Abstract
Purpose
Cisplatin and gemcitabine have single-agent activity in metastatic breast cancer, and preclinical data support synergy of the combination. Two parallel, phase II trials were conducted to evaluate the response rate, response duration, and toxicities of the combination. Genetic polymorphisms were analyzed for correlation with outcomes.
Patients and Methods
Eligible women had measurable disease and heavily or minimally pretreated metastatic breast cancer. The heavily pretreated protocol required prior anthracycline and taxane therapy; cisplatin as part of high-dose therapy was allowed. All patients received cisplatin 25 mg/m2 on days 1 through 4 and gemcitabine 1,000 mg/m2 on days 2 and 8 of a 21-day cycle with prophylactic granulocyte colony-stimulating factor in the heavily pretreated group. Sera from a subset of patients were evaluated by polymerase chain reaction restriction fragment length polymorphism for polymorphisms in 10 genes of interest.
Results
Of 136 women enrolled, 74 were heavily pretreated. Both protocols accrued to their two-stage design. The response rate for both the heavily and minimally pretreated cohorts was 26%, and the median durations of response were 5.3 and 5.9 months, respectively. In a multivariate analysis, hormone receptor–negative disease was associated with a higher response rate. The most common grades 3 or 4 toxicities were thrombocytopenia (71%), neutropenia (66%), and anemia (38%). In a subset of 55 patients, the xeroderma pigmentosum group D (XPD)–751, x-ray cross-complementing group 3 (XRCC3) and cytidine deaminase polymorphisms were significantly associated with clinical outcomes.
Conclusion
Combination cisplatin and gemcitabine is active in metastatic breast cancer regardless of prior therapy. Genetic polymorphisms may tailor which patients benefit from this regimen.
INTRODUCTION
Anthracyclines and taxanes have the highest activity in untreated metastatic breast cancer. However, many patients have been exposed to these agents in the adjuvant setting, which precludes their front-line use.1–3 Although single-agent cisplatin in first-line metastatic breast cancer has resulted in a respectable 47% response rate,4 limited data support its use in previously treated disease.
Gemcitabine, a nucleoside analog, also has single-agent activity in metastatic breast cancer.5 Its mechanism of action is dependent on the cell cycle, and it is metabolized into its inactive compound by cytidine deaminase (CDA) and other enzymes. Preclinical data demonstrate cytotoxic synergy for the combination of cisplatin and gemcitabine. Specifically, gemcitabine may inhibit repair of cisplatin-induced DNA adducts in a schedule-dependent fashion.6,7
Varying response to cisplatin chemotherapy may be partially attributed to altered DNA repair capacity. DNA repair enzymes, such as xeroderma pigmentosum group D (XPD) and excision repair cross-complementation group 1 (ERCC1), are implicated in the nucleotide excision repair pathway, and components of this pathway are thought to be the principal elements in platinum-adduct removal. Other enzymes include the x-ray cross-complementing group 1 (XRCC1) and group 3 (XRCC3), which are implicated in double-stranded break repair.8–10 Polymorphisms in all of these DNA-repair genes that result in altered functional activity have been associated with clinical outcome in various cancer types.11–13
The California Cancer Consortium and Loyola University Chicago designed two phase II trials of combination cisplatin and gemcitabine in two populations, one heavily and one minimally pretreated. The rationale was to explore synergy with these two agents by using a unique dose and schedule to duplicate optimal in vitro conditions.7 The schedule of cisplatin, daily for 4 days, was similar to the front-line single-agent study.4 Gemcitabine was administered midway through the cisplatin schedule, and repeat dosing was given on day 8 to inhibit repair of late-forming adducts and to optimize synergy. Polymorphisms in 10 genes involved in pathways relevant to gemcitabine and cisplatin—specifically to DNA repair, cell cycle control, and drug metabolism—were evaluated for correlation with clinical outcomes in an optional ancillary study.
PATIENTS AND METHODS
Patient Eligibility
Eligible patients were 18 years of age or older and had histologically confirmed, measurable, metastatic or locally recurrent breast cancer not amendable to definitive surgical resection. Patients were enrolled onto two parallel protocols. The heavily pretreated protocol required prior treatment with at least two chemotherapy regimens for metastatic disease or disease progression after bone marrow or hematopoeitic cell transplantation in the adjuvant or metastatic setting. In this study, patients must have received anthracycline or taxane therapy in either the adjuvant or metastatic setting. Prior cisplatin was allowed only as part of high-dose chemotherapy (HDCT). The minimally pretreated protocol limited prior treatment for metastatic disease to no more than one prior regimen, and previous cisplatin or gemcitabine was not allowed. All patients had to have a Karnofsky performance status ≥ 60% and adequate hematologic, renal, and hepatic function, as indicated by an absolute neutrophil count (ANC) ≥ 1,500/μL, platelets ≥ 100,000, creatinine clearance ≥ 50 mL/min, bilirubin ≤ 2 mg/dL, and AST and ALT levels four or fewer times the institutional limit of normal. Patients with a history of brain metastases were allowed if they had been treated, were off of corticosteroids, and were asymptomatic. Patients were excluded if they were pregnant, had another invasive cancer within 2 years, had any prior cancer not in remission, or had prior strontium therapy. Patients were encouraged to participate in the ancillary biomarker study.
All patients were informed of the investigational nature of the study, and they provided voluntary written informed consent in accordance with institutional and federal guidelines. Each protocol was approved by the respective institutional review boards of all participating sites.
