Abstract
Background
The 1918–1919 A/H1N1 influenza pandemic killed ~50 million people worldwide. Historical records suggest that an early pandemic wave struck Europe during the summer of 1918.
Methods
We obtained surveillance data that were compiled weekly, during 1910–1919, in Copenhagen, Denmark; the records included medically treated influenza-like illnesses (ILIs), hospitalizations, and deaths by age. We used a Serfling seasonal regression model to quantify excess morbidity and mortality, and we estimated the reproductive number (R) for the summer, fall, and winter pandemic waves.
Results
A large epidemic occurred in Copenhagen during the summer of 1918; the age distribution of deaths was characteristic of the 1918–1919 A/H1N1 pandemic overall. That summer wave accounted for 29%–34% of all excess ILIs and hospitalizations during 1918, whereas the case-fatality rate (0.3%) was many-fold lower than that of the fall wave (2.3%). Similar patterns were observed in 3 other Scandinavian cities. R was substantially higher in summer (2.0 –5.4) than in fall (1.2–1.6) in all cities.
Conclusions
The Copenhagen summer wave may have been caused by a precursor A/H1N1 pandemic virus that transmitted efficiently but lacked extreme virulence. The R measured in the summer wave is likely a better approximation of transmissibility in a fully susceptible population and is substantially higher than that found in previous US studies. The summer wave may have provided partial protection against the lethal fall wave.
Recent studies of the 1918 “Spanish” influenza pandemic have dramatically altered our understanding of its transmission. The transmissibility of the influenza virus during the main pandemic wave, in the fall of 1918, was found to be far lower than had previously been thought, with a reproductive number (R) of ~2 rather than 22 [1, 2]. R is defined as the average number of secondary cases produced by an infected individual at the onset of the epidemic—a key quantity for disease control. Simulation models predict that social-distancing and medical intervention will substantially reduce attack rates and slow the transmission of influenza during the next influenza pandemic if R is <2, whereas such interventions fail if R is >2 [3– 6].
For influenza epidemics that recur year after year, transmissibility strongly depends on the initial fraction of susceptible people. If some populations were exposed to a variant of the pandemic virus (which conferred some immunity) before the fall 1918 wave, then previous R estimates for the fall wave would reflect the effective transmission in a partially susceptible population and would underestimate the basic reproductive number (R0) in an influenza-naive population [1].
Influenza outbreaks were reported to occur in Europe during the summer of 1918 and are thought to represent a first wave of the pandemic [7, 8]. For studies relying solely on mortality data the transmissibility and mortality impact of these outbreaks is hard to quantify because relatively few deaths occurred. Studies of the “herald waves” of the 1918 pandemic are limited to data on summer mortality in the United Kingdom [7, 9], on summer hospitalizations in Geneva [10], and on spring mortality in New York City [11]. Moreover, the public-health significance of herald pandemic waves has not been addressed in contemporary reviews [12].
A robust public health–surveillance system was in place in Copenhagen, Denmark, during the first decades of the 20th century [13]. In the present study, we analyze the weekly morbidity and mortality data gathered by that system and estimate excess morbidity, excess mortality, and transmissibility associated with the influenza outbreak during the summer of 1918.On the basis of historical mortality data, we describe similar summer waves in 3 other Scandinavian cities, and we discuss the reports from additional European cities, which suggest that summer waves might have been the typical European experience. We close with a discussion of what our findings imply about both the evolution of the 1918 pandemic virus and the mitigation of a future 1918-like pandemic.
METHODS
Copenhagen epidemiological-surveillance system
At least since the turn of the century [13, 14], primary-care physicians in Copenhagen have participated in weekly reporting of cases of epidemic diseases, including influenza. The archived reports include weekly deaths stratified by cause and age, as well as weekly hospitalizations stratified by cause only. We also consulted contemporaneous reports from physicians describing their experience with the summer wave [15].
Morbidity time series
A total of ~350 general practitioners contributed to the surveillance system [13–15].We extracted the weekly number of reported patient visits for influenza-like illnesses (ILIs) for the years 1910–1919. We also tabulated the weekly number of influenza hospitalizations and total hospitalizations (all-cause).
Mortality time series
Weekly deaths due to respiratory diseases (influenza, pneumonia, or bronchitis) and all-cause deaths were tabulated for the years 1910–1919. To investigate age specific mortality patterns, we stratified all causes of death into 4 age groups: <15, 15–44, 45–64, and ≥65 years of age. In all tabulations, we included deaths due to bronchitis, because the steep rise in deaths during October 1918, the peak of the pandemic in Copenhagen, was mostly reported as being due to bronchitis. A more complete account of this assessment is available in a historical British report on the 1918 influenza pandemic, in the section on Denmark [8].
Population data for Copenhagen
On the basis of census data for Copenhagen [16], we linearly interpolated the composition of the age-stratified population for each study year.
Excess mortality and morbidity for each pandemic wave
We applied a Serfling seasonal regression model to each disease outcome (respiratory deaths, all-cause deaths, influenza hospitalizations, all-cause hospitalizations, ILIs), excluding data from epidemic periods, to establish baseline levels in the absence of influenza activity [17–19]. Excess mortality and morbidity above the baseline level during epidemic periods were attributed to influenza. The 3 pandemic waves (summer 1918, fall 1918, and winter 1918–1919) were studied separately, and the epidemic of 1915–1916 was used as a reference interpandemic season. Mortality data were aggregated by 4-week periods to obtain robust estimates. Separate models were fitted to each age group; the model fit was good in all 4 age strata (0.65 ≤ R2 ≤ 0.73).
