Abstract
Melatonin is rhythmically synthesized and released by the avian pineal gland and retina during the night, targeting an array of tissues and affecting a variety of physiological and behavioral processes. Among these targets, astrocytes express two melatonin receptor subtypes in vitro, the Mel1A and Mel1C receptors, which play a role in regulating metabolic activity and calcium homeostasis in these cells. Molecular characterization of chick astrocytes has revealed the expression of orthologs of the mammalian clock genes including clock, cry1, cry2, per2, and per3. To test the hypothesis that pineal melatonin entrains molecular clockworks in downstream cells, we asked whether coculturing astrocytes with pinealocytes or administration of exogenous melatonin cycles would entrain metabolic rhythms of 2-deoxy [14C]-glucose (2DG] uptake and/or clock gene expression in cultured astrocytes. Rhythmic secretion of melatonin from light-entrained pinealocytes in coculture as well as cyclic administration of exogenous melatonin entrained rhythms of 2DG uptake and expression of Gallus per2 (gper2) and/or gper3, but not of gcry1 mRNA. Surprisingly, melatonin also caused a dose-dependent increase in mitotic activity of astrocytes, both in coculture and when administered exogenously. The observation that melatonin stimulates mitotic activity in diencephalic astrocytes suggests a trophic role of the hormone in brain development. The data suggest a dual role for melatonin in avian astrocytes: synchronization of rhythmic processes in these cells and regulation of growth and differentiation. These two processes may or may not be mutually exclusive.
Keywords: astrocytes, avian, circadian, development, melatonin
Introduction
The avian circadian system comprises multiple circadian oscillators, pacemakers, and photosensitive structures [1, 2]. These include the retina [3, 4], the avian homolog of the suprachiasmatic nucleus [SCN; 5-7], and the pineal gland [8, 9]. The pineal gland, and in some species, the retina, influence the system by secreting the neurohormone melatonin during the night [10-12]. However, the molecular mechanisms by which melatonin influences physiology and circadian behavior are not completely known.
Birds express three melatonin receptor subtypes: the Mel1A or MT1 receptor, the Mel1B or MT2 receptor, and the Mel1C receptor [13-15]. Based on receptor binding studies and in situ hybridization of the three receptor subtypes in chick diencephalic astrocytes, our laboratory [14] demonstrated that approximately 25% of these cells express Mel1A receptor mRNA, none express Mel1b receptor mRNA, and nearly all express Mel1C receptor mRNA. Additionally, when melatonin was rhythmically administered to these cultures, rhythms in glucose utilization and in the release of the glycolytic end products pyruvate and lactate were observed, with a circadian phase that corresponds to that of the melatonin cycle [14].
Molecular components responsible for the generation of circadian rhythms have been identified and extensively studied in several model systems [16, 17]. In mammals, a network of ‘clock genes’ forms an interlocking feedback loop that is believed to be responsible for the generation of circadian rhythms [18]. ‘Positive elements’ clock and brain muscle ARNT-like protein (bmal1 and bmal2) are rhythmically transcribed and translated (CLOCK and BMAL, respectively). They then dimerize in the cytoplasm, enter the nucleus and activate the transcription of ‘negative elements,’ period (per), and cryptochrome (cry), which then are translated, dimerize, and feedback to inhibit their own transcription by inhibiting the action of CLOCK/BMAL dimers. Several groups have identified avian orthologs of these clock genes in various species [19-25].
