Abstract
Thiopurines are among the most successful chemotherapeutic agents for treating a number of human diseases including acute lymphoblastic leukemia. The mechanisms through which the thiopurines elicit their cytotoxic effects remain unclear. We postulate that the incorporation of 6-thioguanine into CpG site may perturb the methyltransferase-mediated cytosine methylation at this site, thereby interfering with the epigenetic pathways of gene regulation. To gain biochemical evidence for this hypothesis, we assessed, by using a restriction enzyme digestion coupled with LC-MS/MS method, the impact of 6-thioguanine on cytosine methylation mediated by two DNA methyltransferases, human DNMT1 and bacterial HpaII. Our results revealed that the incorporation of 6-thioguanine into CpG site could affect the methylation of cytosine residue by both methyltransferases and the effect on cytosine methylation is dependent on the position of 6-thioguanine with respect to the cytosine to be methylated. The presence of 6-thioguanine at methylated CpG site enhanced the DNMT1- mediated methylation of the opposing cytosine in the complementary strand, whereas the presence of 6-thioguanine at unmethylated CpG site abolished almost completely the methylation of its 5′ adjacent cytosine by both DNMT1 and HpaII. We further demonstrated that the treatment of Jurkat T cells, which were derived from acute lymphoblastic leukemia, with 6- thioguanine could result in an appreciable drop in the level of global cytosine methylation. These results showed that 6-thioguanine, after being incorporated into DNA, may perturb the epigenetic pathway of gene regulation.
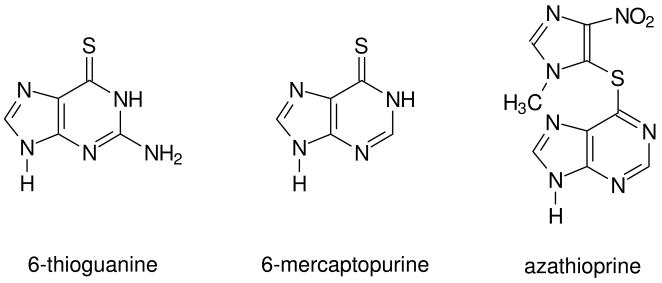
Thiopurine drugs, which include 6-thioguanine (SG) and 6-mercaptopurine (Scheme 1), are commonly used for the treatment of acute lymphoblastic leukemia, inflammatory bowl disease, autoimmune hepatitis and other pathological conditions (1, 2). Azathioprine (Scheme 1), which is converted to 6-mercaptopurine in vivo, is a standard immunosuppressant prescribed to prevent graft rejection in organ transplant patients (1). Although the exact mechanisms underlying the cytotoxic effects of these thiopurines remain elusive, it is known that the activation of the thiopurine drugs requires the formation of thioguanine nucleotides and their incorporation into nucleic acids (1).
Scheme 1.
The chemical structures of 6-thioguanine, 6-mercaptopurine and azathioprine.
Compared to guanine, the increased chemical reactivity of 6-thioguanine may contribute to the cytotoxic effects of the thiopurine drugs. After being incorporated into DNA, 6- thioguanine can be methylated by S-adenosyl-L-methionine (S-AdoMet) to form S6- methylthioguanine (S6mG) (3) or oxidized to guanine-S6-sulfonic acid (SO3HG) upon exposure to UVA light (4). Both S6mG and SO3HG exhibit considerable miscoding properties (3–6), and a recent replication study with single-stranded M13 shuttle vector demonstrated that, in E. coli cells, these two modified nucleosides could lead to G→A transition at frequencies of 94% and 77%, respectively (7). The S6mG:T base pairs arising from DNA replication can trigger the post-replicative mismatch repair system (3, 8), which may account for the cytotoxicity of the thiopurine drugs. Moreover, our recent replication experiments showed that 6-thioguanine is mutagenic in E. coli cells, and it can give rise to G→A transition mutation at a frequency of ~11%, raising the possibility that the replication-produced SG:T base pair may trigger directly the post-replicative mismatch repair pathway (7).
DNA methylation occurs in the genomes of diverse organisms and, in higher eukaryotes, methylation of cytosine occurs predominantly at CpG dinucleotide sites in the promoter and initial exons of genes (9). In this respect, each protein revealed by biochemical experiments to be involved in the establishment, maintenance, or interpretation of genomic methylation pattern in mammals is encoded by an essential gene, underscoring the biological importance of CpG methylation (10). In human cells, the CpG methylation pattern is established and maintained by DNA (cytosine-5)-methyltransferases (10). In addition, a family of human proteins exhibit high binding affinity to DNA sequences bearing the methylated CpG dinucleotide (11); upon binding to methylated DNA, these methyl-CpG binding proteins may further recruit histone-modifying enzymes to promote chromatin condensation and gene silencing (12).
Previous studies revealed that the covalent modifications of nucleobases at CpG dinucleotide site could perturb cytosine methylation by DNA methyltransferases. In this context, earlier studies showed that the replacement of the guanine residue at CpG site with an 8-oxo-7,8- dihydroguanine (8-oxoG) resulted in diminished methylation of its 5′ neighboring cytosine catalyzed by DNA methyltransferases; the substitution of guanine at methylated CpG site with an 8-oxoG or O6-methylguanine (O6mG), however, enhanced the methylation of the opposing cytosine in the complementary strand (13–16). Furthermore, Subach et al. (17) showed that benzo[a]pyrene (B[a]P) guanine adduct (+)-trans-B[a]P-N2-dG inhibited the binding and the methylation by both SssI and HhaI methyltransferases. However, the isomeric (+)-cis-B[a]P-N2- dG adduct allowed for normal binding of methyltransferases and did not result in apparent change in methylation efficiencies. On the other hand, Sowers and coworkers (18) demonstrated recently that 5-chlorocytosine or 5-bromocytosine present at the CpG dinucleotide site behaves like a 5-methylcytosine and directs the methylation of the opposing cytosine residue in the complementary strand.
Previous studies also revealed that the presence of a 6-thioguanine in DNA can perturb a number of enzymatic reactions including T4 DNA ligase-mediated DNA ligation (19), RNase H-induced cleavage of DNA/RNA heteroduplexes (20), and topoisomerase II-catalyzed DNA cleavage (21). In light of these previous findings, we reason that the replacement of the guanine residue at CpG site with a 6-thioguanine may interfere with CpG methylation by DNA methyltransferases, thereby perturbing the epigenetic pathways of gene regulation. As a first step toward testing this hypothesis, we developed an LC-MS/MS method and examined how 6- thioguanine in DNA affects the methylation of cytosine residues at CpG site by two DNA methyltransferases, i.e., the bacterial HpaII methylase and human DNMT1. HpaII methylates the internal cytosine residue at unmethylated or hemimethylated CCGG site (22), and DNMT1 methylates preferentially the cytosine residue at hemi-methylated CpG site (23). Our results revealed that the presence of a 6-thioguanine at CpG site could indeed perturb the cytosine methylation mediated by the two methyltransferases, and the perturbation is dependent on both the nature of the methyltransferase and the location of SG with respect to the cytosine to be methylated. Moreover, we showed that the treatment of Jurkat T cells with 6-thioguanine could lead to an appreciable drop in the level of global cytosine methylation. Together, our results support that the thiopurine drugs may exert their cytotoxic effects by perturbing the epigenetic pathway of gene regulation.