Epidemiological data from other Scandinavian cities
From the historical British report [8], we compiled weekly numbers of ILIs and deaths attributable to influenza, for Gothenburg, Oslo (formerly Christiania), and Stockholm. Because the Scandinavian data in the British report [8] covered only a brief period (June 1918–March 1919), we did not apply a Serfling seasonal regression model but, instead, used the numbers of influenza-attributable cases and deaths directly available in the British report. It was reassuring to note that, for Copenhagen, the figures published in the British report ([8], p. 221) were very similar to the excess ILIs and deaths generated by our Serfling seasonal regression model. The differences in the total numbers of clinical illnesses and deaths were, respectively, 1% and 6% for the summer wave and 3% and 4% for the fall wave.
The case-fatality rate in Stockholm (33.1%) was an outlier, compared with those in the other 3 cities (range, 0.3%–4.7%). The Stockholm surveillance efforts reportedly failed to identify most of the cases of influenza morbidity [8]; hence, the Stockholm data were excluded from morbidity analyses.
Transmissibility (reproduction numbers R and R0)
We estimated R from raw morbidity data (ILI cases and influenza hospitalizations) for Copenhagen, on the basis of the observed growth rate in case numbers during the early ascending phase of the summer and fall waves. We did not estimate R for the third wave, during winter 1918–1919, because it overlapped with the fall wave, thereby precluding identification of the early, ascending phase. The estimates of R for the summer wave closely approximate R0— a measure of transmissibility in a fully naive population. We also used ILI data for Oslo and Gothenburg, but we excluded those for Stockholm because of the incomplete reporting of clinical illnesses.
To identify the early, ascending phase, we used a standard Poisson model and systematically varied the starting week, to find the longest period with exponential growth (see the Appendix, which is available in the online edition of the Journal of Infectious Diseases). For estimates of R based on respiratory and all-cause excess mortality data, we used the simpler method described by Mills et al. [1].
Using the estimated weekly growth rate in the early, ascending phase, we derived R on the basis of data on the time course of a typical influenza infection [20]. In past research [1, 6, 21], the duration of infection has traditionally been set at 6 days. The corresponding serial interval, defined as the time from infection in a primary case to infection in a secondary case, is ~4 days, on average [6]. However, a recent study of influenza transmission in households during a contemporary epidemic indicates that the serial interval may be as short as 2.6 days [4, 22]. Because the serial interval for a pandemic virus is not well understood, we present estimates of R for a short serial interval of 2.6 days (as in reference [4]) and for a longer serial interval of ~4 days (as in references [1, 6]), thereby encompassing the range of values used in recent models of pandemic containment and mitigation [3–6, 23]. We report ranges of R based on estimates for short and long serial interval values, as well as the 95% confidence intervals derived by standard methods (see the Appendix, which is available in the online edition of the Journal of Infectious Diseases).
RESULTS
1. Pandemic Experience in Copenhagen
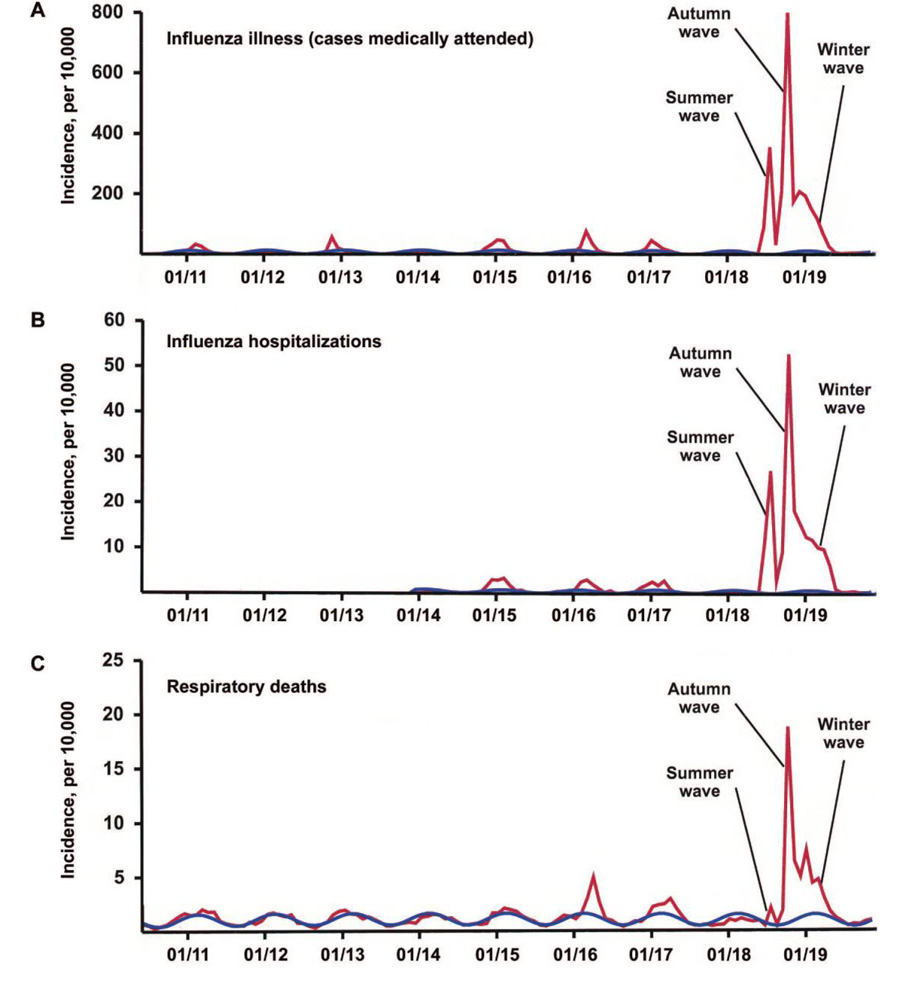
We identified 3 waves of morbidity and mortality during the 1918–1919 influenza pandemic in Copenhagen: a summer wave (June–July 1918), a severe fall wave (September–November 1918), and a winter wave (December 1918–April 1919) (see figure 1 and table 1).
