecular interactions at the center of the fold is highlighted. A close-up of this region, rotated, is shown at the right. The 5′ to 3′ direction of three RNA strands (arbitrarily named “A,” “B,” and “C” for the purposes of this review) is indicated, and nucleotides are numbered. A base pair between bases in strands B and C is colored green, a pairing between bases in strands A and C is colored red, and a pairing between bases in strand A and B is colored yellow. These three pairs continue an unbroken stack on C6099 (cyan).
The position of SL V in the major grove of helix P2.2 is possible because this helix is substantially underwound, which make the major groove wide and accessible (Figure 3b). In addition to this, helix P2.2 either contains or interacts with some of the most highly conserved bases in the structure (Figure 1c,d), which in many cases are forming specific intermolecular interactions within the tightest-packed part of the IRES that likely stabilize the structure of region 2 and perhaps the entire ribosome-binding domain (Figure 3d). Hence, in this part of the structure, a direct structural and functional role for these bases is revealed by the structure: they must interact in a complex and specific way to stabilize the overall fold of the IRES, and in so doing properly position other elements (such as SL IV and SL V ) to recognize the 40S subunit. The solved structure therefore helps to draw a connection from sequence conservation to three-dimensional structure to function.
The underwound helix that facilitates major groove docking is a striking feature of this part of the IRES structure, but it is not unique among RNAs. For example, the full-length hammerhead ribozyme, a self-cleaving RNA first identified in plant viroids, also contains a structure in which one part of the RNA docks into a major groove widened by underwinding (Figure 3b) (Martick and Scott, 2006). The identification of this motif in other RNAs demonstrates its usefulness for stabilizing higher-order RNA structures. Thus, although the 40S subunit-binding region 2 of the IRES is complex, it is built on motifs also used by functionally diverse RNAs.
4.2. Interacting directly with the 60S subunit: L1.1
After 40S subunit binding, the next step in the IGR IRES mechanism is 60S subunit recruitment (Figure 2a). Stable association of the 60S subunit within the 80S ribosome has been shown to depend on internal loop L1.1 of the IGR IRES, which interacts directly with the 60S subunit (Nishiyama et al., 2003; Pfingsten et al., 2006). L1.1 is part of region 1 of the IRES, but despite being highly conserved and necessary for stable ribosomal subunit association, this loop was unstructured or disordered in the crystal structure (as is the adjacent helix P1.1). This disorder could be the result of crystal packing, but examination of cryo-EM reconstructions of the CrPV IGR IRES bound to the 40S subunit support the idea that this part of the IRES is unstructured until the 60S subunit binds (Schuler et al., 2006; Spahn et al., 2004).
What does the disorder observed in this part of the IRES structure mean in terms of function? Because of this disorder, the structure of the unbound form of this highly conserved loop and how this structure relates to its ability to interact with the 60S subunit cannot be analyzed. However, its presence does illustrate the important conclusion that the function of this IRES may depend on both highly structured (such as PK III and P2.2, above) and disordered or dynamic elements whose structure changes in response to interactions with the translational machinery. In other words, it now seems apparent that this part of the IRES may be disordered in the free- and 40S subunit-bound forms, but then becomes ordered in the presence of the 60S subunit, and this restructuring of the IGR IRES might be an important component of function. This idea remains to be fully tested, but is a good illustration of how structure leads to new mechanistic hypotheses.
4.3. Holding regions 1 and 2 together: Pseudoknot II and L1.2
When bound in the ribosome, region 1 interacts with the 60S subunit and region 2 with the 40S subunit, but the two must combine to form the folded ribosome-binding domain within the context of a functioning IGR IRES. Within the crystal structure of the PSIV IGR IRES ribosome-binding domain, the two regions lay adjacent, packed side-by-side with the helical stacks of each roughly parallel (Pfingsten et al., 2006). The two regions are held against one another by the formation of pseudoknot II (PK II) and by interactions from L1.2.