Study Design and Treatment
Protocol treatment consisted of cisplatin 25 mg/m2 intravenously (IV) daily on days 1 through 4 and gemcitabine 1,000 mg/m2 IV on days 2 and 8 of a 21-day cycle. Patients on the heavily pretreated protocol received granulocyte colony-stimulating factor (G-CSF) 5 μg/kg daily on days 5 through 7 and from day 9 until the absolute neutrophil count exceeded 10,000/μL. G-CSF was not used in cycle 1 of the minimally pretreated protocol, but it was allowed in subsequent cycles at the discretion of the treating physician.
Disease Assessment
Measurable disease was defined as bidimensionally measurable lesions that had clearly defined margins by medical photograph or plain x-ray, with at least one diameter of 0.5 cm or greater, excluding bone lesions; computed tomography, magnetic resonance imaging, or other imaging, in which both diameters were greater than the distance between cuts of the study; or palpation, in which both diameters were 1 cm or greater. Baseline radiographic studies were performed within 4 weeks before initial protocol treatment; they were repeated after the initial three cycles and then after every two subsequent treatment cycles. Objective responses were confirmed 6 weeks later with repeat radiographic studies of the area of responsive disease.
Toxicity Assessment
Patients were evaluated for treatment-related toxicity at a minimum of every 21 days, per the National Cancer Institute Common Toxicity Criteria, version 2.0. The worst grade of toxicity per patient was recorded. Dose-modification criteria were specified for subsequent cycles in each protocol.
Genotyping
Serum samples were obtained at time of entry into the treatment protocol from a subset of patients who elected to participate in the biomarker study. Genomic DNA was extracted from approximately 200 μL of serum that was isolated from whole blood by using a rapid DNA extraction from serum method. Briefly, serum was microcentrifuged for 10 minutes, supernatant was discarded, and 20 μL of 50 mmol/L NaOH was added. The solution was boiled for 10 minutes, 2.5 μL of 1M trisHCL was added, and the resulting solution was mixed thoroughly.
All samples were analyzed with a polymerase chain reaction (PCR) restriction fragment length polymorphism technique. The PCR reaction volume was 25 μL. Digested fragments were visualized on 3% agarose gel. Primers and restriction enzymes for each of the 13 polymorphisms are listed in Appendix Table A1.
Statistical Methods
Patients were accrued via a two-stage design on each protocol.14 The primary objectives were to evaluate the response rate and the response duration to combination chemotherapy with cisplatin and gemcitabine. Overall survival (OS) and toxicities also were evaluated. The response rate was defined as complete response (CR) or a partial response (PR) that was confirmed by a second evaluation approximately 3 to 6 weeks later. Response duration was defined as the time from first objective status assessment of CR/PR to the first time of progression or death as a result of any cause. Progression-free survival (PFS) was defined as the time from the first day of treatment to the first observation of disease progression or death as a result of any cause. OS was defined as the time from the first day of treatment to the time of death as a result of any cause.
RESULTS
Patients
A total of 136 women were enrolled at four institutions between August 1998 and August 2003. Both studies completed planned accrual, and 135 patients were eligible. One patient was ineligible on the heavily pretreated study because of nonmeasurable disease. Patient characteristics are listed in Table 1. Seventy-four patients were accrued to the heavily pretreated study, including 13 who had prior HDCT and 43 with four or more prior regimens. Sixty-two patients were enrolled in the minimally pretreated study, of which 38 had no prior chemotherapy for metastatic disease. The median age was 46 years (range, 21 to 70 years); 79% were white; and the median Karnofsky performance status was 90%. The majority of patients had received prior adjuvant chemotherapy, and 46% had known hormone-receptor–positive breast cancer. Tumor human epidermal growth factor receptor 2 (HER-2)/neu status was not collected.
Table 1.
Patient Characteristics
| Characteristic | Protocol |
|
|---|---|---|
| Heavily Pretreated (n = 74) | Minimally Pretreated (n = 62) | |
| Age at diagnosis, years | ||
| Median | 45 | 46 |
| Range | 21-70 | 29-70 |
| Karnofsky performance status, % | ||
| Median | 90 | 90 |
| Range | 60-100 | 60-100 |
| No. of prior treatments for metastatic disease | ||
| 0 | 0 | 38 |
| 1 | 0 | 24 |
| ≥ 2 or prior high-dose chemotherapy | 74* | 0 |
| No. of visceral sites | ||
| 0 | 21 | 14 |
| 1 | 34 | 35 |
| 2 | 17 | 13 |
| ≥ 3 | 2 | 0 |
| Prior adjuvant chemotherapy | ||
| No. | 37 | 39 |
| % | 50 | 63 |
| Tumor hormone receptor status | ||
| ER- and/or PgR-positive | ||
| No. | 37 | 25 |
| % | 50 | 40 |
| ER- and/or PgR-negative | ||
| No. | 22 | 21 |
| % | 30 | 34 |
| ER- and/or PgR-unknown | ||
| No. | 15 | 16 |
| % | 20 | 26 |
Abbreviations: ER, estrogen receptor; PgR, progesterone receptor.
One patient was ineligible on the heavily pretreated arm because of nonmeasureable disease.