Fig. 1.
First 3 waves of the 1918–1919 pandemic in Copenhagen—monthly incidence of medically treated influenza illnesses (A), influenza hospitalizations (B), and respiratory deaths (C), during 1910–1919 (red lines). Serfling seasonal regression model baselines are shown as blue cyclical lines. The summer pandemic wave, peaking during July 1918, is characterized by substantial morbidity and low mortality.
Table 1.
Morbidity and mortality impact of 3 influenza pandemic waves in Copenhagen, 1918–1919.
| 1918 summer wave (12 weeks: 23 June– 8 September 1918) |
1918 fall wave (12 weeks: 15 September– 1 December 1918) |
1918–1919 winter wave (24 weeks: 8 December– 27 April 1919) |
Sum of 3 waves |
|
|---|---|---|---|---|
| Excess influenza illnessesa | 480 | 1170 | 710 | 2360 |
| Excess influenze hospitalizationsa | 40 | 80 | 60 | 180 |
| Excess all-cause hospitalizationsa | 40 | 80 | 50 | 170 |
| Excess all-cause deathsa | 1.7 | 27 | 12 | 41 |
| Case-fatality rate, %b | 0.35 | 2.3 | 1.7 | 1.7 |
NOTE. Data are no. of cases/10,000 individuals, unless otherwise indicated. Estimates are excess rates per 10,000, based on Serfling seasonal regression model.
In excess of seasonal baseline, as derived from Serfling seasonal regression model.
Calculated as ratio of all-cause excess deaths:excess influenza illnesses.
Clinical influenza illnesses (ILI cases)
During June–July 1918, the number of medically treated ILI cases in Copenhagen spiked dramatically, rising ~300-fold above the level of the preceding summers of 1910–1917, to a level nearly half that of the fall peak (see figure 1 and table 1). Nearly 5% of the population sought medical attention for influenza illness in excess of the seasonal baseline during the summer wave, compared with ~12% during the fall wave and ~7% during the winter wave (table 1). This suggests that a total of ~24% of the population became afflicted with medically treated influenza illness during the 3 pandemic waves (an unknown proportion of asymptomatic infectious and unattended illnesses are not taken into account).
Hospitalizations
Severe morbidity, as analyzed on the basis of weekly hospitalizations, revealed a ~300-fold elevation in influenza hospitalizations during the 1918 summer wave, compared with the summers of previous years. Model estimates of excess hospitalizations for the first wave were similar for both hospitalization time series (influenza and all-cause) (table 1). Approximately 0.4% of the Copenhagen population was hospitalized for influenza-related conditions during the 1918 summer wave, compared with 0.8% during the fall wave. Of the total number of excess hospitalizations during 1918–1919, ~24% occurred during the summer wave—a pattern similar to that of ILI cases (table 1).
Mortality
Using the all-cause mortality time series, we detected a small increase of ~90 excess deaths during the summer wave, corresponding to a rate of ~1.7/10,000, an order of magnitude lower than that during the fall wave (~1450 excess deaths, corresponding to a rate of 27/10,000; table 2). Analysis of data on excess mortality due to respiratory diseases gave similar estimates. Excess mortality in the summer wave revealed the “signature” age shift of the 1918 pandemic virus, with most deaths occurring among young adults and no deaths occurring among seniors, similar to the pattern in US data [11, 24, 25]. Overall, the summer wave was responsible for <5% of all influenza-related excess deaths during 1918–1919 in Copenhagen.
Table 2.
Comparison of age-specific excess mortality rates per 10,000 in Copenhagen, for each of the 3 pandemic waves during 1918–1919 and for the 1915–1916 reference.
| 1916 reference epidemic (16 weeks: 6 February– 21 May 1916) |
1918 summer wave (12 weeks: 23 June– 8 September 1918) |
1918 fall wave (16 weeks: 15 September– 29 December 1918) |
1919 winter wave (16 weeks: 5 January– 30 March 1919) |
Total for 3 pandemic waves during 1918–1919 |
|
|---|---|---|---|---|---|
| Excess respiratory deaths, all agesa | 6.6 | 1.4 | 27.0 | 12.0 | 41.1 |
| Excess all-cause deaths, all ages | 18.4 | 1.7 | 27.0 | 12.6 | 41.2 |
| <15 years | 37.5 | 0.2 | 12.2 | 17.3 | 29.7 |
| 15–44 years | 1.4 | 3.1 | 46.7 | 14.9 | 64.7 |
| 45–64 years | 11.4 | 0.3 | 7.7 | 7.3 | 15.5 |
| ≥65+ years | 82.6 | 0 | 0.5 | 0 | 0.5 |
NOTE. The 1915–1916 interpandemic influenza season illustrates the mortality impact of a contemporary severe nonpandemic season.
Estimates of excess mortality due to respiratory disease include deaths attributable to influenza, pneumonia, and bronchitis.