Materials and Methods
Chemicals and enzymes
All chemicals and enzymes, unless otherwise noted, were obtained from Sigma-Aldrich (St. Louis, MO). Reagents for solid-phase DNA synthesis were obtained from Glen Research (Sterling, VA, USA). Unmodified oligodeoxynucleotides (ODNs) used in this study were purchased from Integrated DNA Technologies (Coralville, IA). Human maintenance DNA methyltransferase DNMT1, HpaII methylase, BciVI and uracil-DNA glycosylase (UDG) were from New England Biolabs (Ipswich, WA). Jurkat T (Clone E6–1) acute lymphoblastic leukemia cells, penicillin, streptomycin, and RPMI-1640 media were purchased from ATCC (Manassas, VA, USA).
Preparation of ODN substrates containing 6-thioguanine and 5-methylcytosine
ODNs containing SG and 5-methylcytosine (mC) were synthesized on a Beckman Oligo 1000S DNA synthesizer (Fullerton, CA) at 1-μmol scale. After solid-phase synthesis, the controlled pore glass support was treated with 1.0 M DBU (1,8-diazabicyclo[5.4.0]undec-7-ene) in anhydrous acetonitrile at room temperature for 5 hrs to remove the cyanoethyl protecting group on the thionucleoside. The solid support was subsequently treated with 50 mM NaSH in concentrated NH4OH solution at room temperature for 24 hrs to complete the deprotection. The synthesized 15mer ODNs, i.e., d(ACCmCSGGTGACACACC) and d(ACCmCGSGTGACACACC), were purified by HPLC and their identities were confirmed by electrospray ionization-mass spectrometry (ESI-MS) and tandem MS (MS/MS) measurements. Likewise, we synthesized and characterized the 24-mer SG-bearing ODNs (the top strands for Substrates 2 and 3, Table 1).
Table 1.
Substrates employed for the in-vitro methylation assay.
| 5′-TGG CAG GTA TCC AGU ACC C XY TGA-3′ | 1. XY=GG |
| 3′-ACC GTC CAT AGG TCA TGG GMCC ACT-5′ | 2. XY=SGG |
| 3. XY=GSG | |
|
| |
| 5′-TGG CAG GTA TCC AGT ACC MCXY TGA CAC ACC-3′ | 4. XY=GG |
| 3′-ACC GTC CAT AGG TCA UGG GCC ACT-5′ | 5. XY=SGG |
| 6. XY=GSG | |
The above 15-mer SG-bearing ODNs were ligated with d(TGGCAGGTATCCAGT) in the presence of template DNA to afford d(TGGCAGGTATCCAGTACCmCXYTGACACACC) (“XY” represents SGG and GSG in the top strand for Substrates 5 and 6, respectively. See Table 1). The ligation reaction was performed at 16 °C overnight with 5 nmol each of the above two ODNs along with the template DNA and 1200 units of T4 DNA ligase in a buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT, 1 mM ATP and 25 μg/mL BSA. The ligation products were purified by employing 15%, 1:19 cross-linked polyacrylamide gels containing 8 M urea. The substrates were then isolated from the gel and desalted by ethanol precipitation. The purity of the resulting ODNs was confirmed by polyacrylamide gel electrophoresis (PAGE) analysis and their identities were verified by ESI-MS and MS/MS.
HPLC
The ODNs were purified on a Beckman HPLC system with pump module 125 and a UV detector (module 126). A 4.6 × 250 mm Apollo C18 column (5 μm in particle size and 300 Å in pore size, Alltech Associate Inc., Deerfield, IL) was used. Triethylammonium acetate (TEAA, 50 mM, pH 6.6, Solution A) and a mixture of 50 mM TEAA and acetonitrile (70/30, v/v, Solution B) were employed as mobile phases. The flow rate was 0.8 mL/min. A gradient of 0–20% B in 5 min, 20–45% B in 40 min, and 45–100% B in 5 min was employed for the separation. The purified ODNs were desalted on the same HPLC system with H2O as mobile phase A and acetonitrile as mobile phase B, and a gradient of 0% B in 20 min, 0–50% B in 5 min, and 50% B in 25 min was used.
Methylation of cytosine residues in duplex DNA by DNMT1 and HpaII
Each of the six double-stranded DNA substrates listed in Table 1 was annealed in the DNMT1 reaction buffer, which contained 50 mM Tris-HCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), and 5% glycerol. The resulting hemimethylated duplex DNA (10 pmol) was incubated with DNMT1 (3 units) in the presence of S-AdoMet (20 nmol) at 37 °C for 1, 2 and 4 hrs. The enzyme was subsequently inactivated by heating at 65 °C for 20 min and removed by chloroform extraction. The aqueous layer was dried by using a Savant Speed-Vac (Thermo Savant Inc., Holbrook, NY, USA).
For the HpaII-mediated methylation, ODNs were hybridized in the reaction buffer supplied by the vendor, which contained 50 mM Tris-HCl (pH 7.5), 10 mM EDTA, and 5 mM β- mercaptoethanol. To the annealed duplex solution (10 pmol) were added HpaII (4 units) and S-AdoMet (4.8 nmol), and the reaction mixture was incubated at 37 °C for 10, 20, 40 and 60 min. HpaII was subsequently inactivated by heating the solution to 70 °C for 15 min and removed by chloroform extraction, and the aqueous layer was dried.
BciVI digestion
The dried residues obtained from the above methylation reactions were redissolved in doubly distilled water and incubated with 1 unit of BciVI in a buffer containing 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate and 1 mM DTT at 37 °C for 1 hr. The BciVI was inactivated by heating at 65 °C for 20 min and removed by chloroform extraction. The reaction mixture was dried by using Savant Speed-Vac and reconstituted in doubly distilled water for LC-MS/MS analysis. After the above restriction cleavage, the DNA fragments of interest were liberated as 6- (for Substrates 1–3) or 7-mer ODNs (for Substrates 4–6), which were subjected to LC-MS/MS analysis for the identification and quantification of cytosine methylation.
UDG digestion
Some methylation reaction mixtures were also treated with UDG to produce short ODNs for the LC-MS measurements. In this respect, the products arising from the 10-pmol methylation reaction was treated with UDG (~0.4 unit) in a buffer containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT and 1 mM EDTA at 37 °C for 4 hrs. The UDG cleavage reaction mixture was quenched by adding piperidine until its final concentration reached 0.25 M. The resulting solution was incubated at 60 °C for 1 hr to induce DNA strand cleavage at the UDG-produced abasic site. The mixture was extracted with chloroform to remove the UDG and the aqueous layer was dried. The dried residues were redissolved in water for LC-MS/MS analysis.
