Summary
AMPK is a highly conserved sensor of cellular energy status that is activated under conditions of low intracellular ATP. AMPK responds to energy stress by suppressing cell growth and biosynthetic processes, in part through its inhibition of the rapamycin-sensitive mTOR (mTORC1) pathway. AMPK phosphorylation of the TSC2 tumor suppressor contributes to suppression of mTORC1, however TSC2-deficient cells remain responsive to energy stress. Using a proteomic and bioinformatics approach, we sought to identify additional substrates of AMPK that mediate its effects on growth control. We report here that AMPK directly phosphorylates the mTOR binding partner raptor on two well-conserved serine residues, and this phosphorylation induces 14-3-3 binding to raptor. The phosphorylation of raptor by AMPK is required for the inhibition of mTORC1 and cell cycle arrest induced by energy stress. These findings uncover a novel conserved effector of AMPK that mediates its role as a metabolic checkpoint coordinating cell growth with energy status.
Introduction
The AMP-activated protein kinase (AMPK) is a highly conserved heterotrimeric kinase complex composed of a catalytic (α) subunit and two regulatory (β and γ) subunits. AMPK is activated under conditions of energy stress, when intracellular ATP levels decline and intracellular AMP increases, as occurs during nutrient deprivation or hypoxia (Hardie, 2007). Upon energy stress, AMP directly binds to tandem repeats of crystathionine-β-synthase (CBS) domains in the AMPK γ subunit. Binding of AMP is thought to prevent dephosphorylation of the critical activation loop threonine in the α subunit (Hardie, 2007). The phosphorylation of the activation loop threonine is absolutely required for AMPK activation. Biochemical and genetic analyses in worms, flies, and mice have revealed that the serine/threonine kinase LKB1 represents the major kinase phosphorylating the AMPK activation loop under conditions of energy stress across metazoans (Apfeld, 2004; Shaw et al., 2005; Sakamoto et al., 2005, Mirouse et al., 2007, Lee, J.H. et al., 2007).
LKB1 was originally identified as a human tumor suppressor gene mutated in Peutz-Jeghers syndrome, an autosomal dominant inherited cancer disorder (Hemminki et al., 1998). In addition, LKB1 mutations occur in a large percentage (30–40%) of sporadic non small cell lung cancers (NSCLC) (Sanchez-Cespedes et al., 2002; Ji et al., 2007). Peutz-Jeghers syndrome (PJS) shares a number of clinical features with Cowden’s Disease, which is caused by inactivating mutations in the PTEN tumor suppressor. This phenotypic overlap suggested that LKB1-dependent signaling might negatively regulate some aspect of PI3-kinase signaling, analogous to PTEN function. However, while classic PI3K/Akt signaling is not elevated in LKB1-deficient cells, mTOR signaling is uniquely hyperactivated in LKB1-deficient murine embryonic fibroblasts (MEFs) and liver (Corradetti et al., 2004; Shaw et al., 2004b, 2005). Similarly, mTOR signaling is hyperactivated in hamartomas from LKB1-heterozygous mice (Shaw et al., 2004b) and in LKB1-deficient human lung carcinomas (Carretero et al., 2007).
mTOR (mammalian target of rapamycin), is a highly conserved nutrient responsive-regulator of cell growth found in all eukaryotes (Wullschleger et al., 2006). Whereas AMPK is active under nutrient-poor conditions and inactive under nutrient-rich conditions, mTOR is activated in the inverse pattern. In higher eukaryotes, mTOR activation requires positive signals from both nutrients (glucose, amino acids) and growth factors. mTOR, like its budding yeast orthologs, is found in two biochemically and functionally distinct signaling complexes (Wullschleger et al., 2006). The mTORC1 complex is nutrient-sensitive, acutely inhibited by rapamycin, and functions as a master regulator of cell growth, angiogenesis, and metabolism (Sabatini, 2006). mTORC1 is composed of four known subunits: mTOR, mLST8/Gbl, PRAS40, and the WD40 repeat-containing subunit raptor (Sabatini, 2006; Sancak et al., 2007; Vander Haar et al., 2007). Raptor acts as a scaffold to recruit downstream substrates such as 4EBP1 and ribosomal S6 kinase (p70S6K1), to the mTORC1 complex (Nojima et al., 2004; Schalm et al., 2004).
Genetic studies in Drosophila and mammalian cells identified the Tuberous Sclerosis Complex (TSC) tumor suppressors as critical upstream inhibitors of the mTORC1 complex. TSC2 (also known as tuberin) contains a GTPase activating protein (GAP) domain at its carboxyl terminus that inactivates the Rheb GTPase, which has been shown to associate with and directly activate the mTORC1 complex in vitro (Sancak et al., 2007). Loss of TSC1 or TSC2 therefore leads to hyperactivation of mTORC1. Phosphorylation of TSC1 and TSC2 serves as an integration point for a wide variety of environmental signals that regulate mTORC1 (Sabatini, 2006). Mitogen-activated kinases including Akt, Erk, and Rsk directly phosphorylate TSC2, leading to its inactivation by an unknown mechanism. In addition, another Akt substrate, PRAS40, was recently shown to bind and inhibit the mTORC1 complex. Upon phosphorylation by Akt, PRAS40 no longer inhibits mTORC1 (Sancak et al., 2007; Vander Haar et al., 2007).
In addition to these growth stimulatory cues that activate mTORC1, the complex is rapidly inactivated by a wide variety of cell stresses, thereby ensuring that cells do not continue to grow under unfavorable conditions. One of the unique aspects of the mTORC1 complex is that unlike many of the aforementioned growth factor activated kinases, it is dependent on nutrient availability for its kinase activity. Withdrawal of glucose, amino acids, or oxygen leads to rapid suppression of mTORC1 activity (Shaw and Cantley, 2006). Upon LKB1- and AMP-dependent activation of AMPK by nutrient loss, AMPK directly phosphorylates the TSC2 tumor suppressor on conserved serine sites distinct from those targeted by other kinases, which constitutes one mechanism through which glucose and oxygen control mTORC1 activation (Inoki et al., 2003; Corradetti et al., 2004; Shaw et al., 2004b, Liu et al., 2006).
We have found that cells lacking TSC2 remain responsive to energy stress, albeit less so than wild-type cells, suggesting that additional AMPK substrates may directly or indirectly modulate mTORC1 activity. Moreover, the relationship between glucose inactivation of AMPK and stimulation of TOR is conserved across all eukaryotes, including several that lack TSC2 orthologs such as C. elegans and S. cerevisiae. This suggests that either additional mechanisms exist to coordinate the kinase activity of these two master regulators of cell growth and metabolism, or AMPK must target additional conserved components of the pathway. Here, we find that the critical mTOR binding partner raptor is a direct substrate of AMPK, and that phosphorylation of raptor by AMPK is required for suppression of mTORC1 activity by energy stress. Further, we report that raptor phosphorylation is necessary for the full engagement of a AMPK-mediated metabolic checkpoint. These findings have broad implications for the control of cell growth by nutrients in a number of cellular and organismal contexts.
Results
Peptide library identification of the optimal substrate motif for AMPK reveals novel substrates controlling growth and metabolism
In an effort to find novel substrates of AMPK that may mediate its effects on growth and metabolic control, we determined its consensus phosphorylation motif with the aim of identifying proteins that carry optimal phosphorylation sequences. We utilized a positional scanning peptide library (PSPL) technique in which radiolabelled kinase assays are performed on a spatially arrayed set of peptide mixtures. Each peptide contains one fixed amino acid at a given position relative to a centrally fixed phospho-acceptor (an even mixture of serine or threonine) and degenerate amino acid mixtures at all flanking positions (Hutti et al., 2004). From the relative amount of phosphate incorporated into each peptide mixture, one obtains a quantitative measure of the selectivity for, and against, each individual amino acid residue at each position (Turk et al., 2006). We and our colleagues have previously used this technique to successfully identify optimal substrate motifs for a number of mammalian kinases, including CK2, Erk, PKA, Akt, Pim, Pak, MAP3K, and IKK family kinases (Hutti et al., 2004; Bullock et al., 2005; Bunkoczi et al., 2007; Hutti et al., 2007; Rennefahrt et al., 2007).