Treatment and Efficacy
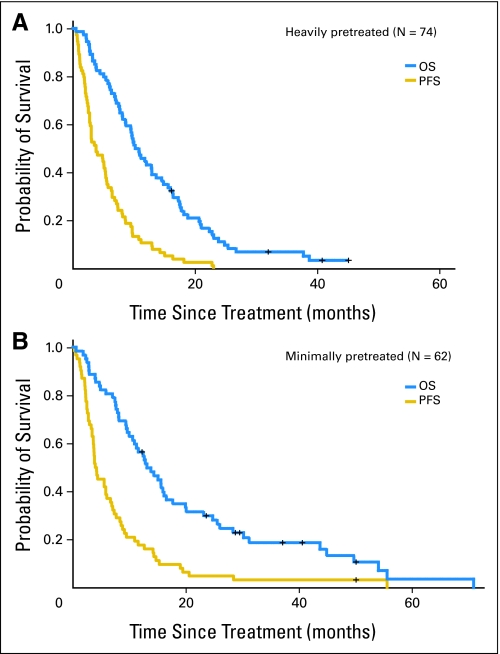
Patients on the heavily and minimally pretreated studies received a median of three (range, one to 17) and four (range, one to 17) cycles of protocol therapy, respectively. Table 2 lists the outcomes of the patients treated with cisplatin and gemcitabine for the intent-to-treat analysis. The overall response rate was 26% for both the heavily pretreated (95% CI, 16% to 37%) and the minimally pretreated (95% CI, 16% to 38%) studies. Durations of response were 5.3 months and 5.9 months in the heavily and minimally pretreated groups, respectively. The median OS rates were 10.8 months (95% CI, 8.6 to 14.5 months) in the heavily pretreated study and 13.1 months (95% CI, 10.9 to 17.6 months) in the minimally pretreated study. The median PFS rates were 3.8 months (95% CI, 2.7 to 5.4 months) and 4.2 months (95% CI, 3.6 to 6.0 months) in the heavily and minimally pretreated studies, respectively. Figures 1A and 1B show the Kaplan and Meier plots for OS and PFS among the heavily pretreated and minimally pretreated groups, respectively, for the intent-to-treat analysis. An unplanned subgroup analysis was performed for response rate (RR) by hormone receptor (HR) status. No difference was seen in the heavily pretreated cohort. However, in the minimally pretreated study, two (8%) of 25 patients with HR-positive disease responded compared with nine (43%) of 21 with HR-negative disease (P < .05). In a multivariate analysis, HR-negative disease was associated with a higher RR independent of the number of prior regimens or visceral sites.
Table 2.
Patient Outcomes
| End Point | Protocol |
|
|---|---|---|
| Heavily Pretreated (n = 74) | Minimally Pretreated (n = 62) | |
| Partial response | ||
| No. | 16 | 13 |
| % | 22 | 21 |
| Complete response | ||
| No. | 3 | 3 |
| % | 4 | 5 |
| Overall response | ||
| % | 26 | 26 |
| 95% CI | 16 to 37 | 16 to 38 |
| Response duration, months | ||
| Median | 5.3 | 5.9 |
| Range | 2.0 to 21.2 | 3.0 to 27.0 |
| Progression-free duration, months | ||
| Median | 3.8 | 4.2 |
| 95% CI | 2.7 to 5.4 | 3.6 to 6.0 |
| Overall survival, months | ||
| Median | 10.8 | 13.1 |
| 95% CI | 8.6 to 14.5 | 10.9 to 17.6 |
Fig 1.
Kaplan-Meier plots of overall survival (OS) and progression-free survival (PFS) in the (A) heavily pretreated and (B) minimally pretreated cohorts. The median OS and PFS rates were 10.8 months (95% CI, 8.6 to 14.5 months) and 3.8 months (95% CI, 2.7 to 5.4 months), respectively, in the heavily pretreated and 13.1 months (95% CI, 10.9 to 17.6 months) and 4.2 months (95% CI, 3.6 to 6.0 months), respectively, in the minimally pretreated cohorts.
Toxicity
All patients were assessable for toxicity. Table 3 lists the grades 3 and 4 toxicities. The most common toxicities for the entire cohort included thrombocytopenia (71% of patients), neutropenia (66%), anemia (38%), fatigue (18%), and nausea and vomiting (15%). Within the heavily pretreated group, 49% developed grade 4 neutropenia, and 3% had grade 4 febrile neutropenia despite the use of prophylactic G-CSF. Of the 74 heavily pretreated patients, 41 (55%) had protocol-specified dose delays, and 11 (15%) discontinued treatment because of toxicity. Electrolyte abnormalities were reported in 12% of the patients, which included one report of grade 4 hypocalcemia and one report of grade 4 hypokalemia. The remainder of the electrolyte abnormalities was grade 3 and included hypokalemia, hyperglycemia, hypomagnesemia, hyponatremia, and hypophosphatemia. More grade 4 toxicities occurred in the heavily pretreated group. There were no treatment-related deaths.
Table 3.