During the course of the study, we noted that the 6-thioguanine-bearing ODN was partially degraded during the UDG digestion and the subsequent hot piperidine treatment, which affected the measurement of the methylation level of cytosine residue that is situated in the same strand as SG. However, the UDG digestion does not affect the measurement of the methylation of cytosine that is in the opposite strand with respect to SG. Most data were obtained from the cleavage with BciVI.
LC-MS/MS analysis and data processing
The extent of cytosine methylation in synthetic duplex DNA was quantified by on-line LC-MS/MS experiments, which were carried out using an Agilent 1100 capillary HPLC pump (Agilent Technologies, Palo Alto, CA) and an LTQ linear ion-trap mass spectrometer (Thermo Electro Inc., San Jose, CA). A 0.5 × 150 mm Zorbax SB-C18 column (particle size 5 μm, Agilent Technologies) was used for the separation of ODNs after the restriction enzyme digestion, and the flow rate was 6.0 μL/min. A gradient of 5 min of 0–20% methanol followed by a 35 min of 20–50% methanol in 400 mM 1,1,1,3,3,3-hexafluoroisopropanol (HFIP, pH was adjusted to 7.0 by addition of triethylamine) was employed (24). The voltage for electrospray was 4.0 kV, and the ion transport tube of the mass spectrometer was maintained at 300 °C to minimize the formation of HFIP adducts of ODNs.
The specific precursor ions for the methylated and unmethylated sequences were chosen for fragmentation to acquire the tandem mass spectra. The integrated peak areas (IAs), which were found in the selected-ion chromatogram (SIC) plotted for the formation of three abundant fragment ions of the ODNs containing a methylated and unmethylated cytosine at CpG site, were used to calculate the percentage of methylation:
Cell culture, drug treatment, and DNA isolation
Jurkat T cells were cultured in RPMI-1640 medium with 10% (v/v) fetal bovine serum (FBS) at 37 °C in a 5% CO2 atmosphere. When the confluence level of the cells reached about 60%, the old media were removed and the cells were cultured, for 24 hrs, in fresh media alone (served as control) or in the same media containing 6-thioguanine (1 or 3 μM) or 5-aza-2′- deoxycytidine (5-aza-dC, 5 μM).
DNA isolation
The above drug- or mock-treated cells (~2 × 107 cells) were harvested by centrifugation, and the cell pellets were washed with phosphate buffered saline (PBS) and resuspended in a lysis buffer containing 10 mM Tris-HCl (pH 8.0), 0.1 M EDTA, and 0.5% SDS. The cell lysates were then treated with 20 μg/mL of RNase A at 37 °C for 1 hr and subsequently with 100 μg/mL of proteinase K at 50 °C for 3 hrs. Genomic DNA was isolated by extraction with phenol/chloroform/isoamyl alcohol (25:24:1, v/v) and desalted by ethanol precipitation. The DNA pellet was redissolved in water and its concentration was measured by UV absorbance at 260 nm.
DNA digestion and HPLC quantification of global cytosine methylation
The above cellular DNA (50 μg) was denatured by heating to 95 °C and chilled immediately on ice. The denatured DNA was subsequently digested with 2 units of nuclease P1 and 0.008 unit of calf spleen phosphodiesterase in a buffer containing 30 mM sodium acetate (pH 5.5) and 1 mM zinc acetate at 37 °C for 4 hrs. To the digestion mixture were then added 12.5 units of alkaline phosphatase and 0.05 unit of snake venom phosphodiesterase in a 50-mM Tris-HCl buffer (pH 8.6). The digestion was continued at 37 °C for 3 hrs, and the enzymes were removed by chloroform extraction. The aqueous DNA layer was dried by using a Speed-vac and the dried DNA was redissolved in water. The amount of nucleosides in the mixture was quantified by UV absorbance measurements.
The above digested nucleoside mixtures were separated by HPLC on an Agilent 1100 capillary pump (Agilent Technologies, Palo Alto, CA) with a UV detector monitoring at 260 nm and a Peak Simple Chromatography Data System (SRI Instruments Inc., Las Vegas, NV, USA). A 4.6×250 mm Polaris C18 column (5 μm in particle size, Varian Inc., Palo Alto, CA) was used for the separation of the enzymatic digestion mixture. A solution of 10 mM ammonium formate (pH 6.3, solution A) and a mixture of 10 mM ammonium formate and acetonitrile (30:70, v/v, solution B) were employed as mobile phases. A gradient of 5 min 0–4% B, 40 min 4–30 % B and 5 min 30–100% B was employed, and the flow rate was 0.80 mL/min. Under these conditions, we were able to resolve 5-methyl-2′-deoxycytidine (5-mdC) from other nucleosides. The global methylation in cells was quantified based on the peak areas of 5-mdC and 2′-deoxycytidine (dC) with the consideration of the extinction coefficients of the two nucleosides at 260 nm (5020 and 7250 L mol−1 cm−1 for 5-mdC and dC, respectively).
The identities and purities of the nucleosides were assessed by LC-MS/MS under the same conditions as described above for the analysis of the methylation reaction mixtures except that different mobile phases and gradient were used (Figure S4). In this respect, 0.1% formic acid in water and 0.1% formic acid in methanol were employed as mobile phases A and B, respectively, and a gradient of 0–20% B in 5 min followed by 20–45% B in 35 min was employed.
Results
Although it is known that the DNA 6-thioguanine is the ultimate metabolite for the thiopurine drugs to exert their cytotoxic effects (1), the molecular mechanisms through which the DNA 6-thioguanine results in cell death remain not very clear. In light of the previous findings that the chemical modifications of cytosine and guanine residues at CpG dinucleotide site can perturb the enzymatic methylation of cytosine at this site (13–18), we postulate that the incorporation of SG into DNA may perturb the epigenetic pathway of gene regulation by altering the DNA methyltransferase-mediated cytosine methylation. To test this hypothesis, we set out to assess how the presence of 6-thioguanine in synthetic duplex DNA affects the methylation of cytosine residues by purified DNA methyltransferases including human DNMT1 and bacterial HpaII methylase.
1. Preparation of ODNs containing 6-thioguanine and 5-methylcytosine
To assess how the presence of 6-thioguanine perturbs the methylation of cytosine residues at CpG sites by DNA methyltransferases, we designed two sets of DNA substrates. The first set has three substrates where the bottom strand contains a CmCGG sequence motif and the top strand carries CCGG, CCSGG or CCGSG (Substrates 1–3 in Table 1). These three substrates allow us to investigate how the presence of SG at CCGG site affects the methylation of the internal cytosine at this site in the same strand. The second set has another three substrates where the bottom strand bears CCGG sequence motif and the top strand contains CmCGG, CmCSGG, or CmCGSG (Substrates 4–6 in Table 1). These three substrates facilitate us to ask how the existence of SG in the methylated strand perturbs the methylation of the internal cytosine at CCGG site in the opposite strand.