PSPL profiling revealed that AMPK is a highly selective kinase, strongly preferring basic residues in the −3 and −4 positions relative to the phospho-acceptor site. In addition, hydrophobic residues including leucine and methionine were strongly selected in the −5 position and the +4 position consistent with previous studies of the optimal peptide substrates for AMPK based on mutagenesis and molecular modeling (Scott et al., 2002; Towlee and Hardie, 2007). In addition, strong selection for polar residues in the +3 position was noted, with asparagine and aspartate being the most highly selected.
Comparing the optimal motif we identified from the peptide library screen to all known well-established in vivo substrates of AMPK shows excellent concordance (Fig. 1B). Each of these substrates contains not only the required basic residue in −3 or −4, and hydrophobic residues in −5 and +4, but they also exhibit strong bias towards the novel secondary selections for serine and valine in the −2 position and polar residues in the +3 position. The strong selectivity for particular residues in at least four out of the 9 flanking residues analyzed makes AMPK one of the most selective mammalian kinases we have examined thus far (out of ~60 kinases profiled to date, B.E.T, unpublished data). The high degree of selectivity at multiple residues substantially reduces the odds that any protein will contain serine residues within this sequence context by random chance, especially when examined for evolutionary conservation. This suggests that proteins that do carry this signature sequence are likely to be authentic substrates of AMPK or related kinases. Thus we used our optimal AMPK substrate motifs to mine protein databases to search for matching sequences - using bioinformatics tools including Scansite (http://mit.scansite.edu) and Prosite (http://ca.expasy.org/prosite/). We focused our efforts on those candidate substrates bearing optimal AMPK motifs in which the target serine and its critical flanking residues that dictate AMPK-dependent substrate specificity were conserved broadly throughout eukaryotes.
Figure 1. Peptide library profiling the optimal substrate motif for AMPK and comparison with known and candidate in vivo phosphorylation sites.
A. A spatially arrayed PSPL was subjected to in vitro phosphorylation with active AMPKα1 and radiolabelled ATP. Each peptide contained one residue fixed at one of nine positions relative to the centrally fixed phospho-acceptor (an equal mix of serine and threonine). Aliquots of each reaction were spotted onto avidin membrane, which was washed, dried and exposed to a phosphor storage screen, providing the array of spots shown in the figure. AMPK displayed strong selectivity at the −5, −4, −3, −2, +3, and +4 positions.
B. The optimal and secondary selections taken from triplicate analyses as in A are displayed. AMPK phosphorylation sites in the best established in vivo substrates of AMPK conform to the substrate motif derived from the peptide library data. All substrates shown were isolated in bioinformatics searches for proteins containing a conserved AMPK phosphorylation motif. These same searches yielded two novel predicted AMPK sites in raptor.
C. The predicted AMPK sites in raptor are highly conserved across evolution.
Raptor is a novel AMPK substrate
We first examined potential AMPK substrates that might underlie the ability of AMPK and its upstream kinase LKB1 to regulate cell growth and tumorigenesis. A number of recent studies have revealed that a key effector of AMPK signaling in the control of cell growth is the suppression of the mTORC1 signaling complex. We and others previously reported that the effect of LKB1 and AMPK to regulate mTORC1 is at least in part via direct phosphorylation of the TSC2 tumor suppressor by AMPK (Corradetti et al., 2004; Shaw et al., 2004b). Indeed Ser1387 of human TSC2 conforms perfectly to the AMPK optimal motif we obtained with our peptide library analysis and this residue and its flanking sequences are conserved across vertebrates and to Drosophila (Fig. 1B, data not shown).
However, two pieces of data suggested that TSC2 could not be the only substrate of AMPK to regulate mTORC1 signaling. First, the inverse regulation of TOR and AMPK by glucose levels is found throughout all eukaryotes examined thus far, including C. elegans and Saccharomyces cerevisiae, although a TSC2 ortholog is not found in either of those species. Secondly, while performing further experiments to examine the role of TSC2 in regulating energy stress, we found that while TSC2 is needed for rapid suppression of mTORC1 by the AMP-mimetic AICAR and the mitochondrial complex I inhibitor phenformin, mTORC1 is still potently inhibited by both of these AMPK activators in TSC2−/− MEFs (Fig. 2A). Similar findings have been made by others using glycolytic inhibitors (e.g. 2-DG) in TSC2-deficient cells (Hahn-Windgassen et al., 2005).
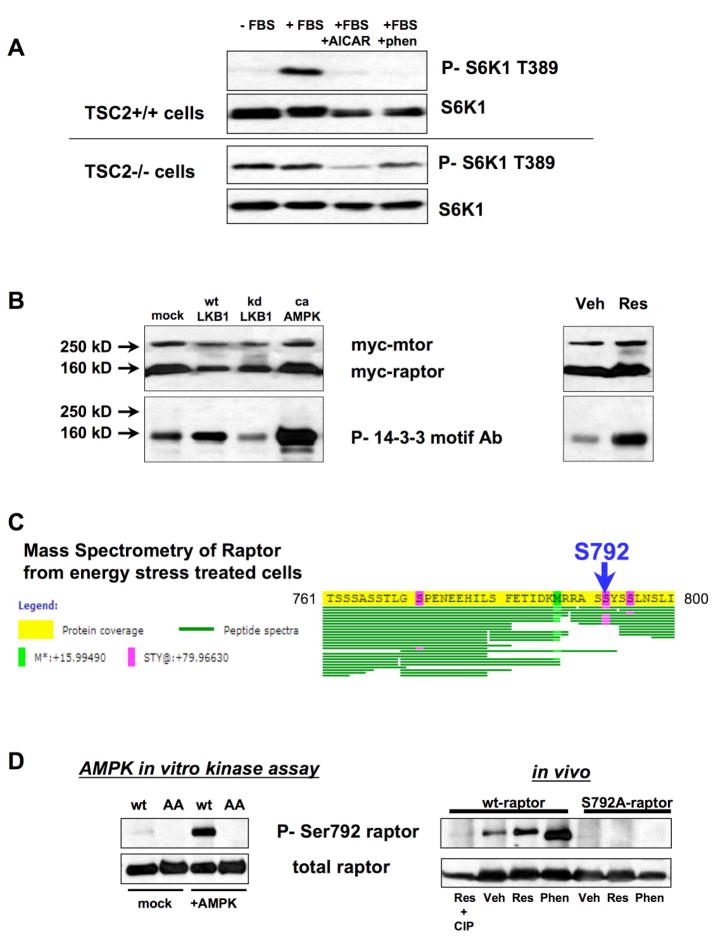
Figure 2. Raptor is phosphorylated in vitro and in vivo by AMPK.
A. mTORC1 signaling in TSC2-deficient cells remains responsive to energy stress. TSC2+/+ or TSC2−/− matched murine embryonic fibroblasts (MEFs) were serum-starved overnight (-ser) and replaced with fresh media containing 10% fetal bovine serum (+FBS) or serum-containing media with 2mM AICAR or 5mM phenformin. Cells were lysed one hour after media replacement. Lysates were immunoblotted for the mTORC1-dependent Thr389 phosphorylation in p70 S6K1 and for total S6K1 protein.
B. Overexpressed raptor is phosphorylated in HEK293 cells in an LKB1- and AMPK-dependent manner. Left panel: myc-tagged mTOR and myc-tagged raptor were co-expressed in HEK293 cells with empty vector, wild-type LKB1, kinase-dead LKB1, or a constitutively active AMPKα1 allele (1–312 truncation). Raptor phosphorylation was detected using the phospho-14-3-3 motif antibody. Right panel: HEK293 cells expressing mTOR and raptor were treated with 50μM resveratrol for 30 min and phosphorylation of raptor was detected with the Phospho-14-3-3 motif antibody.