The case-fatality rate in Copenhagen was ~0.35% during the summer wave, compared with 2.3% during the fall wave (table 1) (P <.0001). Thus, only 0.02% of Copenhagen’s 534,000 inhabitants died of influenza during the summer wave, compared with 0.27% during the fall wave.
Taken together, the data reveal a substantial summer pandemic wave occurring in Copenhagen, resulting in 50% fewer clinical illnesses and hospitalizations than the fall wave. Surprisingly, the case fatality rate in summer was ~7-fold lower than in the fall (table 1). Throughout the first 3 waves in 1918–1919, a total of 0.41% of the Copenhagen population died of pandemic influenza.
2. Experience of Other Scandinavian Cities
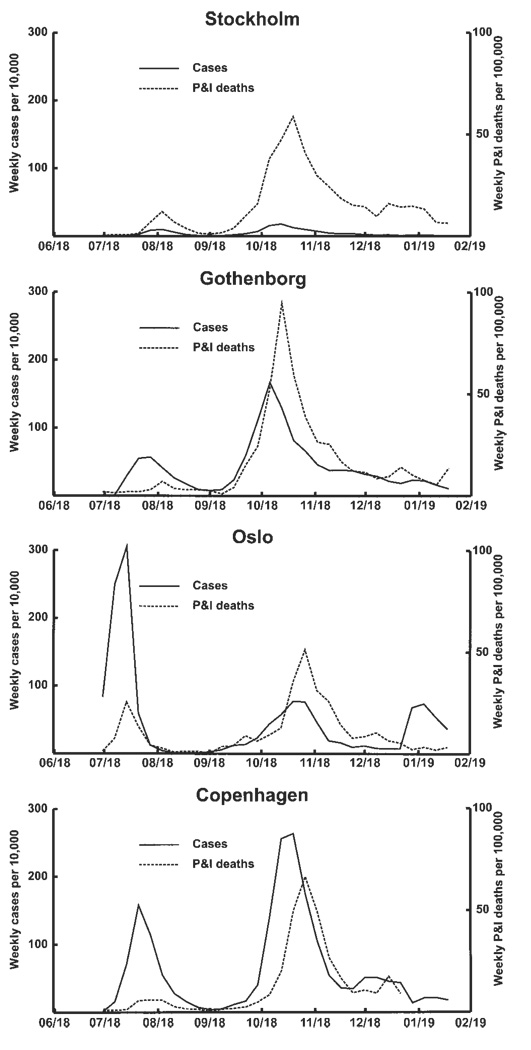
Summary data for Oslo, Gothenburg, and Stockholm were similar to those for Copenhagen, revealing that a substantial summer pandemic wave with a low case-fatality rate had occurred (table 3 and figure 2). The July summer wave in Oslo was most striking, resulting in twice as many ILI cases as the fall wave. In Gothenburg, the summer wave had ~1/3 as many ILI cases as the fall wave. The impact that the pandemic had on the overall mortality rate was low in Oslo, Gothenburg, and Stockholm: 0.18%–0.34% of the populations of those cities died from pandemic influenza during 1918–1919.
Table 3.
Comparison of the 1918 summer and fall pandemic waves in 4 Scandinavian cities: pandemic morbidity and mortality impact, and estimates of the reproduction number R (data from [8]).
| Characteristic | Copenhagena | Gothenburg | Oslob | Stockholmc |
|---|---|---|---|---|
| Population size | 560,000 | 197,000 | 260,000 | 413,000 |
| Summer wave | 30 June– 7 September |
30 June– 7 September |
30 June– 24 August |
30 June– 7 September |
| Cases of influenza illness | 25,360 | 4812 | 18,544 | NA |
| Cases/10,000 individuals, no. | 471 | 244 | 713 | NA |
| Deaths attributable to influenza | 85 | 59 | 146 | 146 |
| Deaths/10,000 individuals, no. | 1.5 | 3.0 | 5.6 | 3.5 |
| Case-fatality rate, % | 0.3 | 1.2 | 0.8 | NA |
| Rd | 2.2–3.0 | 3.1–4.8 | 2.0–3.2 | NA |
| Fall wave | 8 September– 30 November |
8 September– 16 November |
25 August– 16 November |
8 September– 16 November |
| Cases of influenza illness | 61,285 | 14,411 | 9872 | NA |
| Cases/10,000 individuals, no. | 1090 | 732 | 380 | NA |
| Deaths attributable to influenza | 1293 | 673 | 488 | 1119 |
| Deaths/10,000, no. | 23.1 | 34.2 | 18.8 | 27.1 |
| Case-fatality rate, % | 2.1 | 4.7 | 1.9 | NA |
| Rd | 1.2–1.3 | 1.4–1.6 | 1.3–1.5 | NA |
NOTE. NA, not available.
Estimates are based on data from the British report [8, p. 217] and correspond well with those of the present study, which were estimated by use of the Serfling seasonal regression model.
The summer epidemic started ~2 weeks before the date of the first data reported in the present study, so we estimated R0 by assuming that the last week at baseline was either 1 June or 9 June, with exponential growth until 23 June (see the Appendix, which is available in the online edition of the Journal of Infectious Diseases).
Morbidity data are not as reliable as those from the incompleteness of the other 3 cities and were removed from analysis; the overall incidence in any week, including the fall pandemic wave, is surprisingly low, consistent with reporting system (as discussed in [8, p. 205]).
Based on weekly growth rate in cases of influenza illness. Range includes estimates for short and long serial intervals (2.6 and 4 days, respectively) and confidence intervals of point estimates.
Figure 2.