It is worth noting that the top strands of Substrates 4, 5 and 6 were designed originally for protein binding assays, and it turned out to be somewhat difficult to assess, by using LC-MS/MS, the level of methylation in the bottom strand if the 30-mer duplex DNA was used. Thus, we chose to employ a 24mer bottom strand for the methylation experiments. We compared the DNMT1-mediated methylation of Substrate 4 with the corresponding substrate where the top strand does not carry the 6-nucleotide overhang. Our results showed that these two substrates exhibited no significant difference toward the DNMT1-mediated methylation (Data not shown).
2. LC-MS/MS analysis of cytosine methylation at CpG site in ODNs
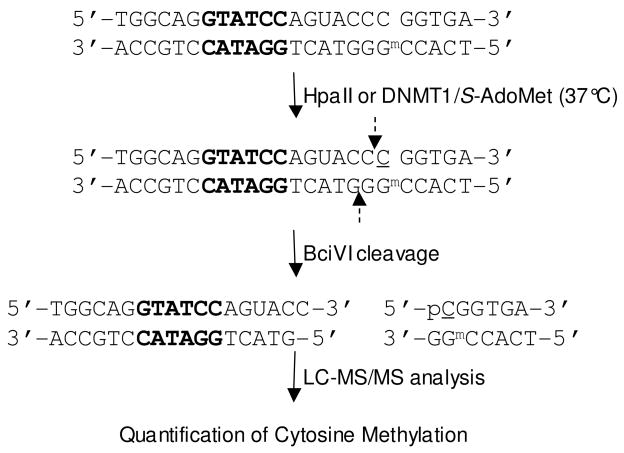
Figure 1 illustrates the method that we employed for examining the effect of 6- thioguanine on the methylation of cytosine at CpG site. In this context, the substrates carry a unique BciVI recognition site, thereby enabling the cleavage of the bottom initially methylated strand to two ODNs, e.g., d(TCACmCGG) and d(pGTACTGGATACCTGCCA) for Substrate 1. The corresponding cleavage of the top strand in Substrate 1 renders d(TGGCAGGTATCCAGUACC) and d(pCGGTGA), where the underlined “C” can be methylated or unmethylated.
Figure 1.
The experimental procedures for assessing the DNA methyltransferase-mediated cytosine methylation in synthetic duplex DNA. The underlined “C” represents unmethylated or methylated cytosine. The recognition site of BciVI is highlighted in bold and the cleavage sites of BciVI are indicated by broken arrows.
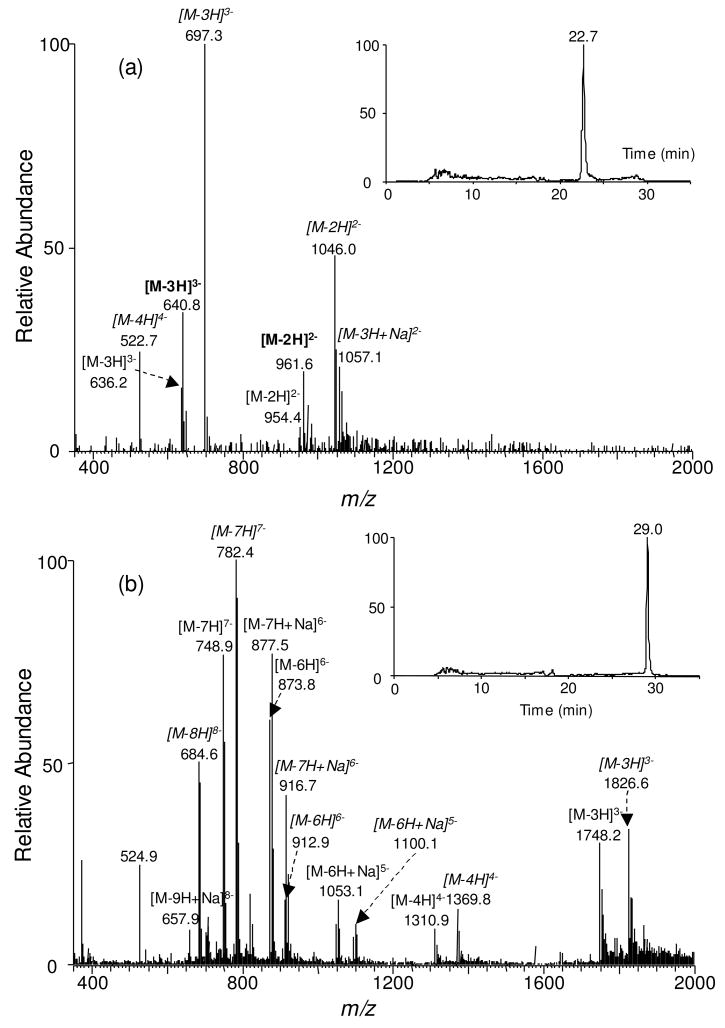
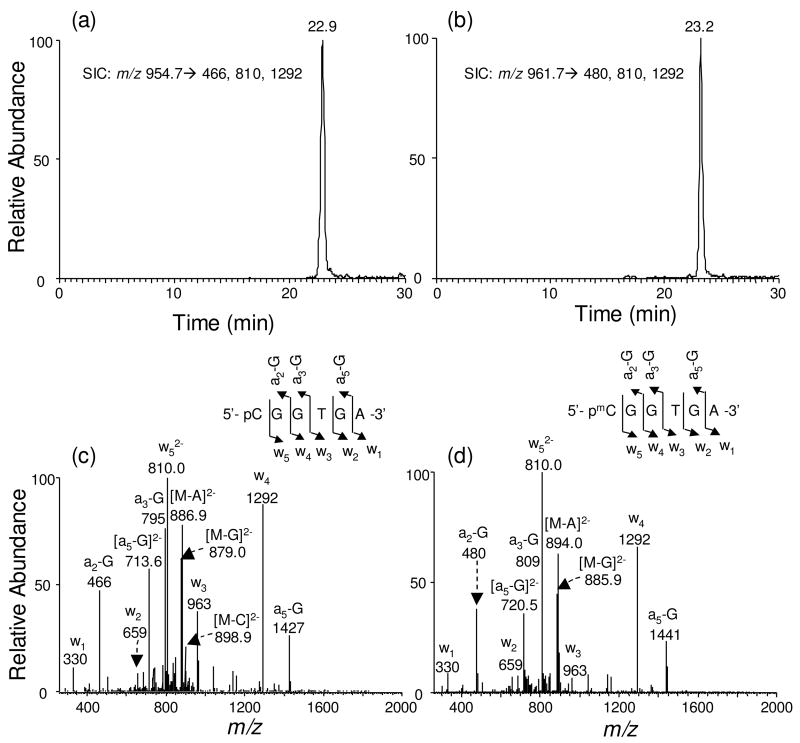
Indeed LC-MS/MS analysis of the BciVI cleavage products of the methylation reaction mixture of Substrate 1 showed the elution of the short 6- and 7-mer ODNs at ~23 min, and the long 17- and 18-mer ODNs at ~29 min (Figure 2). In this respect, the 6-mer methylated product d(pmCGGTGA) and its unmethylated counterpart, i.e., d(pCGGTGA), eluted at 23.2 min and 22.9 min, respectively (Figure 3a&b). The identities of the BciVI-produced ODNs were confirmed by MS/MS analysis. For instance, the production spectra of the [M − 2H]2− ions of the 6-mer ODNs (Figure 3c&d) showed the formation of wn ions, that is, the w1, w2, w3, w4, and w5, and [an-Base] ions, namely, the [a2-G], [a3-G] and [a5-G] ions [Nomenclature for fragment ions follows that described by McLuckey et al. (25). “A”, “G”, and “C” represent adenine, guanine and cytosine, respectively]. The [a2-G], [a3-G] and [a5-G] ions of d(pmCGGTGA) exhibited 14 Da higher in mass than the corresponding fragments formed from the unmethylated d(pCGGTGA). The MS/MS results clearly support the methylation of cytosine residue at CpG site (Figure 3c&d).