Grades 3 and 4 Toxicities
| Description of Toxicity | Toxicity Grade by Protocol Type* |
|||||||
|---|---|---|---|---|---|---|---|---|
| Heavily Pretreated (n = 74) |
Minimally Pretreated (n = 62) |
|||||||
| 3 |
4 |
3 |
4 |
|||||
| No. | % | No. | % | No. | % | No. | % | |
| Neutropenia | 10 | 14 | 36 | 49 | 16 | 26 | 28 | 45 |
| Anemia | 32 | 43 | 7 | 9 | 10 | 16 | 2 | 3 |
| Thrombocytopenia | 21 | 28 | 38 | 51 | 24 | 39 | 13 | 21 |
| Bleeding | 7 | 9 | 0 | 0 | 2 | 3 | 0 | 0 |
| Fatigue | 5 | 7 | 3 | 4 | 15 | 24 | 1 | 2 |
| Nausea or vomiting | 8 | 11 | 0 | 0 | 13 | 21 | 0 | 0 |
| Febrile neutropenia | 3 | 4 | 2 | 3 | 1 | 2 | 0 | 0 |
| Infection | 4 | 5 | 1 | 1 | 2 | 3 | 0 | 0 |
| Sensory neuropathy | 3 | 4 | 0 | 0 | 6 | 10 | 0 | 0 |
| Motor neuropathy | 3 | 4 | 0 | 0 | 1 | 2 | 0 | 0 |
| Lft abnormality | 2 | 3 | 0 | 0 | 5 | 8 | 0 | 0 |
| Electrolyte/metabolic | 6 | 8 | 1 | 1 | 8 | 13 | 1 | 2 |
Abbreviation: LFT, liver function test.
Grade used was the worst recorded per patient.
Genotyping
Serum from 55 patients was available for evaluation of 13 polymorphisms in the 10 genes of interest; 44% of these patients were heavily pretreated. There were no statistically significant differences in baseline characteristics or clinical outcomes among the 55 patients compared with the 81 who did not participate. Genotyping assays for the polymorphisms were successful, as follows: 55 patients for GSTP1; 54 patients for CDA Lyn27Gln, XPD-751, ERCC1 3′UTR, and p53 codon72; 53 patients for XPD-156, XRCC1, and ERCC1-118; 52 patients for p53-13964; 51 patients for GSTM1 and GSTT1; 49 patients for XRCC3; and 48 patients for HER2.
Table 4 lists the XPD and XRCC3 genomic polymorphisms that are significantly associated with clinical outcomes in the subset of 55 patients. The XPD-751 polymorphism was significantly associated with increased OS (P = .008) and PFS (P < .001) among patients who carried the Lys allele compared with those who had the Gln allele. The XRCC3 polymorphism was significantly associated with RR (P = .002) and PFS (P = .03) among patients who carried the Thr/Thr genotype compared with the heterozygous Met/Thr allele. The associations between the XPD-751 and XRCC3 polymorphisms and PFS remained significant in the multivariate Cox model stratified by study and ethnicity (P = .004 and P = .022, respectively, likelihood ratio test; data not shown). The remaining polymorphisms tested did not associate with RR, PFS, or OS (data not shown). Table 5 lists baseline characteristics, genomic polymorphisms, and toxicity (grades ≥ 3 nonhematologic or grade 4 hematologic) among the 55 patients. White patients were more likely to have toxicity from the combination of cisplatin and gemcitabine compared with nonwhite patients (P = .004). CDA polymorphism was also significantly associated with overall toxicity. Patients who carried the Gln variant allele experienced more toxicity than patients who carried the homozygous Lys/Lys genotype (P = .036). However, the association was not significant in the multivariate Cox model stratified by study and ethnicity (P = .90; data not shown). CDA polymorphisms were highly associated with ethnicity, in which white patients (12 of 29) were more likely (P < .01) to carry a variant CDA polymorphism compared with other groups (one of 25). The remaining polymorphisms tested did not associate with toxicity (data not shown).
Table 4.
Genomic Polymorphisms and Clinical Outcome Among 55 Patients
| Polymorphism by Gene | No. | Response* |
Survival Outcomes |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Progression Free |
Overall |
|||||||||||||
| CR and PR |
P† | Duration |
Analysis |
Duration |
Analysis |
|||||||||
| No. | % | Median (months) | 95% CI | Relative Risk | 95% CI | P† | Median (months) | 95% CI | Relative Risk | 95% CI | P† | |||
| XPD 751 | .14 | .0003 | .008 | |||||||||||
| Lys/Lys | 35 | 10 | 29 | 3.8 | 3.0 to 5.7 | 1‡ | 10.5 | 7.7 to 17.2 | 1‡ | |||||
| Lys/Gln | 14 | 7 | 50 | 5.5 | 4.0 to 8.6 | 0.7 | 0.4 to 1.3 | 12.9 | 7.1 to 15.9 | 0.8 | 0.4 to 1.6 | |||
| Gln/Gln | 5 | 0 | 0 | 1.2 | 0.3 to 3.5 | 4.5 | 1.5 to 13.0 | 2.5 | 0.6 to 11.1 | 3.5 | 1.3 to 9.8 | |||
| XRCC3 | .002 | .03 | .19 | |||||||||||
| Met/Met | 18 | 8 | 45 | 6.4 | 1.9 to 8.6 | 1‡ | 11.2 | 3.8 to 18.8 | 1‡ | |||||
| Thr/Met | 26 | 4 | 15 | 3.5 | 2.3 to 4.0 | 1.9 | 1.0 to 3.6 | 9.5 | 4.9 to 13.1 | 1.5 | 0.8 to 2.9 | |||
| Thr/Thr | 5 | 4 | 80 | 6.6 | 5.3 to 20.5 | 0.7 | 0.3 to 1.9 | 13.8 | 10.8 to 54.0 | 0.8 | 0.3 to 2.1 | |||
Five patients who did not receive two courses of treatment and who did return for evaluation were excluded.