Scandinavian 1918 summer wave—weekly incidence of cases of influenza illness and respiratory deaths in 4 Scandinavian cities, during 1918–1919. Mortality data from Stockholm, Oslo, and Gotenborg are based on data reported by Low [8] and depict pneumonia and influenza (P&I) mortality; data from Copenhagen are based on all respiratory deaths, including bronchitis (nearly 90% of deaths during the 1918 fall wave in Copenhagen were coded to bronchitis).
3. Estimates of Transmissibility (R)
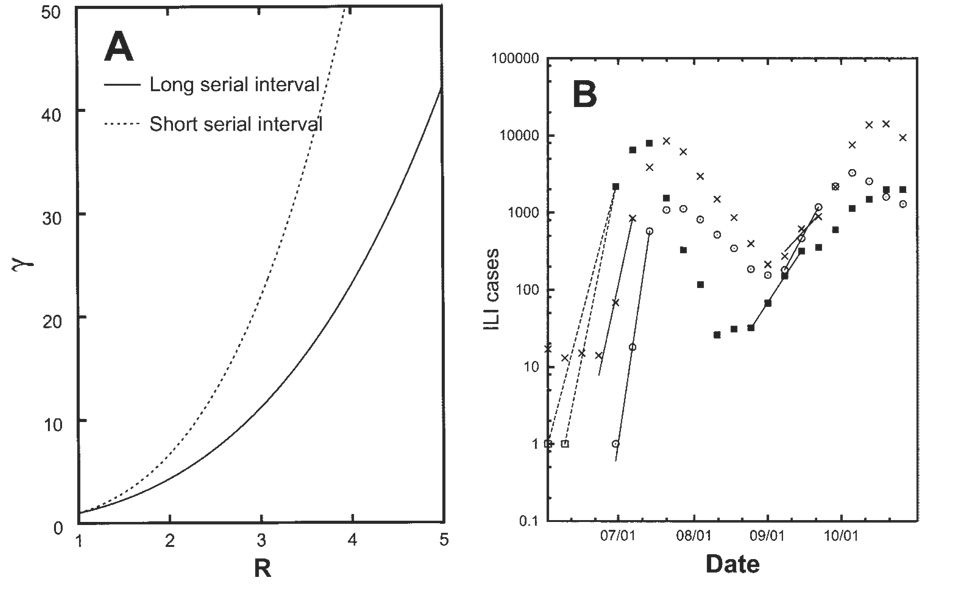
Detailed results of our algorithm for the estimation of R during the summer and fall waves of the 1918 –1919 pandemic in Scandinavian cities are given in the Appendix, which is available in the online edition of the Journal of Infectious Diseases. Figure 3A illustrates the theoretical relationship between the weekly growth rate and R, when short (2.6 days) and long (4 days) serial intervals are assumed; figure 3B illustrates our estimation of the growth rate during the first weeks of the summer and fall waves. In most cases, exponential growth was observed for only 3 weeks (the longest exponential growth lasted 4 weeks during the fall wave in Oslo; see the Appendix, which is available in the online edition of the Journal of Infectious Diseases).
Figure 3.
Algorithm for estimating transmissibility of the 1918–1919 pandemic (R, the reproductive number). A, Relationship between R and the weekly growth factor (γ), for data representing a long, 4-day, serial interval (used by Mills et al. [1]) and a short, 2.6-day, serial interval (used by Ferguson et al. [4]). B, Incidence of clinically treated influenza illness in 3 Scandinavian cities—Copenhagen (×); Gothenborg (○); Oslo (■)—during the summer and fall of 1918. The lines illustrate the fitting procedure to the exponential growth phase for each wave. For Oslo, the first 2 data points (□) suggest the possible timing of the first case in Oslo (see the Appendix, which is available in the online edition of the Journal of Infectious Diseases).
Summer wave
During the summer wave in Copenhagen, the range of R was 2.2–3.0 for data on ILIs and 2.8 –5.4 for data on hospitalizations (table 4). Similarly high R values were estimated from data on ILIs for Oslo and Gothenborg (table 3). The estimation of R for the Oslo summer wave was complicated by the fact that the surveillance data were available starting on 30 June 1918, well into the summer wave. Relying on independent evidence that the epidemic began on 15 June [26, 27], we conducted a sensitivity analysis assuming that the first week of the epidemic started either 2 June or 9 June (table 5), generating a range of R estimates that was 2.0 –3.2—that is, in line with the estimates for the other 2 Scandinavian cities.
Table 4.
Estimates of reproduction number (R), by wave, in Copenhagen.
| Data type | 1918 summer wave | 1918 fall wave | ||
|---|---|---|---|---|
| Rshort (2.6 days) |
Rlong (4 days) |
Rshort (2.6 days) |
Rlong (4 days) |
|
| Cases of clinical influenza | 2.2–2.4 | 2.8–3.0 | 1.22–1.24 | 1.29–1.33 |
| Hospitalizations | 2.8–4.0 | 3.6–5.4 | 1.2–1.3 | 1.3–1.4 |
| Excess respiratory deathsa | NA | NA | 1.4b | 1.6b |
| Excess all-cause deathsa | NA | NA | 1.5b | 1.8b |
NOTE. R is based on 2 serial-interval parameter values—short duration (2.6 days) [4] and long duration (~4 days) [1]. Ranges represent 95% confidence intervals. NA, not available.
R for the summer wave could not be estimated with precision, when mortality data were used, because there were few deaths.
The method for R estimation based on excess mortality does not allow for computation of confidence intervals.
Table 5.