PK II is formed by base-pairing between several nucleotides of the large L1.2 loop in region 1 and the last nucleotides of region 2 and the P-site domain (domain 3) is immediately 3′ of this base-paired element. This base-pairing is mostly ordered in the crystal structure but does not appear to be as stable as other parts of the IRES structure, consistent with biochemical evidence that this pseudoknot is dynamic (Costantino and Kieft, 2005; Jan and Sarnow, 2002; Nishiyama et al., 2003; Pfingsten et al., 2007). Formation of PK II creates a “double-nested pseudoknot,” in which all of region 2 (and PK III) is contained within the pseudoknot formed by the PK II base-pairing (Staple and Butcher, 2005). Unlike PK III, this structure does not follow classic pseudoknot architecture, but involves a complex set of interactions. However, double-nested pseudoknots have been observed in other compact viral RNA structures, including the hepatitis delta virus (HDV) ribozyme (Ferre’-D’Amare’ et al., 1998), where the structure helps create a compact catalytic active site (Figure 3c). In these as well as in other non-viral RNAs, such as the glmS riboswitch/ribozyme (Cochrane et al., 2007; Klein and Ferre-D’Amare, 2006), this nested pseudoknot architecture forms very tightly packed and compact structure and therefore seems to be a fundamental strategy to achieve these types of folds. However, although these RNAs all use a nested pseudoknot architecture to achieve a compact fold, the topologies of the folds and the details of the RNA structures are very different. For example, although the HDV ribozyme and the PSIV IGR IRES both contain a double-nested pseudoknot, the trace of the backbone is very different when the two are compared (Figure 3c). This illustrates the point that viral RNA structures can be built on similar sets of motifs, but that variation on how those motifs are assembled and used leads to large variation in structure and function.
The other interactions between regions 1 and 2 that were revealed in the PSIV IGR IRES crystal structure involved L1.2, which “cradles” helix P2.2 and makes use of several “A-minor” interactions that are adjacent to the PK II interaction described above (Figure 4) (Pfingsten et al., 2006). The A-minor motif can be described simply as docking of the adenosine base into the wide and shallow minor groove of an RNA double helix. Once again, a common RNA folding motif is used. A-minor interactions are ubiquitous in folded RNA structures, found in RNAs as diverse as the P4-P6 domain of the group I intron and the rRNA of the ribosome (Doherty et al., 2001; Nissen et al., 2001), and hence they are a fundamental RNA fold stabilization motif (Battle and Doudna, 2002).
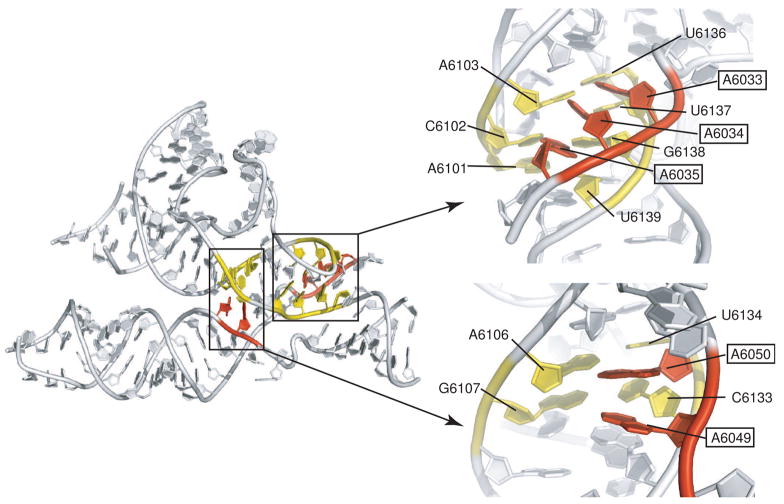
Fig. 4. A-minor interactions between regions 1 and 2 in the PSIV ribosome-binding domain.