One of the great surprises in the discovery of molecular components of circadian oscillators of animal species ranging from Drosophila to mammals [17] was the observation that clock gene rhythmicity is not restricted to pacemaker tissues such as the pineal gland, retina, and SCN. Rather, rhythmic clock gene expression is a global phenomenon raising the current consensus view that the role of pacemaker tissues is to entrain peripheral oscillators rather than to drive them. In Drosophila, for example, the molecular clockworks within olfactory tissues are sufficient to drive rhythms in olfactory sensation [26]. Consistent with this view, certain peripheral tissues including cornea, liver, and lung explanted from transgenic mice containing an mPER2 promoter-driven luciferase gene exhibited rhythmicity in culture for several days before damping out. Furthermore, peripheral tissues from SCN-lesioned mice maintained PER2 rhythms that could be sustained by media exchange, although with some disruption in phase [27]. To differentiate pacemaker from downstream oscillators, our laboratory has previously employed a coculture preparation of immortalized SCN cells and target fibroblast cell lines [28, 29]. Here, we cocultured chick pinealocytes with target diencephalic astrocytes. These studies support the view that the pineal gland regulates target cells through the rhythmic secretion of melatonin, which may involve rhythmic clock gene expression. These data also indicate that pineal melatonin can regulate astrocyte development and mitosis.
Materials and methods
Astrocyte cultures
Astrocytes were harvested from embryonic day 17 chick brains (Hyline Hatcheries, Bryan, TX, USA). Embryos were extracted and the brains were removed. Diencephalic tissue was dissected and placed in Dulbecco’s modified Eagle’s medium (DMEM; Sigma Corp., St. Louis, MO, USA), then dissociated by triturating with 1% trypsin. Cells were filtered through nylon mesh (180 μm pore diameter) and centrifuged for 5 min at 1000 g. The supernatant was removed and the pellet was resuspended in DMEM with 10% fetal bovine serum and a 1% PSN antibiotic cocktail (penicillin, streptomycin, and neomycin). The suspension was filtered again through nylon mesh (77 μm pore diameter) and seeded into 75-cm2 tissue culture flasks. Cultures were maintained in darkness at 37 °C with 95% air and 5% CO2 in a humidified Napco CO2 incubator. Culture media was changed every other day until cell cultures reached confluence. Confluent cultures were processed for immunocytochemical analysis of glial fibrillary acidic protein (GFAP) to determine the purity of the cultures. Cultures were plated on glass-bottom dishes for 5-7 days. Cells were fixed with 4% p-formaldehyde (PFA) for 10 min, washed, and processed for immunocytochemical identification of GFAP (1 : 1000; Incstar, Stillwater, MN, USA) as previously described [30]. Differential interference contrast (DIC) microscopic images of cell cultures were captured using Simple PCI software (C-imaging, Hamamatsu Corp., Sewickley, PA, USA), and dual captures of DIC images and GFAP-immunoreactivity were superimposed to estimate the percentage of cultured cells that were actually GFAP immunoreactive astrocytes. In these parallel cultures, 87.1 ± 4.8% of the cells were immunoreactive to GFAP. The remaining cells may have been fibroblasts or astrocytes that did not react to the antiserum.
Pinealocyte : astrocyte coculture
Confluent astrocyte cultures were passaged into six-well plates (BD Biosciences, San Jose, CA, USA) designed to accommodate insert dishes for coculture and then cultured for 2 days to allow sufficient time to adhere. Pinealocytes were cultured as previously described [31] with modifications. Briefly, newly hatched White Leghorn chicks were killed by decapitation and the pineal glands transferred to sterile Dulbecco’s phosphate-buffered saline (PBS )with d-glucose. After 30 min of digestion in 0.05% trypsin with periodic trituration, the remaining debris was removed from the media and the cells were pelleted at 1000 g for 10 min. The pellet was resuspended in sufficient volume of McCoy’s 5a media supplemented with 1% KCl and 1% PSN to allow for the plating of two glands per well. Pinealocytes were cultured directly onto 0.2 μm mesh inserts (BD Biosciences) which were subsequently returned to their companion well on the six-well plate containing the 2-day-old astrocyte cultures (Fig. 1A, from BD Biosciences Product Insert). Control plates contained inserts without pinealocytes. Astrocytes were maintained in 3 mL of the DMEM-based medium while pinealocytes (and empty inserts) were administered 2 mL of the McCoy’s 5A-based medium. All media and supplements were purchased from Gibco/Invitrogen Corp (Grand Island, NY, USA) unless otherwise stated. Cultures were placed in 12 : 12 light : dark cycles for 7 days, as we have previously determined this to be sufficient time for the pinealocytes to confer rhythmicity to the astrocytes. On the eighth day, cultures were placed in constant darkness and samples taken every 4 hr for 48 hr.