Statistical analysis of the weekly growth factor (γ), based on ILI data from 4 Scandinavian cities, during the summer and fall waves of 1918.
| The table is available in its entirety in the online edition of the Journal of Infectious Diseases. |
Fall wave
We estimated R for the fall wave using the weekly time series of ILI, influenza hospitalizations, excess respiratory deaths, and excess all-cause deaths. During the 1918 fall wave in Copenhagen, the estimates of R were low, ranging between 1.2–1.4 for the morbidity time series and 1.5–1.8 for the excess mortality time series (table 4 and table 5). The estimates of R were similarly low for the other Scandinavian cities (table 3); the between-city variability in estimates ofRwas less for the fall wave than for the summer wave.
Sensitivity analysis
We also explored the likely magnitude of the underestimation of R that would result theoretically from our use of “raw” morbidity time series. Using instead the excess ILI time series from Copenhagen, we found that, depending on the duration of the serial interval, the range of estimates of R for the summer wave was 2.6 –3.6, which is somewhat higher than the 2.2–3.0 range calculated when the “raw” ILI data were used. Thus, our approach generated robust, although slightly conservative, R values. The reason for this robustness is that we used a maximum-likelihood estimator to estimate the growth rate γ, which weighs the weekly observations according to the inverse of their value.
DISCUSSION
The 1918–1919 A/H1N1 influenza pandemic is unique because of its devastating mortality impact and because of the likely purely avian origin of the culprit virus [28]. Experimental infections in macaques have demonstrated that the lethality of the virus was likely attributable to aberrant innate immune responses—a “cytokine storm” [29]. The present population-based epidemiological study complements virological studies by describing the characteristics of the unusual 1918 summer pandemic wave in Copenhagen and other Scandinavian cities, which was marked by high transmissibility, substantial morbidity, and low mortality. Although the unusual characteristics of the summer wave were noted at the time by physicians and public health authorities [8], they have not been quantified or considered in the context of ongoing pandemic preparedness planning. The findings of the present study could guide a search for historical pathological specimens that could illuminate events in the early phases of genetic adaptation of the avian 1918 A/H1N1 virus to the human host.
We were able to characterize the 1918 summer wave in Copenhagen because, in the decades before and after that year, a weekly surveillance system compiled data on respiratory illnesses and deaths. Despite the few deaths attributed to influenza in the summer wave, its general patterns were otherwise characteristic of the 1918 influenza pandemic overall. First, its peak morbidity rate was ~300-fold higher than that of any other summer during 1910–1917 and was ~50% that of the fall wave. Second, it had the 1918-pandemic “signature” mortality age pattern, with the highest mortality rate in young adults and sparing of the elderly [11].
Similar surveillance systems were in place in Oslo, Gothenburg, and Stockholm [8]. Large geographic variations in mortality rate during the 1918 pandemic have recently been reported, with death rates in 26 countries around the world ranging from 0.2% to 8% [30]. The cumulative 1918 –1919 pandemic excess mortality rate in Scandinavian cities fell within the low end of that range, with our estimates ranging from 0.2% to 0.4% of the total population. It is possible that Scandinavia fared better than most other countries because the mild summer wave had immunized a substantial subset of the population—although socioeconomic differences also correlated with mortality rates [30, 31].
Our analysis of weekly morbidity data from Scandinavia paints a consistent picture of a substantial summer 1918 pandemic wave with such intense transmissibility that the virus depleted a substantial number of susceptible hosts during just 3 weeks. We believe that our Scandinavian estimates of R for the summer wave are the closest approximation yet to the true R0 (a measure of transmissibility in a fully naive population) for pandemic influenza, because these estimates are based on morbidity data from the first pandemic wave, in a fully susceptible population.
Using a reasonable range of serial interval values (2.6–4 days), we estimated that the R0 for the summer wave ranged between 2.2–5.4 in Copenhagen and 2.0–4.8 in 2 other Scandinavian cities. These estimates of R0 are substantially higher than contemporaneous pandemic influenza estimates for the United States (R ~ 2) and the United Kingdom (R ~ 2.0 –2.1), in which the authors based their analysis on mortality data from the fall wave [1, 3, 4, 7, 9]. In Scandinavia, transmissibility was much lower during the fall wave (R = 1.2–1.8) than during the summer wave. Interestingly, the low average R ~ 2.0 measured for the fall wave in 45 US cities [1] is consistent with the possibility that an earlier herald wave also occurred in the United States [11]. Mills et al. [1] noted that, if the US population was ~70% susceptible durind the fall, then R0 would have been ~3–4, which is in line with our estimates for the summer wave in Scandinavia.
As discussed by Wallinga and Lipsitch [20], the inference regarding the value of R or R0 depends on specific assumptions about the duration of the serial interval. Different algorithms and underlying data have led investigators to choose different values for the serial interval, ranging from 2.6 [3, 4] to ~4 days [1, 5, 6]. To facilitate comparison with previous studies, we therefore elected to use this parameter range. Most important, this key parameter has not been measured in pandemic-influenza settings, and its value could well be higher than that observed for seasonal influenza, because viral shedding may be prolonged for novel influenza viruses [32].
We could not accurately measure the pandemic attack rate in the present study; certainly, the observed ~5% prevalence of clinically attended ILI cases during the Copenhagen summer wave is an underestimate, because it excludes asymptomatic cases and unattended clinical cases. For a homogeneously mixed population, the theoretical attack rate for R0=3 would be much higher, at ~94%. However, this theoretical estimate does not take into account the complexities of heterogeneous mixing, host-genetic or age-specific differences in susceptibility, or changes in behavior and mixing patterns during the course of the summer wave, such as the fact that the school year ended in early July [31, 33].