At left is the structure of the PSIV IGR IRES ribosome-binding domain, with two locations of A-minor interactions colored and highlighted. At right are close-ups of these two locations presented in an orientation looking into the minor groove of helix P2.2. Nucleotides from region 2 are colored yellow, and adenosines from L1.2 of region 1 are colored red. In both sets of interactions, the Abases contact the minor groove side of the helix. In the interaction at top left, the A-bases are at an angle to the bases of the helix, but contacts are still to the minor groove. In the interactions at lower right, the A-bases are closer to being co-planar with those in the helical stack.
It is interesting to note that outside of the base-pairing of PK II, the crystal structure reveals no other base-pairing interactions between the packed helical stacks of regions 1 and 2 (Pfingsten et al., 2006). Hence, the structure suggests it would be possible for the two regions to move or shift relative to one another, with only a few A-minor interactions being broken or reordered (Figure 4). In other words, it seems possible for P2.2 to “slide” within the cradle formed by L1.2 (Pfingsten et al., 2006), and this may have functional significance as the IRES must undergo conformational changes as the ribosome assembles. Cryo-EM reconstructions of the CrPV IGR IRES bound to 40S subunit and 80S ribosomes show that allosteric changes occur when the IRES contacts the 60S subunit; these involve not only changes locally at the L1.1-60S subunit interaction site, but throughout the IRES (Schuler et al., 2006; Spahn et al., 2004). Could the putative ability of region 1 to move relative to region 2 be a means to transmit allosteric structural changes from one end of the IRES to the other without breaking key ribosomal contacts?
4.4. Mimicking tRNA-mRNA: Pseudoknot I (domain 3)
Within the 80S ribosome, domain 3 of the IRES docks into the P-site of the decoding groove on the 40S subunit and positions the next codon into the A-site, and the structure of this domain explains how this can occur. This P-site domain contains pseudoknot I (PK I), which was shown through functional and biochemical assays to possess some tRNA functional mimicry (Jan et al., 2003; Jan et al., 2001). The structure of the P-site domain was solved recently, revealing that the intramolecular PK I interaction precisely mimics the intermolecular tRNA anticodon-mRNA codon interaction (Figure 5) (Costantino et al., 2008). The codon-anticodon interaction in the P-site and its recognition by the ribosome is fundamental to translation. Thus, this mimicry suggests how this domain can dock into the P-site, position the next codon in the A-site, and perhaps manipulate the ribosome into acting as if it had an authentic codon-anticodon in position in the decoding groove. Furthermore, the modeled position of the P-site domain on the ribosome, combined with cryo-EM reconstructions of the entire IGR IRES on the ribosome, support the idea that the IGR IRES functions by mimicking a P/E-hybrid state tRNA (Costantino et al., 2008; Yamamoto et al., 2007a).
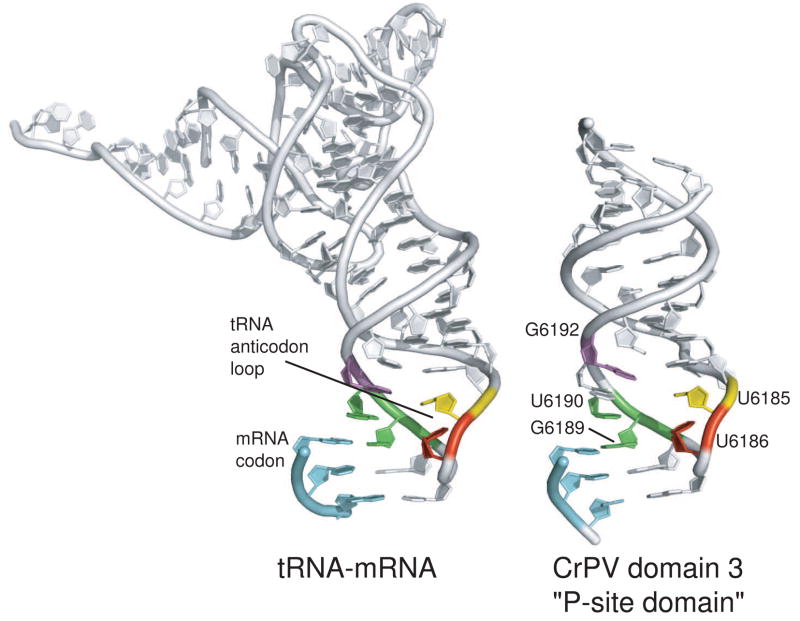
Fig. 5. Comparison of a tRNA-mRNA interaction and the domain 3/P-site domain structure from the CrPV IRES RNA.