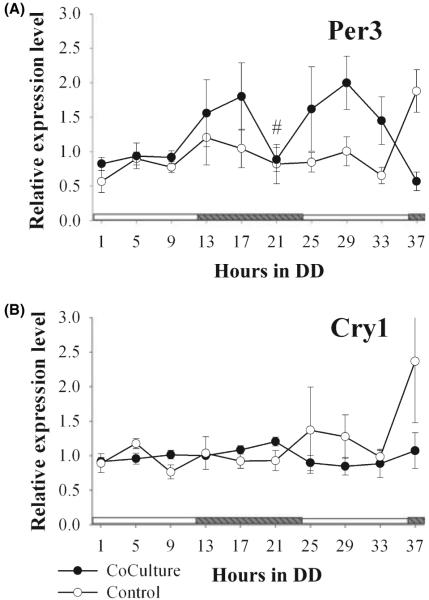
Fig. 1.

Rhythmic melatonin release in entrained pinealocytes confers a rhythm of glucose uptake in cocultured astrocytes. (A) Schematic of coculture design from BD Bioscience webpage illustrating the interaction between cell types in coculture. Here, pinealocytes are cultured onto the insert membranes and then the inserts are subsequently placed in a well with astrocytes cultured on the bottom of the well. (B) Melatonin release from pinealocytes was rhythmic during the first day in constant conditions, but decreasebd in amplitude by the second day. (C) Metabolic activity as measured by radiolabeled 2DG uptake was rhythmic for 2 days in cocultured astrocytes (black circles) as measured by ANOVA. Astrocytes cultured alone (white circles) showed no circadian rhythm in metabolism (n = 6 replicates for each data set). For determined rhythmic cycles, significant differences between peak-to-trough values are indicated by #P < 0.05 or *P < 0.001. Comparisons were made between the first observed peak and trough for each day for each treatment.
2-Deoxy [14C]-glucose collection and quantification
One hour prior to sampling, the cells were incubated with [14C]-2-deoxyglucose (2DG; 0.2 μCi/mL; American Radiolabeled Chemicals, St. Louis, MO, USA). Following incubation, media was removed and saved for melatonin quantification. Cells were rinsed twice with Dulbecco’s PBS (Gibco/Invitrogen Corp.), and harvested in Trizol reagent (Invitrogen Corp.) to extract total cellular RNA and soluble protein. Duplicate aliquots of cell lysate (200 μL) were placed in scintillant and counted on a Beckman scintillation counter. Uptake of 2DG was normalized to protein or total RNA content.
Melatonin quantification
Melatonin was measured using a radioimmunoassay method as in Fraser et al. [32], with modifications. Briefly, media samples from cocultured and control astrocyte cultures were incubated with melatonin antibody (Stockgrand Ltd., Surrey, UK) in tricine buffer (Sigma-Aldrich, Inc., St. Louis, MO) followed by incubation with competing [H3]-melatonin (Perkin Elmer, Waltham, MA, USA). After incubation, free melatonin was separated from bound melatonin using dextran-coated charcoal (Sigma-Aldrich, Inc.) and the bound melatonin counted using the above scintillation counter. Quantities are reported here as relative to the maximum amount of melatonin among all timepoints.
Exogenous melatonin administration
Confluent astrocyte cultures were passaged and plated into 75-cm2 tissue culture flasks, then allowed to grow to confluence (3-4 days). Cultures (n = 4 per treatment) were then placed into constant darkness and subjected to different cycles of melatonin administration. Cultures were given a cycle of melatonin supplemented media (5 nm) during the subjective night and normal media during the subjective day (MN) or an opposing cycle of melatonin supplemented media (5 nm) during the subjective day and normal media during the subjective night (MD), and samples were taken every 4 hr for 48 or 72 hr.