The Scandinavian-summer-wave experience is probably representative of a wider European or even global experience of a mild first pandemic wave. The existence of a summer wave has been documented in several European countries, including the United Kingdom, Belgium, and Spain [8, 10], even though its characteristics apparently differ between countries. For example, a study from Geneva found lower transmissibility during the summer wave (R = 1.5) than during the fall wave (R = 3.8) [10]. Another study, from Madrid, found that most deaths occurred during the summer wave; by contrast, the fall wave was very mild [8]. Furthermore, in the Americas, a herald wave of excess mortality occurred in New York City during the late spring of 1918 [11], and a May 1918 mortality wave was reported in Mexico [34]. However, without the availability of morbidity data, it is not possible to know whether these waves also had low case-fatality rates. Last, a recent study from China reported a June mortality wave in Hong Kong and Shanghai that was similar in magnitude to the fall wave [35].
There are several possible biological explanations for the Scandinavian pattern, which is characterized by a mild summer wave followed by a fall wave less lethal than that experienced in other countries. The “evolutionary” hypothesis is that the pandemic virus acquired transmissibility before summer and only later acquired the virulence factors that caused high mortality. An alternative, “robustness” hypothesis is that Scandinavian populations were more robust during summer months and thus less prone to infection and severe disease outcomes during the first wave [36], suggesting that transmissibility and mortality could have been even higher had the first wave occurred during the colder months. Under both the “evolutionary” and “robustness” hypotheses, the low transmissibility of the fall wave may be explained by partial cross-protection from summer exposure to a related influenza A/H1N1 virus. Unfortunately, neither of these hypotheses fully explains the observed phenomenon of 2 closely spaced pandemic waves during 1918, each with a high attack rate and rapid burnout of susceptibles. We note that the “evolutionary” hypothesis implies that it is possible for a pandemic virus to acquire the ability to be efficiently transmitted between humans first and to become highly virulent later—a counterintuitive evolutionary pattern. Studies of differences between the genomes of viruses circulating during the summer and fall waves—should any viruses from summer 1918 be found— would shed light on this interesting hypothesis.
In conclusion, the Scandinavian 1918 summer-wave experience is immensely interesting in the context of planning for future pandemics. On one hand, our upward reassessment of R0 for the 1918 virus is not good news. Existing simulation models of pandemic influenza assume a low R0, of ≤2, and findings suggest that mitigating a future pandemic would be possible with a combination of medical and nonmedical intervention strategies; but the higher estimates of R0 for the Scandinavian experience would make that assessment overly optimistic. On the other hand, in hindsight, a mild summer wave may actually be something that one would not want to mitigate, because it may afford the population some protection against lethal subsequent waves.
Acknowledgments
We thank our colleagues Robert Taylor, at the National Institutes of Health, and Kaare Mølbak, at the Statens Serum Institute in Copenhagen, for their extremely helpful comments on earlier versions of the manuscript, and Rodolfo Acuna-Soto, at the Autonomous University of Mexico, Mexico City, for sharing his interesting insights into the 1918 pandemic experience in Mexico. We also thank Don Olson, at New York State Health Department, for inspiring this emerging field of “archaeo-epidemiology” with his innovative study of the 1918 pandemic in New York City [11]. Finally, we are indebted to 2 anonymous reviewers for excellent comments.
Financial support: Danish Medical Research Council (grant 271-07-0555 to V.A.); National Institutes of Health, Fogarty International Center (support to C.V.) and Laboratory of Infectious Diseases and Office of Global Research, National Institute of Allergy and Infectious Diseases (support to L.S.).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gog JR, Rimmelzwaan GF, Osterhaus AD, Grenfell BT. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proc Natl Acad Sci USA. 2003;100:11143–11147. doi: 10.1073/pnas.1830296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson NM, Cummings DA, Cauchemez S, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson NM, Cummings DA, Fraser C, Cajka JC, Cooley PC, Burke DS. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–452. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Germann TC, Kadau K, Longini IM, Jr, Macken CA. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci USA. 2006;103:5935–5940. doi: 10.1073/pnas.0601266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longini IM, Jr, Nizam A, Xu S, et al. Containing pandemic influenza at the source. Science. 2005;309:1083–1087. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- 7.Gani R, Hughes H, Fleming D, Griffin T, Medlock J, Leach S. Potential impact of antiviral drug use during influenza pandemic. Emerg Infect Dis. 2005;11:1355–1362. doi: 10.3201/eid1109.041344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low RB. The incidence of epidemic influenza during 1918–1919 in Europe and in the western hemisphere, in Pandemic of Influenza 1918–19. London: HMOS; Reports on Public Health and Medical Subjects. 1920:199–348.