At left is the structure of an initiator methionine tRNA interacting with an AUG start codon in the P-site of the ribosome, extracted from a structure of a bacterial ribosome bound to all three tRNAs and mRNA (Selmer et al., 2006). At right is the crystal structure of the P-site domain (domain 3) of the CrPV IRES. In both structures, some nucleotides have been removed for clarity. The location of the anticodon and codon elements are shown, analogous bases are colored to match each other, and the locations of nucleotides mentioned in the main text are shown.
The conservation of sequence within PK I is partially explained by the crystal structure. Several of the most conserved nucleotides are involved in interactions that appear to stabilize the anticodon loop mimic’s structure. For example, U6186 crosses the loop structure to hydrogen bond with a phosphate, a feature also seen in canonical tRNA anticodon loops. This U is present in 8 of the 9 sequences aligned in figure 1a, and mutation of this loop reduces IRES activity (Costantino et al., 2008). Likewise, mutation of U6185 and G6192, both of which have analogous nucleotides in tRNA, reduce IRES activity and mutation of all three at once eliminated detectable activity (Costantino et al., 2008). Overall, it is clear that this structure has evolved to mimic the tRNA-mRNA intermolecular interaction in a single intramolecular pseudoknot interaction. There are examples of tRNA mimicry in other RNAs, such as bacterial transfer-messenger RNA (tmRNA) (Moore and Sauer, 2007), and at the 3′ ends of some plant viruses (such as Turnip Yellow Mosaic Virus and Brome Mosaic Virus, see chapter by T. Dreher, this edition), but this is the only precise structural mimic of a tRNA-mRNA anticodon-codon interaction reported to date. It will be interesting to see if other RNAs are found that use this motif, and to relate the presence of this motif to function.
5. Conclusion and perspective
The IGR IRESs are remarkable in the sense that they dispense with the need for protein initiation factors, bind directly to the ribosome, and bypass certain key steps in ribosomal subunit association and translation initiation. One might expect the structure of the IGR IRESs to be equally remarkable, and indeed overall it is unlike any other RNA structure yet solved. However, close examination of the structure shows that many of its elements contain versions of structural motifs found in other RNAs, such as classic H-type pseudoknots, major groove docking into underwound helices, A-minor motifs, and an anti-codon loop mimic. The ability of these motifs to combine in a new and complex way within an RNA containing both stable and dynamic elements illustrates the diversity of structure and function that can be achieved by a molecule containing only four different bases. This structure is the basis for the function of the IGR IRESs, and the crystal structures allow new mechanistic models to be proposed.
In addition to providing a foundation for new mechanistic models, the IRES structures provide a means to understand many functional aspects of the IGR IRES function. For example, the ability of SL IV and SL V to work together to bind the 40S subunit is explained by their adjacent positioning, made possible by a compact folded core comprising a pseudoknot and other elements. Another example is in the “division of labor” that exists between regions 1 and 2 (in terms of subunit interactions), which is reflected in the side-by-side packing of these regions, the distribution of stable and dynamic elements, and the apparent ability of these regions to move relative to one another. Finally, a third example is the structure of the P-site domain, whose tRNA-mRNA mimicry provides an explanation for docking of this domain into the P-site and possibly for initiation from the A-site.