Figure 2.
Negative-ion ESI-MS of BciVI fragments from the DNMT1-mediated methylation reaction mixture of Substrate 1. Products corresponding to 5-pmol methylation reaction mixture was injected for LC-MS analysis. (a) ESI-MS of the short BciVI fragments averaged from the 22.7-min peak found in the SIC for monitoring the formation of d(pCGGTGA) (SIC shown in the inset). Multiply charged ions for d(pCGGTGA), d(pmCGGTGA), and d(TCACmCGG) are designated in normal, bold, and italic fonts, respectively. (b) ESI-MS of the longer BciVI fragments averaged from the 29.0-min peak found in the SIC for monitoring the formation of d(pGTACTGGATACCTGCCA) (SIC shown in the inset). Multiply charged ions for d(pGTACTGGATACCTGCCA) and d(TGGCAGGTATCCAGUACC) are labeled in normal and italic fonts, respectively.
Figure 3.
LC-MS/MS for monitoring the restriction fragments of interest with or without methylation at CpG site [i.e., d(pCCGTGA) and d(pmCGGTGA), which were from the BciVI cleavage of Substrate 1 after in-vitro methylation]. Shown in (a) and (b) are the SICs for monitoring the formation of indicated fragment ions of these two ODNs, and illustrated in (c) and (d) are the MS/MS of the [M – 2H]2− ions of the two ODNs (m/z 954.7 and 961.7). Nomenclature for fragment ions follows that described by McLuckey et al. (25)
The LC-MS/MS analysis combined with BciVI digestion also allowed for the monitoring of the methylation of other DNA substrates listed in Table 1, and example LC-MS/MS results are shown in Figures S1–S2 in the Supporting Information. In this context, it is worth emphasizing that our LC-MS/MS results revealed only the expected BciVI cleavage products; no other products could be detected, which underscores the specificity of BciVI cleavage.
3. 6-Thioguanine perturbs cytosine methylation at CpG site by both human DNMT1 and bacterial HpaII
Based on the peak areas found in the SICs (Figure 3a&b), we were able to quantify the extent of cytosine methylation by using the equation described in Materials and Methods. It is worth noting that, in the above quantification studies, the ionization and fragmentation efficiencies for the methylated 6- or 7-mer ODNs were assumed to be the same as their unmethylated counterparts. To assess whether this might introduce substantial error to the quantification, we digested an equi-molar mixture of hemimethylated Substrate 1 and the corresponding fully methylated duplex ODN, in which the internal cytosine at CCGG site in the top strand is also methylated, with BciVI and subjected the digestion mixture to LC-MS/MS analysis under the same experimental conditions as described above. It turned out that the efficiency for forming the three abundant fragment ions from the methylated substrate is approximately 10% greater than that for the unmethylated substrate. Thus, the above assumption does not result in significant error in measuring the level of methylation.
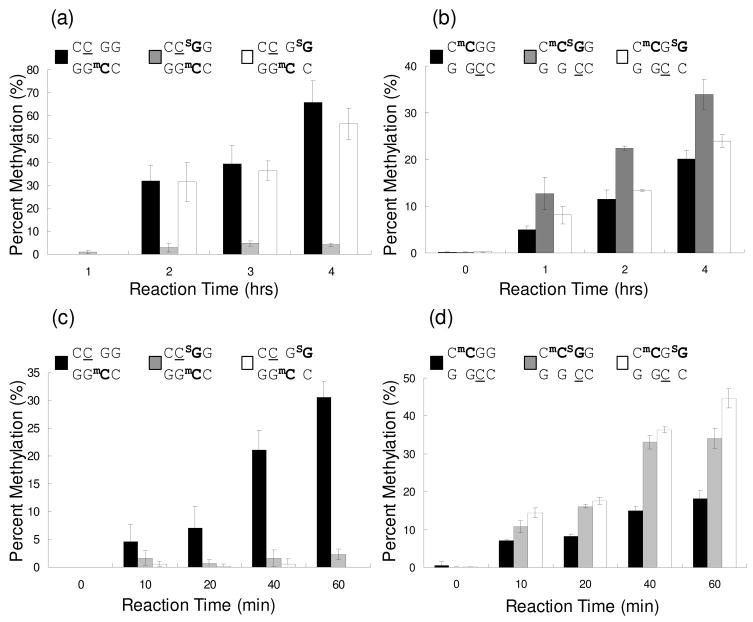
Our quantification data, summarized in Figure 4, revealed that the incorporation of SG into CpG site affected the methylation of nearby cytosine in duplex DNA by both human maintenance DNA methyltransferase DNMT1 and bacterial HpaII methylase. The presence of SG at CpG site suppressed strongly the methylation of its 5′ adjacent cytosine by both DNMT1 and HpaII (i.e., Substrate 2 vs. Substrate 1, Figure 4a and 4c). On the other hand, when SG was located immediately 3′ to the CpG site on the unmethylated strand of a hemimethylated target (Substrate 3 vs. Substrate 1), it did not bear any significant effect on cytosine methylation mediated by DNMT1 (Figure 4a). Substrate 3, however, displayed greatly diminished tendency toward HpaII-mediated cytosine methylation when compared to Substrate 1 (Figure 4c). Interestingly, relative to the control substrate, a higher level of HpaII-mediated methylation was found for substrates where either guanine at CCGG site in the opposing strand was replaced with a SG (compare Substrate 4 with Substrates 5 and 6, Figure 4d). However, the replacement of guanine at CpG site, but not the substitution of the flanking guanine 3′ to the CpG site, stimulated the DNMT1-mediated CpG methylation in the opposing strand (Figure 4b).
Figure 4.
Levels of cytosine methylation in different substrates induced by DNMT1 and HpaII methyltransferases. (a) DNMT1-induced methylation of cytosine at CpG site in Substrates 1, 2 and 3 (Table 1). (b) DNMT1-catalyzed methylation of cytosine at CpG site in Substrates 4, 5 and 6 (Table 1). (c) HpaII-mediated methylation of cytosine at CpG site in Substrates 1, 2 and 3. (d) HpaII-catalyzed methylation of cytosine at CpG site in Substrates 4, 5 and 6. The target cytosine residues to be methylated are underlined. The data represent the means and standard deviation of three independent methylation reactions and LC-MS/MS measurements. All results shown in panel (b) were obtained from UDG digestion, and the rest results were from BciVI cleavage (See Materials and Methods).