Astrocyte growth study
For cell growth studies, astrocytes and pinealocytes were cocultured as before. Images of the astrocytes were taken at zeitgeist time (ZT8) every day for 7 days using an Olympus IX-70 inverted microscope fitted with a Hamamatsu camera controller and processed using Simple PCI software. Quantitative measurements of cell coverage were analyzed using the public domain ImageJ program (developed at the U.S. National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/) and were determined as the ratio of the number of pixels in a specific area of the well covered by cells to the total number of pixels in that area. Six areas per well were quantified. To evaluate the rate of cell growth, confluent astrocyte cultures were passaged into 24 well plates (BD-Falcon, San Jose, CA, USA) and cultured for 2 days in DD. On the third day, and every 24 hr thereafter, the media was replaced at ZT12 with astrocyte media alone, or with media supplemented with 10 μm, 1 μm, 100 nm, 10 nm, 1 nm, 100 pm, or 10 pm melatonin (Sigma Corp.) in DMSO. Six wells per concentration of melatonin were analyzed. Beginning on day 3, samples were taken at the time of media replacement (ZT12) every day for 6 days to observe mitotic growth by DAPI staining. Cells were fixed in 4% PFA overnight at 4°C, washed with PBS, and incubated in DAPI for 20 min. Staining was visualized on the same imaging system fitted with a DAPI filter. Images were analyzed using ImageJ and stained nuclei were counted by hand from six regions per well, each region being 2.025 mm2 in area.
Quantitative real-time polymerase chain reaction
Total RNA, isolated with Trizol as above and subsequently treated with DNase I (Invitrogen Corp.), was primed with random hexamers and cDNA was synthesized by reverse transcription using a Superscript II RT polymerase chain reaction (PCR) kit (Invitrogen Corp.). An additional control reaction was performed in each experimental run by replacing reverse transcriptase enzyme with water for a selected duplicate sample. Relative quantification of selected genes was achieved by performing SYBR green-based real-time PCR using an ABI Prism 7500 Fast Sequence Detection instrument following the standard curve method outlined in the user protocol from Applied Biosystems (Foster City, CA, USA). Primers optimized for SYBR green real-time PCR amplification were designed for selected genes using Primer Express software (Applied Biosystems), and synthesized at our local gene technologies laboratory. Primer sequences used are as follows: gCry1, forward primer: CCGGGAAACGCCCAAA and reverse primer: TGCTCTGCCGCTGGACTT; gPer3, forward primer: CAGAATGGAAACGATCAGCCTAT and gPer3 reverse primer: TCGGGAGAAAACAGGAAGCA; and gPer2 forward primer: CCCCAGTAGTTGGTGCTCACTT; gPer2 reverse primer: GACTGGTGAGCGATACAACACTTT. Standard curves were generated from target gene cDNAs diluted at 1 : 50, 1 : 100, 1 : 250, 1 : 500, and 1 : 1000. Identical standard curves were generated for gCyclophilinA used here as an endogenous control. The cDNA for each timepoint was loaded in triplicate at 1 : 100 dilution, amplified using a preloaded, 9600 emulation protocol, and quantified based on the standard curves generated for each target gene. Target gene expression levels of were first normalized to corresponding endogenous control values, and the resulting value normalized to a calibrator (a mixture of each timepoint cDNA). Each plate included a no template control set (replacing sample with water) and a RT-control set (no reverse transcriptase added) to check against contamination.
Statistical analysis
Changes in 2DG uptake and of relative levels of clock gene mRNA were tested with ANOVA. When significant changes (P < 0.05) were found a Newman-Keuls post hoc test was performed on those data. Astrocyte growth studies were analyzed by one of two methods: for the growth area study, cocultured astrocytes were compared with solo cultures using a Student’s t-test. For the melatonin dose-response study, the number of DAPI-positive cells was plotted over time for each concentration. The resulting data points were fitted to a nonlinear polynomial using SigmaPlot software (Systat Software Inc., Richmond, CA, USA). These regressions yielded equations that contained a coefficient of slope, and the respective slopes at each dose was compared with that of the vehicle treated control by ANOVA. A Holm-Sidak post hoc test was performed on significantly different data sets. All statistical analyses were performed using SigmaStat software (Systat Software Inc.).