C. Raptor is phosphorylated at a high level on serine 792 following resveratrol treatment. Mass Spectrometry was performed on raptor protein purified from resveratrol treated HEK293s as in panel B. Commassie-stained raptor protein was isolated from an SDS-polyacrylamide gel and subjected to chymotryptic digestion prior to analysis by LC-MS/MS. Amino acids 761 – 800 of human raptor are shown here. Each recovered peptide is illustrated by a single green line. Phosphorylated residues are shown in magenta.
D. A phospho-specific antibody against Serine792 of raptor recognizes raptor phosphorylated in vitro by AMPK (left) as well as wild-type, but not S792A mutant, raptor (right) following treatment with 50μM resveratrol or 5mM phenformin in HEK293 cells.
Our bioinformatics analysis revealed that the mTOR binding partner raptor contains two conserved serine sites that match the AMPK consensus motif (serine 722 and serine 792 of human raptor). Importantly, the critical residues flanking raptor Ser792 which were found in the peptide library studies to be important for recognition by AMPK are highly conserved through Drosophila, C. elegans, and Dictyostelium, as well as in both budding and fission yeast (Fig. 1C). Such a high degree of conservation is rare among phosphorylation sites. For example, of the ten best established AMPK substrates shown in Fig. 1B, only two of them are conserved across eukaryotes (ACC1 Ser1216 and HMG CoR Ser862). Moreover, half of the known AMPK substrate proteins, including TSC2, have no orthologs in primitive eukaryotes. The striking conservation in the candidate AMPK sites in raptor suggested it could represent an ancestral AMPK target that dictates the responsiveness of TOR to nutrients across eukaryotes.
To test the possibility that raptor is an AMPK substrate, we first examined whether we could detect phosphorylation of raptor in cultured cells using phospho-motif antibodies. These antibodies broadly recognize phosphorylated serine or threonine residues found within a specific sequence motif (Zhang et al., 2002). Interestingly, we found that the “14-3-3 motif” antibody, which was generated against peptides bearing R-X-X-pS or R-X-X-X-pS sequences, recognized raptor in HEK293 cells. Co-expression with wild-type and kinase-dead LKB1 lead to an increase and decrease, respectively, in reactivity of raptor to the antibody (Fig. 2B). Moreover, co-transfection with a truncated constitutively active allele of AMPKα1 resulted in a dramatic increase in raptor phosphorylation, and activation of endogenous AMPK through the use of the polyphenol compound resveratrol also stimulated acute phosphorylation of raptor (Fig. 2B).
Tandem mass spectrometry was then used to identify the specific sites of phosphorylation of raptor in cultured cells. Epitope-tagged raptor was co-transfected with mTOR in HEK293T cells. Cells were either untreated, or treated with either resveratrol or phenformin, both of which potently activate AMPK in HEK293T cells. Mass spectrometry (MS) of chymotryptic fragments of raptor from resveratrol- and phenformin-treated cells revealed that the Ser792 site was phosphorylated at high stoichiometry in both samples, with 5 of the 7 peptides identified containing this serine residue being phosphorylated with either treatment (Fig. 2C), unlike the untreated sample which revealed 2 of 9 peptides bearing phosphate at raptor Ser792 (data not shown). The region flanking the candidate Ser722 site was not well-represented in our mass spectrometry analysis despite repeated attempts, including digestion with alternative proteases (see complete MS analysis of raptor phosphorylation sites in Fig. S1). Notably, during the course of this study, two large scale analyses of phospho-proteins from rat and mouse liver revealed phosphorylation of endogenous raptor at Ser722, suggesting that it is indeed a bona fide phosphorylation site in vivo (Moser and White, 2006;Villen et al. 2007)
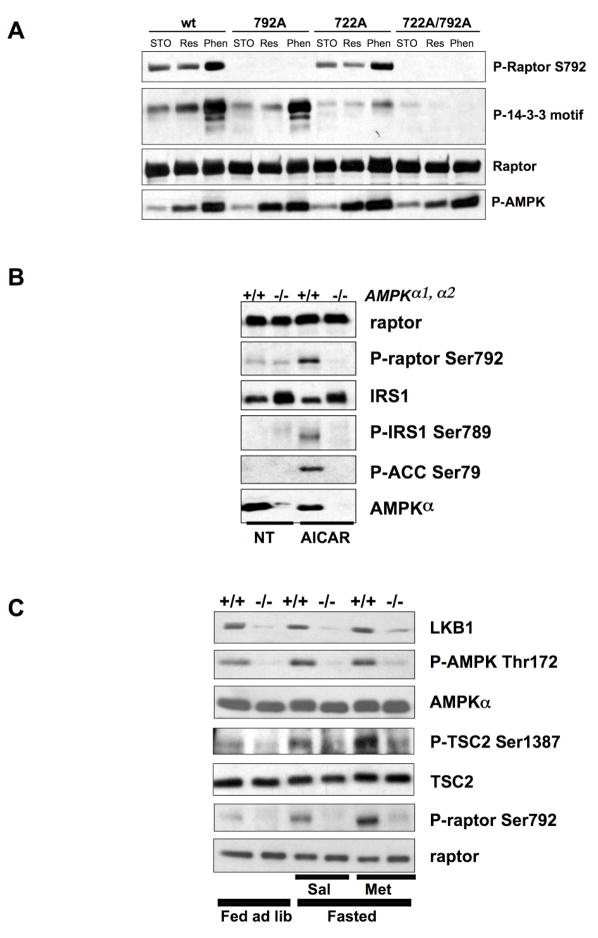
A phospho-specific antibody against the Ser792 site in human raptor was generated and its specificity was assessed using epitope-tagged wild-type or S792A mutant raptor overexpressed in HEK293T cells, under conditions analogous to those employed for the mass spectrometry (Fig. 2D). In addition, we examined whether purified AMPK could directly phosphorylate raptor at Ser792 in vitro. Active AMPK rapidly and potently induced raptor Ser792 phosphorylation in vitro (Fig. 2D). Similarly, employing non-phosphorylatable mutants, we mapped the sites recognized by the 14-3-3 motif antibody. As expected, we found that reactivity to the phospho-Ser792 antibody was unaffected in the S722A mutant. Surprisingly, the AMPK induced reactivity of raptor with the 14-3-3 motif antibody was minimally affected in the S792A mutant but was dramatically reduced in the S722A mutant (Fig. 3A). Reactivity was completely abolished in the S722A/S792A double mutant (henceforce referred to as the “AA mutant”). These results suggest that AMPK activation can induce phosphorylation of both serine 722 and serine 792.
Figure 3. Raptor Ser722 and Ser792 are phosphorylated by AMPK in cultured MEFs and in murine liver in an AMPK- and LKB1-dependent manner.
A. Both Ser722 and Ser792 are phosphorylated in an AMPK-dependent manner in HEK293 cells. HEK293 cells were transfected with wild-type, S722A, S792A, or the double mutant S722A/S792A raptor allele and treated as indicated. Raptor was immunoprecipitated and immunoblotted with the phospho-Ser792, phospho-14-3-3 motif, or anti-myc epitope tag antibody. Phospho-ACC was immunoblotted from the total cell extracts to illustrate the degree of AMPK activation in the cells.
B. Endogenous raptor is phosphorylated at Ser792 in wild-type but not AMPK-deficient (AMPKα1−/−, α2−/−) immortalized MEFs. MEFs were treated with 2mM AICAR for 1h (AICAR) or left untreated (NT) and total cell extracts were immunoblotted with the indicated antibodies.
C. Endogenous raptor is phosphorylated at Ser792 in wild-type but not LKB1-deficient murine liver following fasting and metformin treatment. 8 week old mice were either fed ad libidum (ad lib) or fasted 18h and treated with either 250mg/kg metformin in saline (Met) or saline alone (Sal) for 1h. Total cell extracts made from harvested livers were immunoblotted with the indicated antibodies.