P was based on Fisher's exact test for response and on the log-rank test for progression-free survival and overall survival.
Reference value.
Table 5.
Baseline Characteristics, Genomic Polymorphisms, and Toxicity Among 55 Patients
| Variable | No. of Patients | Toxicity Analysis* |
|||
|---|---|---|---|---|---|
| Relative Risk | 95% CI | Probability at Cycle 1 | P† | ||
| Age, years | .20 | ||||
| ≤ 49 | 28 | 1‡ | 0.57 ± 0.09 | ||
| ≥ 50 | 27 | 0.75 | 0.40 to 1.41 | 0.48 ± 0.10 | |
| Ethnicity | .004 | ||||
| White | 30 | 1‡ | 0.70 ± 0.08 | ||
| Other | 25 | 0.51 | 0.27 to 1.00 | 0.32 ± 0.09 | |
| Performance status | .73 | ||||
| 60-80 | 19 | 1‡ | 0.47 ± 0.12 | ||
| ≥ 90 | 36 | 1.09 | 0.56 to 2.08 | 0.56 ± 0.08 | |
| No. of prior chemotherapy for metastatic disease | .16 | ||||
| 0 | 19 | 1‡ | 0.47 ± 0.11 | ||
| 1 | 12 | 0.73 | 0.28 to 1.86 | 0.33 ± 0.14 | |
| ≥ 2 | 24 | 1.28 | 0.64 to 2.56 | 0.67 ± 0.10 | |
| CDA | .036 | ||||
| Lys/Lys | 41 | 1‡ | 0.41 ± 0.08 | ||
| Lys/Gln | 8 | 1.71 | 0.73 to 4.02 | 0.88 ± 0.12 | |
| Gln/Gln | 5 | 2.05 | 0.76 to 5.51 | 0.80 ± 0.18 | |
Abbreviation: CDA, cytidine deaminase.
Toxicity was defined as grade 3 or greater nonhematologic toxicity or grade 4 hematologic toxicity.
P was based on the log-rank test.
Reference value.
DISCUSSION
We report the results of two parallel, phase II trials in metastatic breast cancer; each completed two-stage accrual in a multi-institution setting. To our knowledge, the trials are the largest published in advanced breast cancer that utilize the combination of cisplatin and gemcitabine and that test a dose and schedule designed to duplicate the optimal conditions of in vitro synergy. The populations were clearly defined, and one study focused on heavily pretreated patients, including patients who had prior HDCT. Both studies demonstrated activity and tolerance.
Other investigators studied alternate dosing schedules of cisplatin and gemcitabine in previously treated patients but allowed varied amounts of prior therapy, which makes these studies difficult to compare directly to each other. Nagourney et al15 used a weekly regimen of the combination and reported an RR of 50% in 30 heavily pretreated patients, but this regimen required modification because of cytopenias. A study of 38 patients with prior treatment with anthracyclines and taxanes reported a 40% overall RR with weekly cisplatin and gemcitabine as second-line therapy for metastatic breast cancer.16 The differences in RR observed among these studies, including this study, most likely reflect patient selection as well as differences in dosing and schedule. Nevertheless, the responses observed in our heavily pretreated study are encouraging and suggest that the combination has clinically meaningful activity.
Regarding our minimally pretreated cohort, the overall RR of 26% is similar to data of prior published studies of cisplatin and gemcitabine combinations in metastatic breast cancer. The North Central Cancer Treatment Group17 conducted a phase II study of weekly cisplatin and gemcitabine in a similar population of 58 patients with metastatic breast cancer and reported an RR of 29% and a median time to progression (TTP) of 6 months. Protocol modification was necessary secondary to a 38% incidence of thrombocytopenia in the first stage of accrual. In a phase II trial of 42 patients conducted in Mexico, an RR of 81% to the combination in the first-line treatment of metastatic breast cancer was reported.18 An unplanned, subgroup, multivariate analysis in this study revealed a higher RR among minimally pretreated patients with HR-negative disease compared with HR-positive disease. Although these results are provacative, they should be interpreted with caution, given the limited sample size.
Since the completion of this study, carboplatin has been combined more recently with gemcitabine in advanced breast cancer,19 given an extensive experience of excellent tolerance in the treatment of other solid tumors, such as lung carcinoma.20,21 Because carboplatin does not require hydration, and because it causes less emesis, renal toxicity, ototoxicity, and neurotoxicity, it has replaced cisplatin in recently reported and ongoing trials in breast cancer. The schedule in this study, although well tolerated, is less practical because of the daily ×4 cisplatin infusion. However, there are no direct comparisons of cisplatin or carboplatin with gemcitabine reported in advanced breast cancer.
Toxicities on this trial were manageable, and most grades 3 and 4 toxicities were hematologic. This is likely the result of our dosing schedule, which was chosen to optimize synergy and avoid treatment-limiting toxicities reported by trials that used the weekly schedule.15,17 Prophylactic growth factors were a required element of protocol therapy for the heavily pretreated patients. In the minimally pretreated patients, the most common grade 4 toxicities were neutropenia in 45% and thrombocytopenia in 21% of patients. American Society of Clinical Oncology guidelines regarding use of growth factors should be followed in minimally treated patients who receieve this regimen.