Results
The rhythm of melatonin release from the pinealocytes in pineal-astrocyte cocultures persisted for 2 days of constant darkness (Fig. 1B). As expected, the first peak in melatonin levels occurred at CT 17, near the middle of subjective night. Two factors impeded a clear resolution of the second peak in melatonin production: the melatonin levels did not return to baseline levels after the initial peak, and the rhythm damped out by the end of the second day. This may be due to the static nature of the culture as the media was not changed/removed during the course of the sampling. 2DG uptake from astrocytes also exhibited a circadian rhythm when cocultured with pinealocytes, but not in astrocytes cultured alone (Fig. 1C). On the first cycle, peak uptake coincides with increasing melatonin levels, but by the second day in DD, 2DG uptake and melatonin levels were near antiphase to each other (29 hr in DD, Fig. 1B and C). Interestingly, the basal metabolic rate was higher in astrocytes cocultured with pinealocytes than in astrocytes cultured alone. The expression of two clock genes was profiled from astrocytes cultured alone and in coculture with pinealocytes (Fig. 2). Unlike the clear metabolic rhythm, real-time PCR analysis of gper3 and gcry1 revealed differential effects of pineal-secreted melatonin on astrocyte gene expression. Gper3 expression in cocultured astrocytes entrained to pineal-secreted melatonin for one cycle, with mRNA levels peaking between 13 and 17 hr and subsequently dropping before the onset of the second day - (21 hr into DD, Fig. 2A). Surprisingly, gcry1 expression in cocultured astrocytes did not differ significantly from monocultures (Fig. 2B).
Fig. 2.
Clock gene expression in astrocytes cocultured with pinealocytes was not rhythmic. (A) gper3 expression in cocultured astrocytes (black circles) showed significant variation for one cycle over time when compared with solo cultures (white circles), but no circadian rhythm was detected. (B) gcry1 expression in cocultured astrocytes did not differ significantly from control cultures (n = 6 replicates for each data set). For determined rhythmic cycles, significant differences between peak-to-trough values are indicated by #P < 0.05 or *P < 0.001. Comparisons were made between the first observed peak and trough for each day for each treatment.
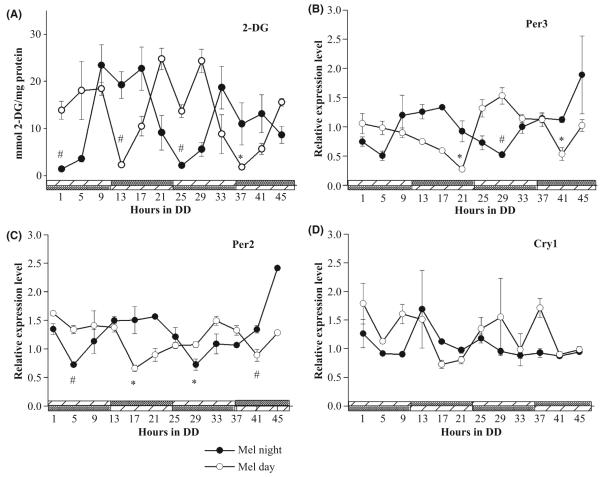
To further characterize the effect of melatonin on target cells cultured astrocytes were administered purified melatonin in a rhythmic fashion. Under constant darkness, astrocyte cultures subjected to cycles of melatonin administration exhibited statistically significant rhythms in 2DG uptake. These metabolic rhythms were 180° out of phase with each other when melatonin was administered during the subjective day (MD) rather than during the subjective night (MN). In the MN regime, peak uptake began after 9 hr in DD, which preceded the time at which melatonin was present (Fig. 3A). A similar anticipatory effect was observed in cells exposed to the MD regime (Fig. 3A). Real-time PCR analysis of gcry1, gper2, and gper3 expression revealed a complex pattern in which melatonin cycles affected the expression of some but not all of the geneexpression patterns. Gper3 mRNA expression entrained to melatonin cycles (Fig. 3B) and the rhythm correlated with those exhibited under coculture conditions. When melatonin was present during the subjective night (MN; 12-24 hr into DD), gper3 expression predominated during the subjective night, and when melatonin was present during subjective day only (MD), gper3 expression was highest when melatonin was present, 180° antiphase from the MN series (Fig. 3B). Similarly, the expression of gper2 mRNA in astrocytes incubated in 24 hr cycles of melatonin varied significantly over time. Cells maintained in MN and MD clearly expressed gper2 mRNA 180° in antiphase with peak-to-peak intervals of 24 hr (Fig. 3C). This strongly indicates the gper2 mRNA rhythm was entrained to the melatonin cycle. However, as the coculture experiments demonstrated, there was no observable variation in gcry1 mRNA levels that was significant for the astrocytes in MD cycles (Fig. 3D). There was a significant variation in the levels of gcry1 expression in MN. However, the amplitude of this variation was very low.