To determine whether AMPK is the physiological kinase for phosphorylation of endogenous raptor at Ser792, immortalized wild-type or AMPK-deficient MEFs (bearing a targeted disruption of both AMPK α genes) were treated with the AMP-mimetic AICAR, followed by immunoblotting for phospho-Ser792 raptor. As controls, we also examined phosphorylation of two well-established AMPK substrates (ACC1/2 Ser79 and IRS1 Ser789). As seen in Fig. 3B, raptor Ser792 is phosphorylated following AICAR treatment in wild-type but not AMPK-null MEFs, precisely paralleling phospho-ACC and phospho-IRS1, thereby indicating that raptor Ser792 is a bona fide AMPK site in vivo. To further define the physiological conditions under which raptor Ser792 phosphorylation is modulated by AMPK, we examined raptor Ser792 phosphorylation in liver extracts from wild-type or LKB1-liver specific KO mice fed ad libitum, fasted, or treated with the biguanide diabetes therapeutic metformin. We have previously shown that metformin rapidly activates AMPK in murine liver in manner completely dependent on LKB1 (Shaw et al., 2005). Likewise, we found that raptor Ser792 phosphorylation in murine liver required LKB1, was slightly potentiated in fasted mice, and was dramatically increased by metformin treatment (Fig. 3C). These results were further extended in isolated primary hepatocytes from wild-type and LKB1-deficient liver (Fig. S2). Taken altogether, these results indicate that endogenous raptor Ser792 is phosphorylated in multiple mammalian tissue types in an LKB1- and AMPK-dependent manner following energy stress.
Raptor phosphorylation is required for inhibition of mTORC1 by AMPK
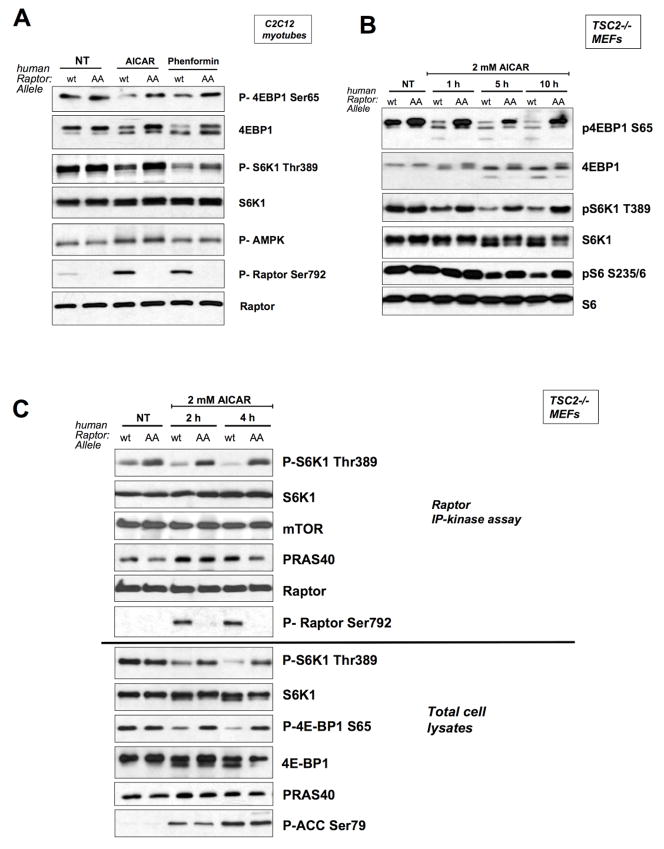
To examine the requirement for raptor phosphorylation in the regulation of mTORC1 activity by energy stress, we utilized the non-phosphorylatable AA mutant in which both Ser722 and Ser792 were replaced by alanine. To assess the physiological role of raptor Ser722 and Ser792 phosphorylation in mTORC1 regulation, we replaced the endogenous raptor by creating cell lines stably expressing low levels of human wild-type or AA raptor using retroviral expression, followed by subsequent knockdown of the endogenous murine raptor utilizing a lentivirally- expressed shRNA that does not target the human raptor sequence. In this manner, we replaced endogenous raptor with human wild-type or AA mutant in three murine cell lines: C2C12 myoblasts, TSC2+/+ p53−/− MEFs, and TSC2−/− p53−/− MEFs. In murine raptor lentiviral shRNA infected cultures lacking reconstitution with human raptor, we observed functional suppression of raptor levels and mTORC1 signaling (see Fig. S3). We then examined the requirement for raptor Ser722 and Ser792 phosphorylation in mTORC1 suppression following AICAR or phenformin treatment in these stable cell lines. Mutation of these sites prevented AMPK agonists from fully suppressing mTORC1, both in cells with normal mTOR signaling (C2C12, TSC2+/+ MEFs) (Fig. 4A, S4) and in cells lacking TSC2 (Fig. 4B, S4). Despite the elevated basal levels of mTORC1 activity in TSC2−/− MEFs, AICAR or phenformin treatment potently suppressed mTORC1 signaling, an effect that was almost fully abolished by reconstitution with the AA raptor allele. In AA raptor mutant expressing TSC2−/− MEFs, mTORC1 activity levels were dramatically elevated compared to TSC2−/− MEFs expressing wild-type raptor at all timepoints following AICAR treatment (Fig. 4B). Similar results were seen with phenformin, which activates AMPK via distinct mechanism (AICAR is an AMP mimetic, phenformin is mitochondrial complex I inhibitor); notably each may have additional distinct effects on signaling independent of the LKB1/AMPK pathway. Altogether our data demonstrate that raptor phosphorylation on serines 722/792 is required for full mTORC1 suppression by AMPK agonists in all cell types we examined. Furthermore, these findings indicate that TSC2 and raptor represent the major targets of AMPK required for the suppression of mTORC1 in mouse embryonic fibroblasts.
Figure 4. Phosphorylation of serine 722 and 792 is required to inhibit mTORC1 following energy stress in a variety of cell types.
A. C2C12 cells in which endogenous raptor has been knocked down were stably reconstituted with human wild-type or AA raptor (see Fig S3), and were treated with 1mM AICAR or 1mM phenformin for 1 hour as indicated. Total cell extracts were immunoblotted with indicated antibodies to examine mTORC1 signaling.
B. TSC2−/−, p53−/−, raptor knockdown MEFs stably reconstituted with wild-type or AA raptor were treated with 2mM AICAR as indicated and immunoblotted with indicated antibodies to examine mTORC1 signaling.
C. TSC2−/−, p53−/−, raptor knockdown MEFs stably reconstituted with wild-type or AA raptor were treated with 2mM AICAR as indicated. Raptor was immunoprecipitated in CHAPS buffer and assayed for mTORC1 kinase activity using purified S6K1 as a substrate as previously described (Sancak et al., 2007). Top: IP-kinase assays were immunoblotted for phosphorylation of purified S6K1 substrate using Phospho-Thr389 S6K1 antibody as well as for level of immunoprecipitated raptor, mTOR, and PRAS40. Bottom: 5% of the total cell extracts that raptor was immunoprecipitated from were immunoblotted with indicated antibodies.