4. 6-Thioguanine treatment leads to decrease in global cytosine methylation in Jurkat T cells
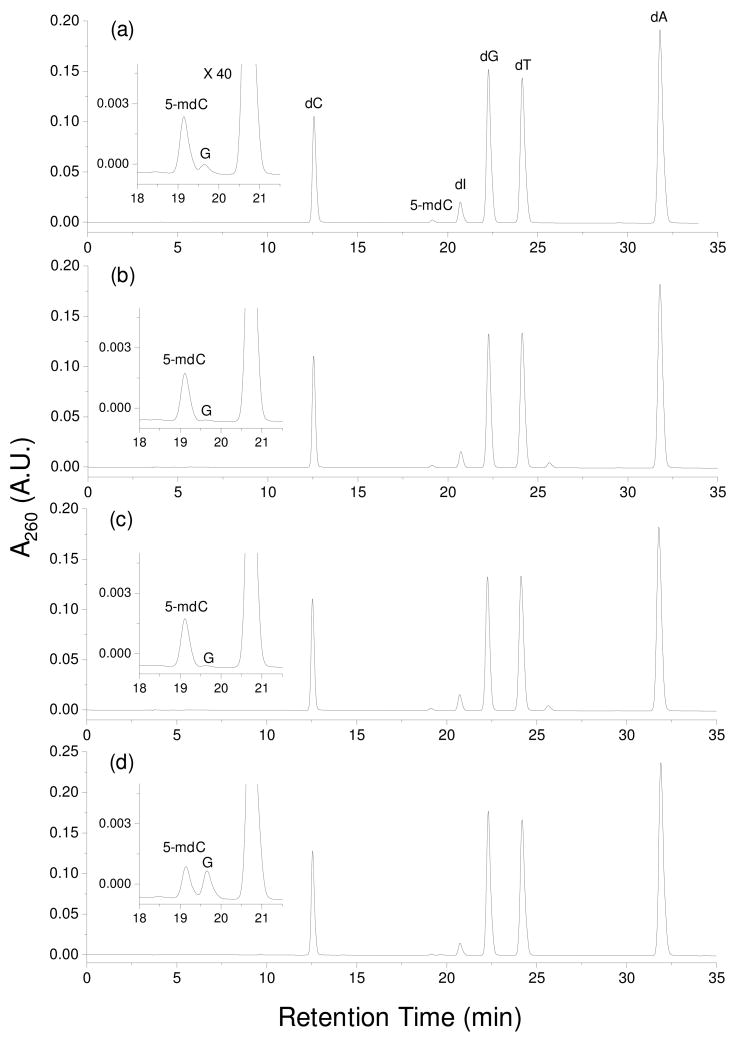
After having demonstrated the inhibitory effect of 6-thioguanine on the methylation of its neighboring 5′ cytosine by DNA methyltransferases, we next assessed whether 6-thioguanine can act as an inhibitor for cytosine methylation in human cells. On the grounds that thiopurine drugs have been successfully employed for treating acute lymphoblastic leukemia (ALL) (1) and aberrant cytosine methylation has been detected for a number of genes in bone marrow samples from ALL patients and two ALL cell lines (Jurkat-T and NALM-6 cells) (26), we chose to employ Jurkat T cells for this experiment. To this end, we treated Jurkat T cells with 6- thioguanine (at a concentration of 1 or 3 μM) or 5-aza-2′-deoxycytidine (5 μM) for 24 hrs. We then isolated the genomic DNA from the cells, digested the DNA with nuclease P1 and alkaline phosphatase, and assessed the level of cytosine methylation by HPLC analysis of the resulting nucleoside mixtures. Under optimized HPLC conditions (See Materials and Methods), we were able to resolve dC and 5-mdC from each other and from other nucleosides (Figure 5 and Figures S3–S4). From the integrated peak areas for dC and 5-mdC observed in the HPLC traces (Figure 5) and the extinction coefficients for the two nucleosides at 260 nm, we were able to quantify the percentage of cytosine methylation in cells that are untreated or treated with 6-thioguanine or 5- aza-2′-deoxycytidine.
Figure 5.
The HPLC traces for the separation of nucleoside mixtures produced from the digestion of genomic DNA isolated from Jurkat T cells that were: (a) untreated; (b) treated with 1 μM SG; (c) treated with 1 μM SG; (d) treated with 5 μM 5-aza-dC. Shown in the insets are the expanded chromatograms to visualize better the 5-mdC peak. “dI” and “G” represent 2′- deoxyinosine and guanosine, respectively. The former arises from the deamination of 2′- deoxyadenosine during enzymatic digestion. It is worth noting that the amount of RNA present in the isolated DNA varied from sample to sample, which resulted in different amounts of G in the nucleoside mixtures.
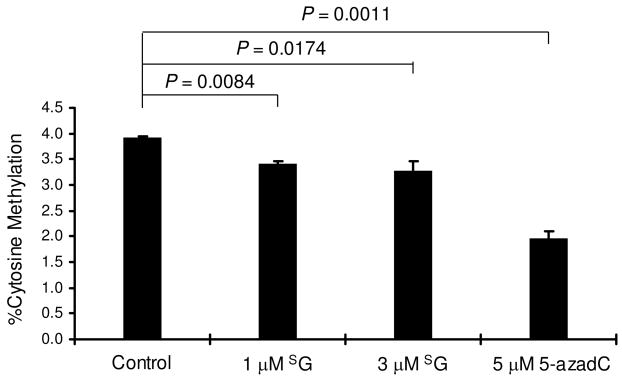
It turned out that the culturing of Jurkat T cells in a medium containing 1 or 3-μM of 6- thioguanine for 24 hrs led to appreciable decreases in global cytosine methylation, i.e., the percentage of cytosine methylation decreased from 3.92 ± 0.04% in untreated cells to 3.42 ± 0.04% and 3.39 ± 0.18% in cells treated with 1 and 3-μM of 6-thioguanine, respectively (Figure 6). Similar treatment with 5-μM of 5-aza-2′-deoxycytidine, a well-known inhibitor for cytosine methylation (27), resulted in the decrease of the percentage of cytosine methylation to 1.96 ± 0.15% (Figure 6).
Figure 6.
6-Thioguanine treatment results in decreased cytosine methylation in human cells. Plotted are the percentages of global cytosine methylation in genomic DNA isolated from Jurkat T cells that were untreated or treated with SG or 5-aza-dC. The data represent the means and standard deviations of results from three independent drug treatments and HPLC measurements. The P values were calculated by using paired t-test.