Fig. 3.
Exogenous melatonin imposes metabolic and clock gene rhythms in diencephalic astrocytes. (A) Under constant conditions, opposing cycles of rhythmic melatonin (nm) administration, either during the night or day, elicited rhythmic uptake of glucose in the cultures, such that 2DG uptake was higher during the time which melatonin was not present (n = 4). (B) The gPer3 mRNA rhythm under the MN cycle (n = 3 sample replicates) was 180° antiphase with respect to the rhythm generated under the MD cycle (n = 3 sample replicates). (C) The gPer2 mRNA rhythm under the MN cycle (n = 3 sample replicates) was 180° antiphase with respect to the rhythm generated under the MD cycle (n = 3 sample replicates). (D) No clear pattern of gCry1 expression was evident under either condition (n = 3 sample replicates each). For determined rhythmic cycles, significant differences between peak-to-trough values are indicated by #P < 0.05 or *P < 0.001. Comparisons were made between the first observed peak and trough for each day for each treatment.
It was noted throughout the course of the entrainment studies that the astrocytes cocultured with pinealocytes grew to confluence much more quickly than did monocultured astrocytes. When confluence of the cultures was compared over time (Fig. 4A), an increase in astrocyte density was observed as early as 2 days after pinealocyte introduction. When compared with control cultures, the increase in density remained statistically greater from days 3 to 6 at which point the monolayer of astrocytes in coculture reached 100% confluence, defined here as when cultures begin to lift off the plate for lack of space. We hypothesized that melatonin may be responsible for this increased growth rate. To test this hypothesis and determine whether the increased cell density was due to increased cell size or an increase in cell number, different doses of melatonin were administered to primary astrocytes daily at ZT12. Several cultures treated with different concentrations of melatonin were stained with DAPI and compared (Fig. 4B). The number of astrocytes increased in a dose-dependent fashion (Fig. 4C). While picomolar and 1 nm concentrations of melatonin had no effect on mitotic growth when compared with DMSO-treated controls (data not shown), the growth rate in cells treated with 10 nm, 1 μm, and 10 μm melatonin differed significantly from control cultures, as measured by regression analysis (Fig. 4C, 3 treatments shown for clarity). The rate of growth of the astrocytes, as measured by the slopes calculated from Fig. 4C, was also dependent on the concentration of melatonin administered throughout the 7 days of sampling (Fig. 4D).
Fig. 4.
Exogenous melatonin stimulates mitotic growth in astrocytes. (A) Cocultured astrocytes (n = 6 replicates) grew to confluence faster than monocultures, as measured by the average percent of cell coverage over six regions of interest per replicate; *P < 0.001, #P < 0.05 using t-test. (B) Representative images of DAPI stained astrocytes administered 10 μm melatonin (top panel), 10 nm melatonin (middle panel), or vehicle control (bottom panel). (C) Mitotic growth in astrocytes administered 10 μm melatonin (black circles), 10 nm melatonin (black squares), or DMSO control (white circles) as measured by number of DAPI stained cells on a given day; *P < 0.001 comparing slopes to vehicle control. (D) Growth rates, calculated as the coefficients of slope of the respective line equations from (C), compared among cells given increasing doses of melatonin; *P < 0.001 compared with control (no melatonin), #P < 0.05 compared with control, and aP < 0.05 compared with 1 nm melatonin. Other doses are mentioned in the Materials and methods section.