AMPK phosphorylation of raptor induces 14-3-3 binding
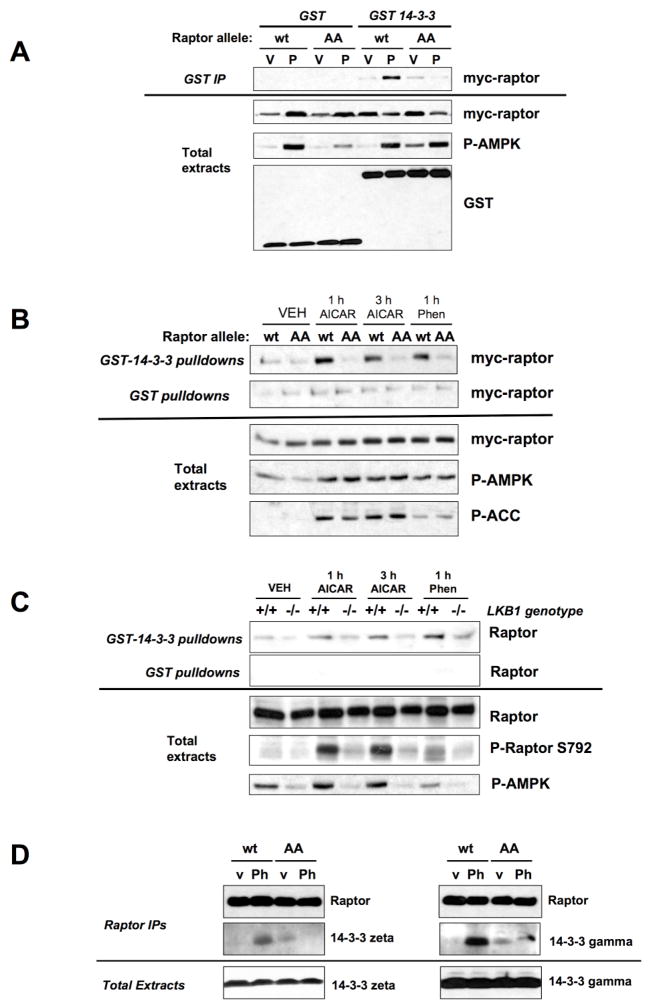
We next considered the mechanism by which AMPK-mediated raptor phosphorylation leads to inactivation of the mTORC1 kinase complex in vivo. We investigated the possibility that phosphorylation of raptor leads to the specific association or disassociation of proteins with the mTORC1 complex. Mass spectrometry was utilized to identify proteins co-immunoprecipitating with overexpressed wild-type raptor following phenformin treatment in HEK293 cells. Among the few novel proteins identified co-precipitating with raptor were two isoforms of 14-3-3 (Fig. S5). A common mechanism for phosphorylation-based inactivation of target proteins is through direct phosphorylation-dependent binding to the 14-3-3 family of proteins (Bridges and Moorhead, 2005). As AMPK-mediated phosphorylation of raptor also created an epitope recognized by the 14-3-3 binding motif antibody (Fig. 2B, 3A), we more closely examined the possibility that phosphorylation of serine 722 and 792 may induce 14-3-3 binding to raptor. Exhaustive peptide library screening and proteomic analyses have revealed that 14-3-3 proteins generally interact with R-X-X-pS/pT-X-P or R-X-X-X-pS/pT-X-P target sequences. Raptor Ser722 and Ser792 both contain the required upstream arginine residue, however neither site contains a proline residue in the +2 position, although several well-established 14-3-3 binding sites also lack proline at +2 (Cbl, IRS-1, PRAS40). Moreover, both Ser722 and Ser792 in raptor have residues at +1 and +2 that arose as secondary selections in peptide library experiments (Yaffe et al., 1997; Rittinger et al., 1999).
We first examined whether 14-3-3 bound to raptor when co-expressed, in an AICAR- or Serine 722/Ser792-dependent manner. Co-expressed GST-14-3-3, but not GST alone, immunoprecipitated with wild-type, but not AA mutant raptor, when cells were subjected to AICAR (Fig. 5A). In addition, recombinant GST-14-3-3 protein fixed to beads precipitated wild-type but not AA raptor in lysates from the MEF stable cell lines treated with AICAR or phenformin (Figure 5B). Furthermore, endogenous raptor co-precipitated with recombinant 14-3-3 protein from wild-type but not LKB1-deficient MEFs following treatment (Fig. 5C). Finally, consistent with the original mass spectometry data (Fig. S5), endogenous 14-3-3 zeta and gamma isoforms co-immunoprecipitated with wild-type but not AA raptor in a phenformin-dependent manner (Fig. 5D). However, it is worth noting that little specificity has been demonstrated for 14-3-3 isoforms other than 14-3-3 sigma, and many of the isoforms can form heterodimers with each other (Gardino et al., 2006; Wilker et al., 2005). Thus, we expect the 14-3-3 isoforms that bind AMPK-phosphorylated raptor may vary between cell types based on expression levels.
Figure 5. AMPK Phosphorylation of Raptor induces 14-3-3 association.
A. Wild-type but not AA mutant raptor complexes with 14-3-3 only under energy stress conditions. HEK293 cells were co-transfected with pEBG or pEBG-14-3-3 with wild-type or AA mutant raptor then complexes were precipitated on glutathione beads. Beads or total cell extracts were immunoblotted with the indicated antibodies. Cells were treated with V, vehicle (DMEM) or P, 5mM phenformin for 1h.
B. Wild-type but not AA mutant raptor precipitates with recombinant GST-14-3-3 protein in extracts from energy stress treated TSC2−/− MEFs stably reconstituted with human raptor alleles. GST protein pulldowns or total cell extracts were immunoblotted with the indicated antibodies.
C. Endogenous raptor binds to immobilized recombinant GST-14-3-3 protein, but not recombinant GST protein, from extracts cells treated with energy stress in an LKB1-dependent manner. GST protein pulldowns or total cell extracts were immunoblotted with the indicated antibodies.
D. Myc-tagged raptor immunoprecipitates from phenformin (Ph) or vehicle (v) treated cells were eluted with myc peptide and immunoblotted for endogenous 14-3-3 isoforms as indicated.
AMPK phosphorylation of raptor regulates mTORC1 IP-kinase activity
14-3-3 has been shown to regulate its best studied binding partners through three distinct mechanisms, each involving allosteric conformational changes that: 1) induce changes in protein catalytic activity; 2) trigger a disruption of existing protein-protein interactions; or 3) cause changes in subcellular localization. Analyses of crystal structures of 14-3-3 isoforms bound to phosphopeptides suggest that 14-3-3 regulates the activity of many of its binding partners via allosteric stabilization of unfavorable states (“the molecular anvil” hypothesis) (Yaffe, 2002). We first examined whether we could detect suppression of mTORC1 IP-kinase activity by immunoprecipitating raptor from AICAR treated MEFs. Using conditions that were recently reported to reconstitute insulin-dependent stimulation of mTORC1 IP-kinase activity (Sancak et al., 2007), we found that raptor immunoprecipitates from AICAR treated cells showed a time-and dose dependent suppression of mTORC1 kinase activity towards purified S6K1 protein that paralleled the inhibition of mTORC1 signaling by AMPK activation in vivo (Fig. 4C). We subsequently examined the IP-kinase activity of raptor complexes containing the Serine 722A/S792A double mutant. As seen in Fig. 4C, mTORC1 complexes containing AA raptor were refractory to the inhibition of kinase activity seen in mTORC1 complexes containing wild-type raptor. Critically, the amount of mTOR found in association with raptor was not affected by mutation of raptor serines 722/792 or by AMPK activation (Fig. 4C). These data indicate that immunoprecipitates containing the same amount of complexed mTOR and raptor show differences in mTORC1 kinase activity dependent on serine 722/792 phosphorylation by AMPK.
We further examined the association of endogenous mTOR and endogenous raptor from wild-type, LKB1-deficient, and AMPKα-deficient MEFs. The amount of raptor and mTOR co-immunoprecipitating was constant in all contexts examined (Fig. S6). We also examined whether AICAR induced changes in the amount of endogenous PRAS40 co-immunoprecipitating with raptor. As seen in Figure 4C, AICAR treatment induced greater immunoprecipitation of PRAS40 with raptor, which was modestly suppressed in cells expressing the non-phosphorylatable raptor. However, the levels of PRAS40 immunoprecipitating with raptor do not strictly correlate with mTORC1 IP-kinase activity or with raptor phosphorylation, suggesting that PRAS40 association is not the key event dictating the impact of raptor phosphorylation on mTORC1 IP kinase activity. AMPK phosphorylation of raptor may lead to changes in the amount of both 14-3-3 and PRAS40 bound, which collectively act to suppress raptor-associated mTOR kinase activity. Finally, the subcellular localization of each of the raptor alleles with and without AMPK activation in the reconstituted C2C12 myoblasts, TSC2+/+ MEFs, or TSC2−/− MEFs was unchanged (e.g. Fig. S7).