Polymorphisms in genes relevant to cisplatin and gemcitabine were analyzed in a subset of 55 patients treated on both protocols. The 10 genes included those involved in DNA repair, drug metabolism, and cell-cycle control. Polymorphisms in XPD-751 and XRCC3, both involved in DNA repair, were significantly associated with PFS in the patients evaluated. Our data are consistent with data reported by de las Penas22, which demonstrated that XRCC3 and XPD were associated with PFS and OS in patients with non–small-cell lung cancer who were treated with gemcitabine and cisplatin. Additionally, in this study, CDA polymorphism was associated with increased toxicity among those with Lys/Gln and Gln/Gln variants, although this did not retain significance in multivariate analysis. Interestingly, white patients were more likely to sustain toxicities on this combination regimen compared with nonwhite patients, and they were also more likely to carry the CDA polymorphism. These data suggest that differences of efficacy and toxicity may be due to the frequency of polymorphisms in metabolic genes. Although the polymorphism data are interesting, these results should be interpreted with caution, as only 40% of the patients had determined genetic polymorphisms, which created a limited population that had potential for selection bias and underpowered analyses.
Our findings implicate the potential importance of DNA repair enzymes and drug metabolism enzymes in the prediction of clinical outcome to chemotherapy in metastatic breast cancer. The utility of these enzymes as prognostic and predictive factors requires additional study and underscores the importance of other validated markers and patient characteristics in breast cancer. Establishment of a genetic profile of patients with advanced breast cancer may lead to a tailored chemotherapy for the individual patient and may optimize therapeutic benefit, as suggested by other studies in metastatic breast cancer.23 However, because of the small size and patient selection of these studies, our findings should be validated in a larger clinical trial setting.
These two trials are the largest published studies on the combination of cisplatin and gemcitabine in metastatic breast cancer. The combination has activity, particularly in a heavily pretreated population with limited options, and presents a choice in the therapeutic menu for women with advanced disease. In the minimally pretreated study, RRs from our multi-institution study are within the range reported in other phase II trials of the combination, including a cooperative group study. However, the high RR in HR-negative disease supports use of this regimen in this population as well. Our data on gene polymorphisms has the potential to identify prognostic and predictive markers that may additionally tailor treatment and anticipate toxicities.
Appendix
The Appendix is included in the full-text version of this article, available online at www.jco.org. It is not included in the PDF version (via Adobe® Reader®).
Statistical methods.
After the first 16 patients were enrolled, an interim analysis was mandated for the minimally pretreated group on the basis of the percentage of patients without prior regimens, before enrollment continued to the planned total of 47 patients. The resulting decision table was based on Simon's Optimal design that maintained greater than 80% power and less than 20% type-I error, when a discouraging rate of 40% and 20% for zero-prior and one-prior regimen, respectively, were assumed and when an encouraging rate of 60% and 40%, respectively, were assumed. In the heavily pretreated arm, more than one response was required to proceed at 24 patients, and six (11%) of 55 were required at the final stage to declare the regimen worthy of additional consideration. This was based on Simon's Optimal design that had 95% power and 5% type-I error, to distinguish between a discouraging response rate (RR) of 5% and an encouraging RR of 20%. As both protocols were deemed encouraging, enrollment was extended to permit additional specimen collection.
The association of each polymorphism with overall survival, progression-free survival, and cycles' to developing grade 3 or greater toxicity was analyzed individually by using Kaplan-Meier plots and the log-rank test. In the univariate analyses, the Pike estimate of relative risk with 95% CIs was based on the log-rank test (Berry G, Kitchin RM, Mock PA: Stat Med 10:749-755, 1991). The associations of each polymorphism with tumor response were summarized by using contingency tables and Fisher's exact test. Cox proportional hazards regression model with stratification factors was fitted to re-evaluate the association among polymorphisms and the time to event variables, which considered the imbalances in the distributions of baseline characteristics. The polymorphism analysis is exploratory, and no attempt was made to adjust for multiple comparisons. The analysis is meant to identify germline variations associated with clinical outcome for which additional independent studies may be conducted. With the small sample size, the two protocols were combined for this analysis. There was insufficient statistical power to detect small or moderate differences in clinical outcome by genomic polymorphisms. However, the patterns of the association between genomic polymorphisms and clinical outcome were examined.
All tests of statistical significance were two-sided. The analyses were performed by using the SAS statistical package (version 9.0; SAS Institute Inc, Cary, NC) and Epilog Plus (version 1.0; Epicenter Software, Pasadena, CA).
Table A1.