Discussion
In the present study, rhythmic exposure to melatonin synchronized rhythms in glucose uptake, corroborating and extending our previous observations that timed melatonin administration entrains and/or drives 2DG uptake rhythms [14]. Exogenous melatonin cycles also synchronized rhythms in clock gene expression and induced an increase in growth rate in cultured chick astrocytes (Figs 3 and 4). Consistent with the hypothesis that melatonin is an ‘internal Zeitgeber’ for downstream oscillators within the circadian system and in peripheral tissues [33], precise melatonin cycles synchronized rhythms of gper3 and gper2 mRNA levels (Fig. 4). This is the first demonstration that melatonin affects clock gene expression in birds, in spite of the fact that melatonin is known to profoundly affect circadian patterns of behavior and physiology in several vertebrate groups, such as birds, reptiles, and mammals, including humans [2, 33]. These data stand in sharp contrast to two studies that report no role for melatonin in the regulation of clock gene expression. First, Yasuo et al. [34] showed that while Japanese quail that received intra-peritoneal implants of crystalline melatonin in Silastic capsules had elevated or constantly high levels of blood melatonin, there was no effect on per2 or per3 expression in the medial SCN (mSCN) of the mediobasal hypothalamus. These authors suggested that the circadian clock within the quail mSCN was ‘tightly coupled,’ making it immune to external signals. Secondly, Abraham et al. [21] have shown in house sparrows that hypothalamic expression of per2 is not affected by pinealectomy after 1 day in constant darkness. However, pinealectomized house sparrows exhibit rhythmic locomotor behavior and brain glucose utilization after 1 day in DD [35], and neither of these processes become arrhythmic until 10 days in DD.
Although the present study appears to contradict the previous findings, the previous studies’ direct relevance to the present work is only marginal. While it is true that melatonin can acutely affect rhythmic processes in behavior [36] and physiology [37, 38], most of its effects on circadian and seasonal rhythms require multiple days or even weeks of rhythmic administration in vivo [2, 36] and in vitro [14].
The fact that gper3 expression rhythms are entrained by exogenous melatonin cycles in astrocytes is intriguing for several reasons. First, in spite of the fact that per3 does not appear to contribute to circadian rhythm generation in mice [39], recent studies have shown that polymorphisms in the human per3 gene are associated with several sleep disorders, including delayed sleep phase syndrome [40]. Secondly, the long transcript allele in this polymorphism is associated with preference for diurnal activity [41]. Along these lines, it is interesting that astrocytes are the sole source for brain glycogen biosynthesis, which has been shown to increase during sleep and anesthesia [42]. Since we have previously shown that exogenous melatonin administration increases glycogen biosynthesis in addition to synchronizing metabolic coupling in chick astrocytes [14], it is intriguing to speculate that regulation of this particular clock gene by melatonin is linked to birds’ sleep wake cycle.
Whether or not astrocyte metabolism and clock genes are affected by melatonin in vivo is still under investigation. Several areas of the brain, such as the visual SCN, exhibit 2DG uptake rhythms such that uptake is higher during the day during the night [34] when melatonin is present [10]. Pinealectomy abolishes 2DG uptake rhythms in house sparrows [35], and rhythmic administration of melatonin to arrhythmic sparrows restores daily patterns of both locomotor behavior and brain 2DG uptake [43]. Finally acute administration of melatonin to both house sparrows [44] and chicks [45] inhibits hypothalamic 2DG uptake. Since vertebrate astrocytes are known to mediate most glucose uptake and glycolysis in the central nervous system [46], it is likely that these effects are at least partially mediated by these cells.