AMPK phosphorylation of raptor engages a metabolic checkpoint and prevents apoptosis
Activation of AMPK by energy stress causes a metabolic checkpoint, in which cells with intact AMPK signaling undergo cell cycle arrest, while those cells defective for AMPK activation (e.g. LKB1-deficient) or key components of the AMPK pathway (e.g. TSC2- or p53-deficient) continue cycling and subsequently undergo apoptosis (Inoki et al. 2003; Corradetti et al., 2004; Shaw et al., 2004a,b; Jones et al., 2005; Buzzai et al., 2007). A failure to downregulate mTORC1 under conditions of energy stress preferentially induces cells to undergo accelerated apoptosis.
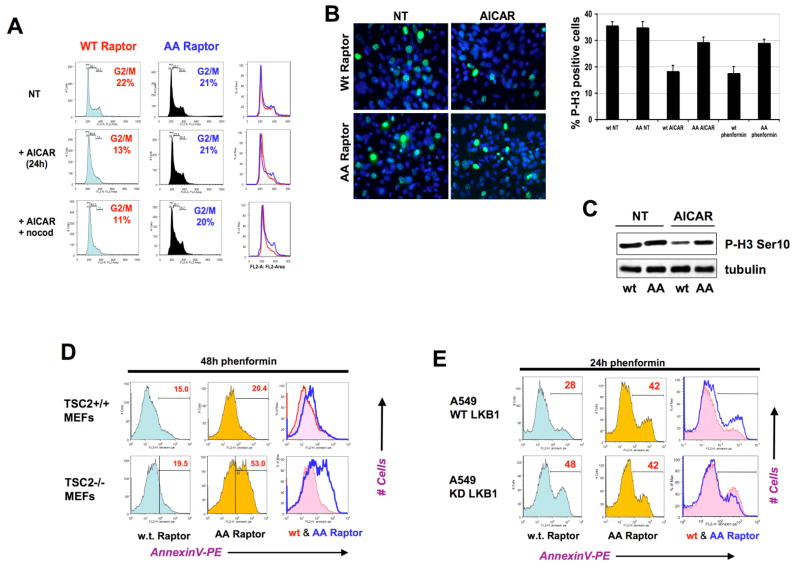
We therefore wished to determine whether phosphorylation of raptor by AMPK is required for full activation of this metabolic checkpoint, and whether the inability to phosphorylate raptor would affect the ability of cells to undergo growth arrest or apoptosis following energy stress. To exclude effects of AMPK regulation of TSC2 and p53 in this process, we utilized MEFs lacking both genes that were suppressed for endogenous raptor and reconstituted with human wild-type raptor or AA raptor (as discussed previously – see Fig. 4). Importantly, under standard growth conditions these cells grew at comparable rates and displayed no differences in viability or proliferation. We examined the response of these cells to several AMPK activating agents, analyzing their DNA content and cell cycle profile by propidium iodide and flourescence-activated cell sorting (FACS). MEFs lacking TSC2 and p53 but expressing wild-type raptor undergo a significant growth arrest in the G1 and S phases of the cell cycle following treatment with AICAR, depending on the time point examined (Fig. 6A, S9, data not shown). This was most readily observed as a decrease in the fraction of cells progressing into G2/M as quantified by DNA content (Fig. 6A, S8). Cells expressing wild-type raptor undergo a significant arrest (13% in G2/M as compared to 22% in the untreated cells) whereas the cells expressing AA raptor do not. Consistent with engagement of a cell cycle checkpoint following energy stress, the reduction in the cycling G2/M peak in AICAR- treated cells expressing wild-type raptor was paralleled by a decrease in the levels of the mitotic marker phospho-histone H3 Serine 10, as detected by immunoblotting (Fig. 6C). In parallel cultures expressing the non-phosphorylatable AA mutant raptor, the cells continued cycling, as observed by a complete absence of reduction in the G2/M population and a similar lack of suppression of phospho-histone H3 levels by AICAR (Fig. 6A, B, C). The suppression of mitotic cells was also observed using the phospho-histone H3 Ser10 antibody for immunocytochemistry on AICAR and phenformin treated cell populations (Fig. 6B). The percentage of wild-type raptor expressing cells arresting prior to G2/M and the percentage of AA raptor expressing cells failing to arrest were concordant in the DNA content FACS analysis, P-Histone H3 immunocytochemistry, and P-Histone H3 immunoblotting. By all three assays, AICAR and phenformin led to a similar suppression of mitotic cells in cells expressing wild-type raptor but not the AA mutant raptor.
Figure 6. Phosphorylation of Raptor at S722 and S792 dictates a metabolic checkpoint controlling growth arrest and apoptosis in response to energy stress.
A. TSC2−/−, p53−/− MEFs expressing wild-type raptor undergo G1/S arrest following AICAR treatment, while those expressing AA mutant raptor do not. Cells were left untreated (NT) or treated with 2 mM AICAR, or treated with 2 mM AICAR and 3h later exposed to nocodazole (+nocod) to arrest any cycling cells in G2/M. At 24h after AICAR treatment, all cells were fixed and analyzed for DNA content using propidium iodide and FACS analysis. The percentage of cells in the G2/M phase of the cell cycle are highlighted in each population.
B. Cells analogous to those in panel A were plated on coverslips and the next day left untreated (NT) or treated with 2mM AICAR and fixed 18h later. Cells were processed for phospho-histone H3 Ser10 immunocytochemistry to visualize the cells actively going through mitosis at the time the cells were fixed. DAPI was used as a nuclear counterstain. Histogram quantifies Phospho-Histone H3 immunocytochemistry on indicated cells treated with 5mM phenformin, or 2mM AICAR for 18h. At least 300 cells were scored for each condition.
C. Cell extracts from parallel plates to the ones analyzed in panel A were immunoblotted for phospho-histone H3 Ser10 as a marker of the percentage of cells in mitosis.
D. TSC2+/+, p53−/− MEFs or TSC2−/−, p53−/− MEFs expressing AA mutant raptor undergo apoptosis to a greater extent than those expressing WT raptor at later timepoints following energy stress treatment. Cell populations of indicated genotypes were treated with 5mM phenformin and at 48h the percentage of cells undergoing apoptosis was quantified using AnnexinV staining and FACS analysis. Histograms of cells expressing wild-type raptor (red trace) and AA raptor (blue trace) are overlaid in the right-most panel. The percentage of apoptotic cells in the annexin V-positive population is indicated in red at the upper right hand corner of each histogram.
E. Upstream AMPK signals from LKB1 are needed for the protective effect of WT raptor on apoptosis following energy stress. A549 human lung adenocarcinoma cells, which are null for LKB1, were stably reconstituted with wild-type LKB1 (WT) or mutant kinase dead K78I (KD) LKB1 expressing retroviruses. These cells were subsequently stably infected with retroviruses expressing wild-type or AA raptor. Each of the four resulting populations were treated with 5mM phenformin and analyzed for apoptosis as above.
In addition to AMPK phosphorylation dictating cell cycle arrest, profound effects on apoptosis were observed at later times following energy stress. Previously, in cells lacking LKB1, AMPK, or TSC2 function, inappropriate hyperactivation of mTORC1 was found to promote apoptosis under conditions of energy stress and rapamycin treatment led to suppression of the apoptotic response (Corradetti et al., 2004; Shaw et al, 2004b; Jones et al., 2005; Lee, C.H. et al., 2007). TSC2+/+ MEFs expressing human AA raptor underwent a modest increase in apoptosis in response to prolonged (48h) treatment with phenformin compared to identical cells expressing human wild-type raptor (48h). Strikingly, in cells lacking TSC2 that express the AA raptor mutant, and are thereby severely attenuated in their ability to downregulate mTORC1 following energy stress (see Fig. 4B), the percentage of cells undergoing apoptosis following phenformin more than doubled when compared to cells lacking TSC2 and expressing the human wild-type raptor (Fig. 6D).