- 9.Viboud C, Tam T, Fleming D, Handel A, Miller MA, Simonsen L. Transmissibility and mortality impact of epidemic and pandemic influenza, with emphasis on the unusually deadly 1951 epidemic. Vaccine. 2006;24:6701–6707. doi: 10.1016/j.vaccine.2006.05.067. [DOI] [PubMed] [Google Scholar]
- 10.Chowell G, Ammon CE, Hengartner NW, Hyman JM. Transmission dynamics of the great influenza pandemic of 1918 in Geneva, Switzerland: assessing the effects of hypothetical interventions. J Theor Biol. 2006;241:193–204. doi: 10.1016/j.jtbi.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 11.Olson DR, Simonsen L, Edelson PJ, Morse SS. Epidemiological evidence of an early wave of the 1918 influenza pandemic in New York City. Proc Natl Acad Sci USA. 2005;102:11059–11063. doi: 10.1073/pnas.0408290102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morens DM, Fauci AS. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis. 2007;195:1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 13.Hansen S. Ugelisterne. Ugeskr Læger. 1920;82:268–270. [Google Scholar]
- 14.Anonymous. Ugentlig oversigt over fødsler sygdomme og dødsfald i København (weekly overview over births, diseases, and deaths in Copenhagen, in Danish and French) Stadslægen, Københavns Kommune, The Medical Officer, The City of Copenhagen; 1910–1919. [Google Scholar]
- 15.Ulrik A. Influenzaepidemien. Ugeskr Læger. 1918;30:1712–1714. [Google Scholar]
- 16.Anonymous. Statistiske Oplysninger for København og Frederiksberg. Statistical year book for Copenhagen and Fredriksberg. Copenhagen: The City of Copenhagen; 1911–1921. [Google Scholar]
- 17.Serfling RE, Sherman IL, Houseworth WJ. Excess pneumonia-influenza mortality by age and sex in three major influenza A2 epidemics, United States, 1957–58, 1960 and 1963. Am J Epidemiol. 1967;86:433–441. doi: 10.1093/oxfordjournals.aje.a120753. [DOI] [PubMed] [Google Scholar]
- 18.Simonsen L, Reichert TA, Viboud C, Blackwelder WC, Taylor RJ, Miller MA. Impact of influenza vaccination on seasonal mortality in the US elderly population. Arch Intern Med. 2005;165:265–272. doi: 10.1001/archinte.165.3.265. [DOI] [PubMed] [Google Scholar]
- 19.Viboud C, Grais RF, Lafont BA, Miller MA, Simonsen L. Multinational impact of the 1968 Hong Kong influenza pandemic: evidence for a smoldering pandemic. J Infect Dis. 2005;192:233–248. doi: 10.1086/431150. [DOI] [PubMed] [Google Scholar]
- 20.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci. 2007;274:599–604. doi: 10.1098/rspb.2006.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elveback LR, Fox JP, Ackerman E, Langworthy A, Boyd M, Gatewood L. An influenza simulation model for immunization studies. Am J Epidemiol. 1976;103:152–165. doi: 10.1093/oxfordjournals.aje.a112213. [DOI] [PubMed] [Google Scholar]
- 22.Cauchemez S, Carrat F, Viboud C, Valleron AJ, Boelle PY. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Stat Med. 2004;23:3469–3487. doi: 10.1002/sim.1912. [DOI] [PubMed] [Google Scholar]
- 23.Glass RJ, Glass LM, Beyeler WE, Min HJ. Targeted social distancing design for pandemic influenza. Emerg Infect Dis. 2006;12:1671–1681. doi: 10.3201/eid1211.060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonsen L, Olson DR, Viboud C, Heiman E, Miller MA, Reichert TA. The threat of pandemic influenza: are we ready? Forum on Microbial Threats series. Washington, DC: Institute of Medicine; 2004. Pandemic influenza and mortality: past evidence and projections for the future. 1-26-46. [Google Scholar]
- 25.Simonsen L, Clarke MJ, Schonberger LB, Arden NH, Cox NJ, Fukuda K. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J Infect Dis. 1998;178:53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- 26.Boza T. Spanskesyken i Noreg 1918–19. Tidsskr Nor Lægeforen. 2001;121:3551–3554. [PubMed] [Google Scholar]
- 27.Hansson R. Litt om influenza før og nu. Tidsskr Nor Lægeforen. 1919;39:268–272. 345–348. [Google Scholar]
- 28.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 29.Kobasa D, Jones SM, Shinya K, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 30.Murray CJ, Lopez AD, Chin B, Feehan D, Hill KH. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918–20 pandemic: a quantitative analysis. Lancet. 2006;368:2211–2218. doi: 10.1016/S0140-6736(06)69895-4. [DOI] [PubMed] [Google Scholar]
- 31.Bootsma MC, Ferguson NM. The effect of public health measures on the 1918 influenza pandemic in U.S. cities. Proc Natl Acad Sci USA. 2007;104:7588–7593. doi: 10.1073/pnas.0611071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishiura H. Time variations in the transmissibility of pandemic influenza in Prussia, Germany, from 1918–19. Theor Biol Med Model. 2007;4:20. doi: 10.1186/1742-4682-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acuna-Soto R, Castaneda L. Options for the control of influenza VI (Toronto) United Kingdom: International Society for Influenza and Other Respiratory Virus Diseases; 2007. Reconstruction of influenza epidemics in Mexico [abstract P1511] p. 320. [Google Scholar]
- 35.Cheng KF, Leung PC. What happened in China during the 1918 influenza pandemic? Int J Infect Dis. 2007;11:360–364. doi: 10.1016/j.ijid.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Cannell JJ, Vieth R, Umhau JC, et al. Epidemic influenza and vitamin D. Epidemiol Infect. 2006;134:1129–1140. doi: 10.1017/S0950268806007175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haberman SJ. The analysis of frequency data. Chicago: University of Chicago Press; 1974. [Google Scholar]
- 38.Nguyen-van-Tam JS, Hampson AW. The epidemiology and clinical impact of pandemic influenza. Vaccine. 2003;21:1762–1768. doi: 10.1016/s0264-410x(03)00069-0. [DOI] [PubMed] [Google Scholar]
- 39.Chowell G, Nishiura H, Bettencourt LMA. Comparative estimation of the reproduction number for pandemic influenza from daily case notification data. J R Soc Interface. 2007;4:155–166. doi: 10.1098/rsif.2006.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]