Primer Sequences of the Germ-Line Polymorphisms in Genes of Interest
| Gene | Primer Sequences | Restriction Enzymes | Polymorphisms |
|---|---|---|---|
| GSTM1 | M+/M− (deletion) | ||
| Forward | 5′-GAACTCCCTGAAAAGCTAAAGC-3′ | ||
| Reverse | 5′-GTTGGGCTCAAATATACGGTGG-3′ | ||
| GSTT1 | T+/T− (deletion) | ||
| Forward | 5′-TTCCTTACTGGTCCTCACATCTC-3′ | ||
| Reverse | 5′-TCACCGGATCATGGCCAGCA-3′ | ||
| GSTP1-105 | BsmAI | Ile/Val (exon 5) | |
| Forward | 5′-ACCCCAGGGCTCTATGGGAA-3′ | ||
| Reverse | 5′-TGAGGGCACAAGAAGCCCCT-3′ | ||
| ERCC1-118 | BsrDI | C/T (exon 118) | |
| Forward | 5′-GCAGAGCTCACCTGAGGAAC-3′ | ||
| Reverse | 5′-GAGGTGCAAGAAGAGGTGGA-3′ | ||
| ERCC1 3′UTR | C/A | ||
| Forward | 5′-TGAGCCAATTCAGCCACT-3′ | ||
| Reverse | 5′-TAGTTCCTCAGTTTCCCG-3′ | ||
| XRCC3 | NcoI | Thr241Met (exon 7) | |
| Forward | 5′-GCCTGGTGGTCATCGACTC-3′ | ||
| Reverse | 5′-ACAGGGCTCTGGAAGGCACTGCTCAGCTCACGCACC-3′ | ||
| XRCC1 | NciI | Arg/Gln (exon 10) | |
| Forward | 5′-GCATCGTGCGTAAGGAGTG-3′ | ||
| Reverse | 5′-CCTTCCCTCATCTGGAGTAC-3′ | ||
| HER2 | BsmAI | Ile/Val | |
| Forward | 5′-AGAGCGCCAGCCCTCTGACGTCCAT-3′ | ||
| Reverse | 5′-TCCGTTTCCTGCAGCAGTCTCCGCA-3′ | ||
| XPD-156 | TfiI | C/A (exon 6) | |
| Forward | 5′-GCAGTACCAGCATGACACCA-3′ | ||
| Reverse | 5′-TTGGCATGCAGGATTGAGTA-3′ | ||
| XPD-751 | MBOII | Lys/Gln (exon 23) | |
| Forward | 5′-CCCTCTCCCTTTCCTCTGTT-3′ | ||
| Reverse | 5′-GGCAAGACTCAGGAGTCACC-3′ | ||
| p53 (13964GC) | MspI | G/C (intron 6) | |
| Forward | 5′-CTTGCCACAGGTCTCCCCAA-3′ | ||
| Reverse | 5′-TGTGCAGGGTGGCAAGTGGC-3′ | ||
| p53 (codon 72) | BstUI | Arg/Pro (G/C) | |
| Forward | 5′-ACAAGGGTTGGGCTGGGACCTGGAG-3′ | ||
| Reverse | 5′-TGAGGGTGTGATGGGATGGATAAAAGC-3′ | ||
| CDA | Lys/Gln | ||
| Forward | 5′-TGAAGCCTGAGTGTGTCCAG-3′ | ||
| Reverse | 5′-GTGCCCACCTTTACCTTTGA-3′ |
Abbreviation: CDA, cytidine deaminase.
Footnotes
Supported in part by Award No. N01-CM17101 to the California Cancer Consortium and by a grant from Eli Lilly.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: David R. Gandara, Eli Lilly (C); Kathy S. Albain, Eli Lilly (C) Stock Ownership: None Honoraria: Kathy S. Albain, Eli Lilly Research Funding: Helen K. Chew, Eli Lilly; Heinz-Josef Lenz, Eli Lilly; David R. Gandara, Eli Lilly Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: James H. Doroshow, Paul Frankel, Heinz-Josef Lenz, Merry L. Tetef, Kathy S. Albain
Administrative support: David R. Gandara
Provision of study materials or patients: Helen K. Chew, James H. Doroshow, Kim A. Margolin, George Somlo, Heinz-Josef Lenz, Michael Gordon, Christy Russell, Darcy Spicer, Tim Synold, Robert Bayer, Alexander Hantel, Patrick J. Stiff, David R. Gandara, Kathy S. Albain
Collection and assembly of data: Helen K. Chew, James H. Doroshow, Paul Frankel, Heinz-Josef Lenz, Michael Gordon, Wu Zhang, Dongyun Yang, Tim Synold, Kathy S. Albain
Data analysis and interpretation: Helen K. Chew, James H. Doroshow, Paul Frankel, Kim A. Margolin, George Somlo, Heinz-Josef Lenz, Michael Gordon, Wu Zhang, Dongyun Yang, Tim Synold, Kathy S. Albain
Manuscript writing: Helen K. Chew, James H. Doroshow, Paul Frankel, Kim A. Margolin, George Somlo, Heinz-Josef Lenz, Michael Gordon, Wu Zhang, Dongyun Yang, Kathy S. Albain
Final approval of manuscript: Helen K. Chew, James H. Doroshow, Paul Frankel, Kim A. Margolin, George Somlo, Heinz-Josef Lenz, Michael Gordon, Wu Zhang, Dongyun Yang, Christy Russell, Darcy Spicer, Tim Synold, Robert Bayer, Alexander Hantel, Patrick J. Stiff, Merry L. Tetef, David R. Gandara, Kathy S. Albain
REFERENCES
- 1.Henderson IC, Berry DA, Demetri GD, et al. Improved outcomes from adding sequential paclitaxel but not from escalating doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J Clin Oncol. 2003;21:976–983. doi: 10.1200/JCO.2003.02.063. [DOI] [PubMed] [Google Scholar]
- 2.Mamounas EP, Bryant J, Lembersky B, et al. Paclitaxel after doxorubicin plus cyclophosphamide as adjuvant chemotherapy for node-positive breast cancer: Results from NSABP B-28. J Clin Oncol. 2005;23:3686–3696. doi: 10.1200/JCO.2005.10.517. [DOI] [PubMed] [Google Scholar]
- 3.Martin M, Pienkowski T, Mackey J, et al. Adjuvant docetaxel for node-positive breast cancer. N Engl J Med. 2005;352:2302–2313. doi: 10.1056/NEJMoa043681. [DOI] [PubMed] [Google Scholar]