To ensure that this differential apoptosis was due to signals coming from AMPK signaling and not a gain of function effect of the AA mutant, we examined whether the AA raptor mutant sensitized cells to apoptosis in a manner dependent on intact upstream AMPK signals. To test this hypothesis, we utilized A549 lung adenocarcinomas cell lines, which bear LKB1 missense mutations and are null for LKB1 protein expression. A549 cell lines stably reconstituted with wild-type or kinase-dead LKB1 were subsequently infected with retroviruses expressing wild-type or AA raptor. Stable cell lines expressing each raptor allele in combination with each LKB1 allele were then treated with phenformin and as before apoptotic rates were quantified using annexin V FACS sorting. As seen in Fig. 6E, wild-type but not AA mutant raptor conferred protection from phenformin-induced apoptosis only in cells expressing wild-type LKB1. In cells expressing kinase-dead LKB1 and hence unable to activate AMPK, we observed no difference in the percentage of cells undergoing cell death between those expressing wild-type and those expressing AA mutant raptor. This observation suggests that the survival signal requires both wild-type LKB1 and wild-type raptor, consistent with the maximal suppression of mTORC1 in these cells. Furthermore, the extent of apoptosis observed from overexpressing the AA raptor mutant in cells with wild-type LKB1 (42%) was equivalent to the degree of apoptosis in cells expressing kinase-dead LKB1 (42–48%). These results indicate at least in A549 cells, raptor phosphorylation is a key control point in the response to energy stress, and other targets of LKB1/AMPK signals such as TSC2 or p53 are not sufficient to induce effective growth arrest and prevent apoptosis in these cells. Taken together with the cell cycle analysis, these data suggest that cells unable to inhibit mTORC1 through LKB1-AMPK-raptor signaling continue to proliferate inappropriately under energy stress conditions, ultimately leading to increased rates of apoptosis.
Discussion
A fundamental requirement of all cells is that they couple the availability of nutrients to signals emanating from growth factors to drive proliferation only when nutrients are in sufficient abundance to guarantee successful cell division. We show here that the direct phosphorylation of the mTOR binding subunit raptor by AMPK under conditions in which ATP levels are low represents a biochemical mechanism by which eukaryotic cells couple their nutrient status to a central regulator of cell growth and proliferation.
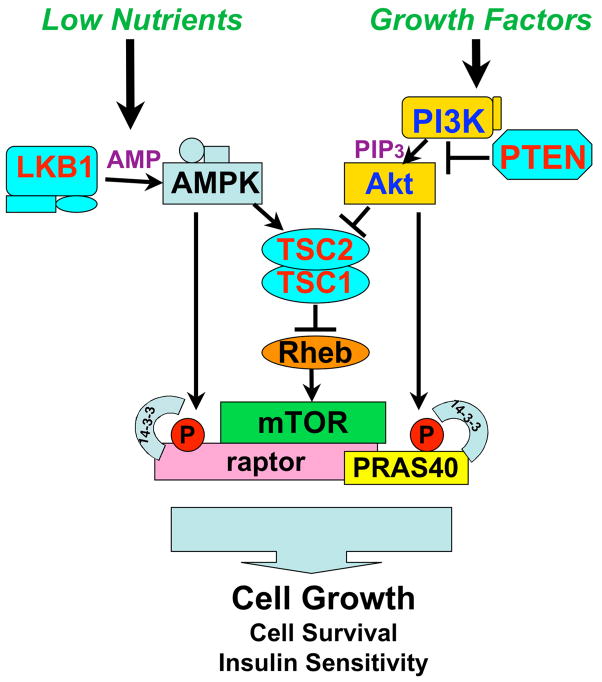
Taken together with previous studies, the findings reported here suggest that energy stress results in LKB-dependent activation of AMPK, which directly phosphorylates both TSC2 and raptor to inhibit mTORC1 activity by a dual-pronged mechanism (Fig. 7). It recently has become apparent that Akt-mediated activation of mTORC1 is also controlled via phosphorylation of two substrates by Akt: TSC2 and a novel mTORC1 inhibitor, PRAS40 (Sancak et al., 2007; Vander Haar et al., 2007). In parallel opposing pathways, AMPK-mediated phosphorylation of raptor induces 14-3-3 binding and inhibition of mTORC1, while Akt-mediated phosphorylation of PRAS40 induces its binding to 14-3-3 and activation of mTORC1.
Figure 7. Nutrients and Growth Factors control mTORC1 activity though common (TSC2) and unique (raptor, PRAS40) downstream targets.
Strikingly, both AMPK-mediated suppression of raptor and Akt-mediated suppression of PRAS40 involve the phosphorylation sites in each protein binding to 14-3-3, resulting in the inactivation of those targets. Inherited mutations in LKB1, TSC1. TSC2, and PTEN all result in hamartoma syndromes in humans indicating that hyperactivation of mTORC1 is a common biochemical mechanism underlying these genetic disorders.
We have demonstrated here that the AMPK phosphorylation sites in raptor play a key role in the function of AMPK as a metabolic checkpoint. This metabolic checkpoint is fully analogous to the DNA damage checkpoint, with kinases serving as sensors of the stress (here ATP loss), and then initiating a response to correct the pathological damage from the stress (stimulating creation of ATP) and halting cell cycle progression while the damage is being corrected. This metabolic checkpoint function of AMPK has been shown to be critical in a variety of cell types under conditions of low glucose, hypoxia, and following acute treatments with mitochondrial or glycolytic inhibitors, or AMP-mimetics (Inoki et al. 2003; Corradetti et al., 2004; Shaw et al., 2004a, b; Jones et al., 2005; Liu et al., 2006; Buzzai et al., 2007; Lee, C.H, et al., 2007). Inactivation of mTORC1 has previously been demonstrated to be critical for the ability of AMPK to enforce a metabolic checkpoint (Inoki et al., 2003; Shaw et al, 2004b). When mTORC1 cannot be inactivated under energy stress conditions, we show here that cells continue through the cell cycle and ultimately undergo apoptosis.
Recent evidence suggests the AMPK mediated metabolic checkpoint on cell growth is widely conserved across eukaryotes. Hyperactivation of AMPK suppressed cell proliferation in both Drosophila and Dictyostelium mutants with defective mitochondrial function (Mandal et al., 2005; Bokko et al., 2007). In C. elegans, AMPK (aak-2) and LKB1 (par-4) orthologs are required for the extended cell cycle arrest of germ cells in dauer worms (Narbonnay and Roy, 2006) as well as the arrest of L1 stage V lineage cells under starvation conditions (Baugh and Sternberg, 2006). In both lineages, AMPK or LKB1 loss causes inappropriate proliferation under nutrient poor conditions. In addition, AMPK activation is required in C. elegans for lifespan extension by daf-2, heat shock, and glycolytic inhibitors (Apfeld et al., 2004; Schulz et al., 2007). In budding yeast (SNF1) and Arabidopsis (KIN10/11), AMPK orthologs play key roles in regulating growth and lifespan in response to diverse nutrient and environmental stresses (Ashrafi et al., 2000; Baena-Gonzalez et al., 2007; Hong and Carlson, 2007; Thelander et al., 2004). Given these conserved functions for AMPK, it will be interesting to determine if the predicted AMPK phosphorylation sites in raptor orthologs in lower organisms play a role in these nutrient dependent controls on cell growth, aging, and stress response.
Taken altogether, our findings indicate that AMPK utilizes multiple targets in mammalian cells to effectively suppress mTORC1 signaling. The integral role that raptor plays in mTORC1 function and the remarkable conservation of the AMPK sites across eukaryotes suggest that raptor phosphorylation by AMPK orthologs may be an ancestral mechanism for coupling cell growth to nutrient status. The phosphorylation of raptor by AMPK could also play a physiological role in other mammalian processes that both AMPK and mTORC1 regulate including autophagy, angiogenesis, insulin sensitivity, mitochondrial metabolism, and specific transcriptional responses. In addition, the existence of this direct regulation of mTORC1 by AMPK suggests that widely used diabetes therapeutics such as metformin, which act through AMPK activation, or environmental factors such as exercise and diet that contribute to physiological AMPK activation, may modulate tumorigenesis through this previously unappreciated signaling route. The direct phosphorylation and inhibition of raptor function by AMPK also suggests a possible therapeutic window for the use of AMPK agonists to treat tumors arising in patients with Tuberous Sclerosis Complex or tumors exhibiting hyperactivation of mTOR via other genetic lesions.