It is worth noting that the extents of decrease in cytosine methylation are modest, by 13–16% relative to the control untreated cells; however, the difference in the level of global cytosine methylation between SG-treated and untreated cells is statistically significant (Figure 6). In addition, the above drop in methylation level reflects the decrease in cytosine methylation in newly synthesized DNA in the background of previously methylated DNA that is not expected to change during the drug treatment.
Discussion
Thiopurine drugs have been available to medical practitioners as effective anticancer and immunosuppressive agents for more than half a century (1, 2). Although the biochemical mechanisms through which the thiopurines exert their cytotoxic effects remain elusive, the formation of thioguanine nucleotides and their subsequent incorporation into nucleic acids are known to be essential for the drugs to be effective (1). In this context, SG in DNA is chemically more reactive than guanine; it can be methylated by S-AdoMet to give S6mG and the resulting S6mG:T base pair may trigger the post-replicative mismatch repair pathway, which may account for the therapeutic effects of the thiopurine drugs (3). In addition, a recent study suggested that 6-thioguanine, because of its miscoding potential, may trigger the post-replicative mismatch repair pathway without being converted to S6mG (7).
In higher eukaryotes, cytosine methylation at CpG dinucleotide site is known to be important in gene regulation and essential for embryonic viability (10), and aberrant cytosine methylation is associated with cancer development (28, 29). We hypothesize that the thiopurine drugs may exert their anti-neoplastic effect via perturbing the methylation of cytosine at CpG site by DNA methyltransferases, thereby interfering with the epigenetic pathways of gene regulation.
To gain experimental evidence for supporting this hypothesis, we first developed an LC MS/MS coupled with restriction enzyme digestion method and assessed the human DNMT1- and HpaII-mediated methylation of cytosine in synthetic duplex DNA where a guanine residue at CCGG site was replaced with a SG. Our quantification results illustrated that the substitution of the guanine residue at CpG site with a SG inhibited greatly the methylation of the adjoining 5′ cytosine by DNMT1; however, little effect on the methylation of CpG dinucleotide was observed when the guanine 3′ neighboring to CpG site was replaced with a SG. In addition, the substitution of either guanine at CCGG site with a SG inhibited the HpaII-catalyzed methylation of the internal cytosine residue in the same strand. By contrast, the replacement of either guanine residue at CmCGG site leads to enhanced CpG methylation in the opposing strand by HpaII methylase, and only the change of the guanine residue at methylated CpG site with a SG stimulated the DNMT1-mediated methylation at CpG site in the opposing strand. The above results with the two different methyltransferases are consistent with the previous findings that the CpG dinucleotide is sufficient for DNMT1 binding and methylation (23), whereas HpaII binding and methylation requires the CCGG sequence motif (22).
The mechanism for the different effects of SG on cytosine methylation remains unclear. Nevertheless, previous X-ray structural studies with M. HhaI methyltransferase demonstrated that the methylation necessitated the flipping of cytosine residue out of DNA double helix and the fitting of this nucleobase into the catalytic pocket of the methyltransferase (30). On the basis of the increased atomic radius, bond length and the altered hydrogen bonding property of sulfur (31), we speculate that, when 6-thioguanine lies on the 3′ side of the target cytosine to be methylated, it may result in compromised binding between DNA and the methyltransferase, thereby resulting in abolished cytosine methylation. When the guanine residing on the opposite strand of the target CpG site was replaced with an SG, the weakened hydrogen bonding interaction between SG and cytosine (5) may render the facile flipping of the target cytosine from the double helix structure thereby enhancing the methylation of cytosine residue.
Our results are consistent with the previous studies about the effect of other guanine modifications on the methyltransferase-mediated cytosine methylation. In this regard, it was found that the substitution of guanine at CpG site with an 8-oxoG inhibited the methylation of its vicinal 5′ cytosine (13, 14). It was also observed that replacing the guanine at methylated CpG site with an 8-oxoG or O6mG stimulated the methylation of the opposing cytosine in the complementary strand (13–16).
The results from the above in-vitro biochemical experiments suggest that the incorporation of SG into the nascent DNA strand during DNA replication may result in diminished maintenance cytosine methylation, thereby perturbing the epigenetic pathway of gene regulation. By quantifying the level of global cytosine methylation with a direct HPLC method, we further showed that the treatment of Jurkat T cells with 1 or 3 μM 6-thioguanine for 24 hrs could lead to a significant decrease in global cytosine methylation. Along this line, by measuring the 3H/14C ratio in DNA isolated from cells pulse-labeled with L-[methyl-3H]methionine and [2-14C]thymidine, Hogarth, De Abreu and their coworkers (32, 33) showed that the treatment of MOLT-F4 human malignant lymphoblastic cells with SG or 6-mercaptopurine could give rise to a lower level of cytosine methylation in newly synthesized DNA. In addition, Hogarth et al. (33) observed that the treatment of MOLT-F4 cells with SG or MP could lead to a decrease in the enzymatic activity of DNMT1 in the whole cell lysate and a drop in the level of the DNMT1 protein. The decreased level of DNMT1 protein was also observed in cells treated with 5-aza-dC and this decrease was attributed to the degradation of DNMT1 by the proteasomal pathway (34). Although it remains unexplored whether the SG-induced decrease in DNMT1 protein level also arises from the proteasomal pathway, it is highly plausible that this pathway may also play an important role. 5-azacytidine and 5-aza-dC are in clinical trials for the treatment of various hematopoietic and solid malignancies but they are considered too toxic and mutagenic for long-term preventive therapy (35), whereas mercaptopurine along with methotrexate has been routinely used in the maintenance therapy for childhood ALL (2). The more potent inhibition of cytosine methylation exerted by 5-aza-dC than by thiopurines may account for the different pharmacological performances for the two types of drugs.
Taken together, the results from the present study support that the thiopurine drugs may exert their cytotoxic effects, at least in part, by perturbing the epigenetic pathway in vivo. In this context, it is worth noting that, in DNA samples isolated from peripheral blood mononuclear cells of patients receiving 6-mercaptopurine or azathioprine, approximately 0.01–0.1% of guanine are replaced with 6-thioguanine (36, 37). This level is orders of magnitude greater than the levels of typical guanine modification products induced by reactive oxygen species or alkylating agents.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by the National Institutes of Health (R01 CA101864).