- 4.Sledge GW, Jr, Loehrer PJ, Sr, Roth BJ, et al. Cisplatin as first-line therapy for metastatic breast cancer. J Clin Oncol. 1988;6:1811–1814. doi: 10.1200/JCO.1988.6.12.1811. [DOI] [PubMed] [Google Scholar]
- 5.Carmichael J, Possinger K, Phillip P, et al. Advanced breast cancer: A phase II trial with gemcitabine. J Clin Oncol. 1995;13:2731–2736. doi: 10.1200/JCO.1995.13.11.2731. [DOI] [PubMed] [Google Scholar]
- 6.Bergman AM, Ruiz van Haperen VW, Veerman G, et al. Synergistic interaction between cisplatin and gemcitabine in vitro. Clin Cancer Res. 1996;2:521–530. [PubMed] [Google Scholar]
- 7.van Moorsel CJ, Kroep JR, Pinedo HM, et al. Pharmacokinetic schedule finding study of the combination of gemcitabine and cisplatin in patients with solid tumors. Ann Oncol. 1999;10:441–448. doi: 10.1023/a:1008301522349. [DOI] [PubMed] [Google Scholar]
- 8.Bishop DK, Ear U, Bhattacharyya A, et al. XRCC3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 9.Liu N, Lamerdin JE, Tebbs RS, et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 10.Pierce AJ, Johnson RD, Thompson LH, et al. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoehlmacher J, Park DJ, Zhang W, et al. A multivariate analysis of genomic polymorphisms: Prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344–354. doi: 10.1038/sj.bjc.6601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allan JM, Smith AG, Wheatley K, et al. Genetic variation in XPD predicts treatment outcome and risk of acute myeloid leukemia following chemotherapy. Blood. 2004;104:3872–3877. doi: 10.1182/blood-2004-06-2161. [DOI] [PubMed] [Google Scholar]
- 13.Gu J, Zhao H, Dinney CP, et al. Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin Cancer Res. 2005;11:1408–1415. doi: 10.1158/1078-0432.CCR-04-1101. [DOI] [PubMed] [Google Scholar]
- 14.Simon R. Optimal two-stage designs for phase II clinical trials. Control Clin Trials. 1989;10:1–10. doi: 10.1016/0197-2456(89)90015-9. [DOI] [PubMed] [Google Scholar]
- 15.Nagourney RA, Link JS, Blitzer JB, et al. Gemcitabine plus cisplatin repeating doublet therapy in previously treated, relapsed breast cancer patients. J Clin Oncol. 2000;18:2245–2249. doi: 10.1200/JCO.2000.18.11.2245. [DOI] [PubMed] [Google Scholar]
- 16.Heinemann V, Stemmler HJ, Wohlrab A, et al. High efficacy of gemcitabine and cisplatin in patients with predominantly anthracycline- and taxane-pretreated metastatic breast cancer. Cancer Chemother Pharmacol. 2006;57:640–646. doi: 10.1007/s00280-005-0093-5. [DOI] [PubMed] [Google Scholar]
- 17.Burch PA, Mailliard JA, Hillman DW, et al. Phase II study of gemcitabine plus cisplatin in patients with metastatic breast cancer: A North Central Cancer Treatment Group Trial. Am J Clin Oncol. 2005;28:195–200. doi: 10.1097/01.coc.0000144815.54746.d0. [DOI] [PubMed] [Google Scholar]
- 18.Fuentes H, Calderillo G, Alexander F, et al. Phase II study of gemcitabine plus cisplatin in metastatic breast cancer. Anticancer Drugs. 2006;17:565–570. doi: 10.1097/00001813-200606000-00011. [DOI] [PubMed] [Google Scholar]
- 19.Nasr FL, Chahine GY, Kattan JG, et al. Gemcitabine plus carboplatin combination therapy as second-line treatment in patients with relapsed breast cancer. Clin Breast Cancer. 2004;5:117–122. doi: 10.3816/cbc.2004.n.015. discussion 123-124. [DOI] [PubMed] [Google Scholar]
- 20.Zatloukal P, Petruzelka L, Zemanova M, et al. Gemcitabine plus cisplatin vs. gemcitabine plus carboplatin in stage IIIb and IV non–small-cell lung cancer: A phase III randomized trial. Lung Cancer. 2003;41:321–331. doi: 10.1016/s0169-5002(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 21.Kosmidis PA, Kalofonos HP, Christodoulou C, et al. Paclitaxel and gemcitabine versus carboplatin and gemcitabine in patients with advanced non–small-cell lung cancer: A phase III study of the Hellenic Cooperative Oncology Group. Ann Oncol. 2008;19:115–122. doi: 10.1093/annonc/mdm430. [DOI] [PubMed] [Google Scholar]
- 22.de las Penas R, Sanchez-Ronco M, Alberola V, et al. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non–small-cell lung cancer patients. Ann Oncol. 2006;17:668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 23.Bewick MA, Conlon MS, Lafrenie RM. Polymorphisms in XRCC1, XRCC3, and CCND1 and survival after treatment for metastatic breast cancer. J Clin Oncol. 2006;24:5645–5651. doi: 10.1200/JCO.2006.05.9923. [DOI] [PubMed] [Google Scholar]