As the response to a shortage of environmental nutrients and resultant loss in cellular energy represents one of the most fundamental pathological events of all organisms, we anticipate that further investigation of the downstream targets of AMPK will provide great insight into the emerging nexus of cancer, diabetes, and lifespan extension controlled by this ancestral signaling pathway.
Experimental Procedures
Materials
Antibodies to phospho-AMPK (T172), AMPK alpha, phospho-ACC (S79), ACC, raptor, mTOR, PRAS40, phospho-S6K1(T389), S6K1, phospho-ribosomal protein S6 (S235/236), ribosomal protein S6, eIF4E, phospho-Akt (S473), phospho-Erk (T202/Y204), phospho 4E-BP1(S65), 4E-BP1, phospho IRS1 (S789), 14-3-3 zeta, 14-3-3 gamma, myc epitope (9B11), myc epitope polyclonal, and phospho-14-3-3 substrate motif antibodies were obtained from Cell Signaling Technology. Anti-IRS1 and anti-phospho histone H3 (S10), and active recombinant AMPK were obtained from Millipore. Anti-Flag antibodies (M2 monoclonal and Flag polyclonal), phenformin, resveratrol, and AMP were obtained from Sigma. HA probe polyclonal and mTOR (N19) antibodies and protein A/G sepharose obtained from Santa Cruz Biotechnology. AICAR was obtained from Toronto Research Chemicals. Glutathione-sepharose was obtained from Amersham Pharmacia. Colloidal blue stain and Supersensitive ECL kits were obtained from Pierce. Additional experimental procedures can be found in the supplementary material online.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. John Asara of the Beth Israel Deaconess Medical Center Mass Spectometry Core (Boston, MA) for all mass spectrometry analysis. Authors thank Qingyuan Ge, Jianxin Xie, Thortsen Wiederhold, and Roberto Polakiewicz at Cell Signaling Technology for collaboration on the generation of the Raptor phospho-Ser792 and TSC2 phospho-Ser1387 antibodies. The authors want to thank Keith Laderoute and Benoit Viollet for their generous donation of the isogenic SV40-immortalized willd-type and AMPKa1, a2 double deficient MEFs and Renaud Dentin for assistance in generating the primary murine hepatocytes. The authors wish to thank Katja Lamia, Karen Cichowski, and Brendan Manning for critical reading of the manuscript. The authors would also like to thank David Sabatini and Lewis Cantley for encouragement and advice on this project. The work was supported in part from grants from the NIH to R.J.S. (R01 DK080425 and P01 CA120964) and B.E.T. (GM079498) and American Cancer Society (R.J.S.). D.B.S. was supported by training grant T32 CA009370 to the Salk Institute Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Bokko PB, Francione L, Bandala-Sanchez E, Ahmed AU, Annesley SJ, Huang X, Khurana T, Kimmel AR, Fisher PR. Diverse cytopathologies in mitochondrial disease are caused by AMP-activated protein kinase signaling. Mol Biol Cell. 2007;18:1874–1886. doi: 10.1091/mbc.E06-09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 2005;2005:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- Bullock AN, Debreczeni J, Amos AL, Knapp S, Turk BE. Structure and substrate specificity of the Pim-1 kinase. J Biol Chem. 2005;280:41675–41682. doi: 10.1074/jbc.M510711200. [DOI] [PubMed] [Google Scholar]
- Bunkoczi G, Salah E, Filippakopoulos P, Federov O, Müller S, Sobott F, Parker SA, Zhang H, Min W, Turk BE, Knapp S. Structural and functional characterization of the human protein kinase ASK1. Structure. 2007;15 doi: 10.1016/j.str.2007.08.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F, Viollet B, Thompson CB. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67:6745–6752. doi: 10.1158/0008-5472.CAN-06-4447. [DOI] [PubMed] [Google Scholar]
- Carretero J, Medina PP, Blanco R, Smit L, Tang M, Roncador G, Maestre L, Conde E, Lopez-Rios F, Clevers HC, Sanchez-Cespedes M. Dysfunctional AMPK activity, signalling through mTOR and survival in response to energetic stress in LKB1-deficient lung cancer. Oncogene. 2007;26:1616–1625. doi: 10.1038/sj.onc.1209951. [DOI] [PubMed] [Google Scholar]
- Corredetti MN, Inoki K, Bardeesy N, DePinho R, Guan KL. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18:1533–8. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardino AK, Smerdon SJ, Yaffe MB. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin Cancer Biol. 2006;16:173–182. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Hahn-Windgassen A, Nogueira V, Chen CC, Skeen JE, Sonenberg N, Hay N. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J Biol Chem. 2005;280:32081–32089. doi: 10.1074/jbc.M502876200. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- Hong SP, Carlson M. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem. 2007;282:16838–16845. doi: 10.1074/jbc.M700146200. [DOI] [PubMed] [Google Scholar]
- Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- Hutti JE, Turk BE, Asara JM, Ma A, Cantley LC, Abbott DW. IKK{beta} phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF{kappa}B pathway. Mol Cell Biol. 2007 doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, Liang MC, Cai D, Naumov GN, Bao L, Contreras CM, Li D, Chen L, Krishnamurthy J, Koivunen J, Chirieac LR, Padera RF, Bronson RT, Lindeman NI, Christiani DC, Lin X, Shapiro GI, Janne PA, Johnson BE, Meyerson M, Kwiatkowski DJ, Castrillon DH, Bardeesy N, Sharpless NE, Wong KK. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Lee CH, Inoki K, Karbowniczek M, Petroulakis E, Sonenberg N, Henske EP, Guan KL. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. Embo J. 2007;26:4812–4823. doi: 10.1038/sj.emboj.7601900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee SH, Shong M, Kim JM, Kim J, Chung J. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–1020. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia- induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Mirouse V, Swick LL, Kazgan N, St Johnston D, Brenman JE. LKB1 and AMPK maintain epithelial cell polarity under energetic stress. J Cell Biol. 2007;177:387–392. doi: 10.1083/jcb.200702053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moser K, White FM. Phosphoproteomic analysis of rat liver by high capacity IMAC and LC-MS/MS. J Proteome Res. 2006;5:98–104. doi: 10.1021/pr0503073. [DOI] [PubMed] [Google Scholar]
- Narbonne P, Roy R. Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development. 2006;133:611–619. doi: 10.1242/dev.02232. [DOI] [PubMed] [Google Scholar]
- Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- Rennefahrt UE, Deacon SW, Parker SA, Devarajan K, Beeser A, Chernoff J, Knapp S, Turk BE, Peterson JR. Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J Biol Chem. 2007;282:15667–15678. doi: 10.1074/jbc.M700253200. [DOI] [PubMed] [Google Scholar]
- Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, Gamblin SJ, Yaffe MB. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, McCarthy A, Smith D, Green KA, Grahame Hardie D, Ashworth A, Alessi DR. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. Embo J. 2005;24:1810–1820. doi: 10.1038/sj.emboj.7600667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose Restriction Extends Caenorhabditis elegans Life Span by Inducing Mitochondrial Respiration and Increasing Oxidative Stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Scott JW, Norman DG, Hawley SA, Kontogiannis L, Hardie DG. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J Mol Biol. 2002;317:309–323. doi: 10.1006/jmbi.2001.5316. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004a;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004b;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Ras, PI(3)K, and mTOR signaling controls tumor cell growth. Nature. 2006;441:424–30. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H. Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. Embo J. 2004;23:1900–1910. doi: 10.1038/sj.emboj.7600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- Turk BE, Hutti JE, Cantley LC. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide substrates. Nat Protoc. 2006;1:375–379. doi: 10.1038/nprot.2006.57. [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EW, Grant RA, Artim SC, Yaffe MB. A structural basis for 14-3- 3sigma functional specificity. J Biol Chem. 2005;280:18891–18898. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yaffe MB. How do 14-3-3 proteins work?-- Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, Polakievicz RD, Comb MJ. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.