Abbreviations
- S-AdoMet
S-adenosyl-L-methionine
- DTT
dithiothreitol
- HFIP
1,1,1,3,3,3- hexafluoroisopropanol
- TEAA
triethylammonium acetate
- SG
6-thioguanine
- 5-aza-dC
5-aza-2′- deoxycytidine
- mC
5-methylcytosine
- S6mG
S6-methylthioguanine
- SO3HG
guanine-S6- sulfonic acid
- 8-oxoG
8-oxo-7,8-dihydroguanine
- O6mG
O6-methylguanine
- B[a]P
benzo[a]pyrene
- UDG
uracil-DNA glycosylase
- ODN
oligodeoxynucleotides
- PAGE
polyacrylamide gel electrophoresis
- ESI-MS
electrospray ionization-mass spectrometry
- MS/MS
tandem mass spectrometry
- SIC
selected-ion chromatogram
Footnotes
Supporting Information Available. LC-MS/MS data for monitoring the level of cytosine methylation in synthetic DNA and for confirming the presence of 5-mdC and guanosine in the nucleoside mixture arising from the hydrolysis of genomic DNA. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Elion GB. The purine path to chemotherapy. Science. 1989;244:41–47. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Evans WE. Acute lymphoblastic leukemia. New Engl J Med. 1998;339:605–615. doi: 10.1056/NEJM199808273390907. [DOI] [PubMed] [Google Scholar]
- 3.Swann PF, Waters TR, Moulton DC, Xu YZ, Zheng QG, Edwards M, Mace R. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 4.O’Donovan P, Perrett CM, Zhang XH, Montaner B, Xu YZ, Harwood CA, McGregor JM, Walker SL, Hanaoka F, Karran P. Azathioprine and UVA light generate mutagenic oxidative DNA damage. Science. 2005;309:1871–1874. doi: 10.1126/science.1114233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu C, Wang Y. In vitro replication and thermodynamic studies of methylation and oxidation modifications of 6-thioguanine. Nucleic Acids Res. 2007;35:3693–3704. doi: 10.1093/nar/gkm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang XH, Jeffs G, Ren XL, O’Donovan P, Montaner B, Perrett CM, Karran P, Xu YZ. Novel DNA lesions generated by the interaction between therapeutic thiopurines and UVA light. DNA Repair. 2007;6:344–354. doi: 10.1016/j.dnarep.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Yuan B, Wang Y. Mutagenic and cytotoxic properties of 6-thioguanine, S6-methylthioguanine, and guanine-S6-sulfonic acid. J Biol Chem. 2008;283:23665–23670. doi: 10.1074/jbc.M804047200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters TR, Swann PF. Cytotoxic mechanism of 6-thioguanine: hMutS alpha, the human mismatch binding heterodimer, binds to DNA containing S-6- methylthioguanine. Biochemistry. 1997;36:2501–2506. doi: 10.1021/bi9621573. [DOI] [PubMed] [Google Scholar]
- 9.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 10.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 11.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Turk PW, Laayoun A, Steven SS, Weitzman SA. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of Human DNA methyltransferase. Carcinogenesis. 1995;16:1253–1255. doi: 10.1093/carcin/16.5.1253. [DOI] [PubMed] [Google Scholar]
- 14.Weitzman SAT, Patrick W, Milkowski Deborah Howard, Kozlowski Karen. Free radical adducts induce alterations in DNA cytosine methylation. Proc Natl Acad Sci USA. 1994;91:1261–1264. doi: 10.1073/pnas.91.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan NW, Li BF. Interaction of oligonucleotides containing 6-Omethylguanine with human DNA (cytosine-5-)-methyltransferase. Biochemistry. 1990;29:9234–9240. doi: 10.1021/bi00491a018. [DOI] [PubMed] [Google Scholar]
- 16.Smith SS, Kan JL, Baker DJ, Kaplan BE, Dembek P. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J Mol Biol. 1991;217:39–51. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- 17.Subach OM, Maltseva DV, Shastry A, Kolbanovskiy A, Klimasauskas S, Geacintov NE, Gromova ES. The stereochemistry of benzo[a]pyrene-2′-deoxyguanosine adducts affects DNA methylation by SssI and HhaI DNA methyltransferases. FEBS J. 2007;274:2121–2134. doi: 10.1111/j.1742-4658.2007.05754.x. [DOI] [PubMed] [Google Scholar]
- 18.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 19.Ling YH, Chan JY, Beattie KL, Nelson JA. Consequences of 6-thioguanine incorporation into DNA on polymerase, ligase, and endonuclease reactions. Mol Pharmacol. 1992;42:802–807. [PubMed] [Google Scholar]
- 20.Krynetskaia NF, Krynetski EY, Evans WE. Human RNase H-mediated RNA cleavage from DNA-RNA duplexes is inhibited by 6-deoxythioguanosine incorporation into DNA. Mol Pharmacol. 1999;56:841–848. [PubMed] [Google Scholar]
- 21.Krynetskaia NF, Cai X, Nitiss JL, Krynetski EY, Relling MV. Thioguanine substitution alters DNA cleavage mediated by topoisomerase II. FASEB J. 2000;14:2339–2344. doi: 10.1096/fj.00-0089com. [DOI] [PubMed] [Google Scholar]
- 22.Quint A, Cedar H. In vitro methylation of DNA with Hpa II methylase. Nucleic Acids Res. 1981;9:633–646. doi: 10.1093/nar/9.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- 24.Apffel A, Chakel JA, Fischer S, Lichtenwalter K, Hancock WS. Analysis of oligonucleotides by HPLC-electrospray ionization mass spectrometry. Anal Chem. 1997;69:1320–1325. doi: 10.1021/ac960916h. [DOI] [PubMed] [Google Scholar]
- 25.McLuckey SA, Vanberkel GJ, Glish GL. Tandem mass-spectrometry of small, multiply charged oligonucleotides. J Am Soc Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 26.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, Rahmatpanah FB, Sjahputera O, Caldwell CW. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007;67:2617–2625. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- 27.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 28.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 29.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 30.O’Gara M, Horton JR, Roberts RJ, Cheng X. Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nat Struct Biol. 1998;5:872–877. doi: 10.1038/2312. [DOI] [PubMed] [Google Scholar]
- 31.Platts JA, Howard ST, Bracke BRF. Directionality of hydrogen bonds to sulfur and oxygen. J Am Chem Soc. 1996;118:2726–2733. [Google Scholar]
- 32.Lambooy LH, Leegwater PA, van den Heuvel LP, Bokkerink JP, De Abreu RA. Inhibition of DNA methylation in malignant MOLT F4 lymphoblasts by 6-mercaptopurine. Clin Chem. 1998;44:556–559. [PubMed] [Google Scholar]
- 33.Hogarth LA, Redfern CP, Teodoridis JM, Hall AG, Anderson H, Case MC, Coulthard SA. The effect of thiopurine drugs on DNA methylation in relation to TPMT expression. Biochem Pharmacol. 2008;76:1024–1035. doi: 10.1016/j.bcp.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 34.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci USA. 1997;94:4681–4685. doi: 10.1073/pnas.94.9.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren DJ, Andersen A, Slordal L. Quantitation of 6-thioguanine residues in peripheral-blood leukocyte DNA obtained from patients receiving 6- mercaptopurine-based maintenance therapy. Cancer Res. 1995;55:1670–1674. [PubMed] [Google Scholar]
- 37.Cuffari C, Li DY, Mahoney J, Barnes Y, Bayless TM. Peripheral blood mononuclear cell DNA 6-thioguanine metabolite levels correlate with decreased interferon-gamma production in patients with Crohn’s disease on AZA therapy. Dig Dis Sci. 2004;49:133–137. doi: 10.1023/b:ddas.0000011614.88494.ee. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.