What about other IRES structures? Do they use structural motifs found in other RNAs? Studies of the HCV IRES RNA shows that it too contains at least one motif found in other RNAs (loop E motif; Klinck et al., 2000; Lukavsky et al., 2000), but analysis of its most folded portions remains incomplete as we do not yet have three-dimensional structural information for all of the IRES. In the case of the more complex picornavirus IRESs, extensive probing and mutational studies both in vitro and in vivo lend great insight into the structure of the IRES RNA and how that structure drives function (see chapter by O. Fernandez-Miragall, this edition for a summary of these studies for FMDV). For example, in the FMDV IRES there is evidence of a tetraloop-tetraloop receptor motif (Fernandez-Miragall et al., 2006), perhaps similar to that found in the P4-P6 domain of the group I intron (Cate et al., 1996), and it is likely that more folded motifs that are present in other functional RNAs are used in these large IRES RNAs. To date the high-resolution structures of the three-dimensional folds of picornaviruses or other IRES that require ITAFs have not been solved, and hence the atomic-resolution details of many of the structural motifs within these RNAs remain unexplored. Future studies aimed at solving these IRES RNAs and IRES RNPs likely will be an important part of ongoing efforts to understand the link between RNA structure and cap-independent translation initiation.
Acknowledgments
I would like to thank D. Barton, D.A. Costantino, J.S. Pfingsten and Q. Vicens for reading of this manuscript, and all the members of the Kieft Lab for discussions about IRES RNA structure. IRES RNA studies in the Kieft Lab are supported by grants R01 GM072560 and R03 AI072187 from the National Institutes of Health and Research Scholar Grant 0805801GMC from the American Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Battle DJ, Doudna JA. Specificity of RNA-RNA helix recognition. Proc Natl Acad Sci U S A. 2002;99:11676–11681. doi: 10.1073/pnas.182221799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles SS. Vaccinia virus transcription. J Gen Virol. 2003;84:2293–2303. doi: 10.1099/vir.0.18942-0. [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Cevallos RC, Sarnow P. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J Virol. 2005;79:677–683. doi: 10.1128/JVI.79.2.677-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier AJ, Gallego J, Klinck R, Cole PT, Harris SJ, Harrison GP, Aboul-Ela F, Varani G, Walker S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat Struct Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 129–153. [Google Scholar]
- Elroy-Stein O, Merrick WC. Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey J, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 155–172. [Google Scholar]
- Engelhardt OG, Fodor E. Functional association between viral and cellular transcription during influenza virus infection. Rev Med Virol. 2006;16:329–345. doi: 10.1002/rmv.512. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miragall O, Ramos R, Ramajo J, Martinez-Salas E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre’-D’Amare’ AR, Zhou K, Doudna JA. Crystal Structure of a Hepatitis Delta Virus Ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Hatakeyama Y, Shibuya N, Nishiyama T, Nakashima N. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA. 2004;10:779–786. doi: 10.1261/rna.5208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbers CW, Michiels PJ, Heus HA. New developments in structure determination of pseudoknots. Biopolymers. 1998;48:137–153. doi: 10.1002/(SICI)1097-0282(1998)48:2<137::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci U S A. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- Jan E, Thompson SR, Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harb Symp Quant Biol. 2001;66:285–292. doi: 10.1101/sqb.2001.66.285. [DOI] [PubMed] [Google Scholar]
- Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7:266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008 doi: 10.1016/j.tibs.2008.04.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferre-D’Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul-Ela F. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels PJ, Versleijen AA, Verlaan PW, Pleij CW, Hilbers CW, Heus HA. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J Mol Biol. 2001;310:1109–1123. doi: 10.1006/jmbi.2001.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T, Lorsch JR, Hellen CU. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J Mol Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Kieft JS. RNA structure-based ribosome recruitment: Lessons from the Dicistroviridae intergenic region IRESes. RNA. 2008 doi: 10.1261/rna.987808. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnbrand R, Thiviyanathan V, Kaluarachchi K, Lemon SM, Gorenstein DG. Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J Mol Biol. 2004;343:805–817. doi: 10.1016/j.jmb.2004.08.095. [DOI] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci U S A. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM Visualization of a Viral Internal Ribosome Entry Site Bound to Human Ribosomes; The IRES Functions as an RNA-Based Translation Factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000a;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000b;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J Biol Chem. 2007a;282:7770–7776. doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]