The mechanisms by which melatonin entrains gper3 and gper2 mRNA rhythms (Fig. 4B and C) are less clear as we know little about the promoter regions of these genes in this species. In mammals, the mper1 and mper2 promoters contain bona fide 3′,5′-cyclic adenosine monophosphate (cAMP)-responsive elements (CREs) that link gene expression to dynamic changes in cellular cAMP [47]. Since the known melatonin receptors in chicks are associated with Gi-guanosine triphosphate-binding proteins, [13], it is possible that activation of the Mel1A and Mel1C receptors found on chick astrocytes [14] may directly affect gper3 expression by decreasing cellular cAMP levels. However, the mammalian mper3 promoter does not contain a known CRE, and the times at which both gper2 and gper3 mRNA peak in a melatonin cycle coincide with peak melatonin levels, making a direct regulation of these clock genes’ expression through these inhibitory receptors unlikely. Alternatively, there is some controversial evidence that melatonin may also act via binding to a retinoid-related orphan receptor (RZR/ROR) intracellularly [48]. This is interesting, since ROR has recently been implicated as a regulatory element controlling mammalian Bmal1 [49], which controls the expression of the per genes in all animal species studied [17].
Another question that arises from these studies is whether mammalian astrocytes are similarly sensitive to melatonin. Exogenous melatonin inhibits 2DG uptake within the rat SCN in vivo in a dose- and phase-dependent fashion [38], and the rat SCN is replete with astrocytes [50, 51]. Furthermore, our previous demonstration of melatonin receptors in mammalian astrocytes, [52] coupled with other studies demonstrating that the response of mammalian astrocytes to oxidative stress is ameliorated by melatonin [53, 54], indicate a possible function for melatonin in the regulation of various astrocyte functions, which could thereby affect metabolic activity in the SCN. However, Poirel et al. [55] have shown that a single injection of melatonin to rats in the late subjective day, which is known to phase shift circadian activity patterns [34, 56], has no effect on SCN clock gene expression. These authors suggested that melatonin might only affect posttranscriptional events within the clock.
Remarkably, both pineal secreted and exogenously administered melatonin also affected the development of astrocytes by increasing mitotic growth in a dose-dependent manner (Fig. 4). This observation contrasts with previous studies that report an inhibitory effect of melatonin on mitotic division of various tumor cells [57, 58, and reviewed in 59]. Perhaps, the effect of melatonin is different between neoplastic cells and diencephalic astrocytes. However, a role for melatonin as a potential developmental factor has been implicated in several systems, most notably in a mouse neural stem cell line, which expresses a functional MT1 receptor and GFAP [60]. Exposure to physiological doses of melatonin in these cells induced glial-derived neurotrophic factor mRNA expression, implicating a direct link between melatonin and astrocytes in a developmental context. Another explanation for these contradictory data may lie in the concentrations of melatonin used. Sainz et al. [61] showed that melatonin reduces cell proliferation in Chinese hamster ovarian cells in a dose-dependent fashion, such that only millimolar concentrations were sufficient to inhibit mitotic growth. Indeed, in our hands low physiogical concentrations of melatonin (10 fm and 10 pm) increased resting calcium levels in chick diencephalic astrocytes, whereas pharmacological doses of melatonin decreased resting calcium levels [30 and Peters, unpublished data]. More recently, nanomolar concentrations of melatonin were shown to increase the rate of in vitro development of porcine embryos [62]. Perhaps, the stimulatory effects we see here are linked to a global role for the pineal hormone in the maintenance of astrocyte function and the temporal organization of the avian brain during development.
References
- 1.Cassone VM, Menaker M. Is the avian circadian system a neuroendocrine loop? J Exp Zool. 1984;232:539–549. doi: 10.1002/jez.1402320321. [DOI] [PubMed] [Google Scholar]
- 2.Cassone VM. Effects of melatonin on vertebrate circadian systems. Trends Neurosci. 1990;13:457–464. doi: 10.1016/0166-2236(90)90099-v. [DOI] [PubMed] [Google Scholar]
- 3.Ebihara S, Uchiyama K, Oshima I. Circadian organization of the pigeon, Columba livia: role of the eyes and pineal gland. J Comp Physiol A. 1984;154:59–69. [Google Scholar]
- 4.Underwood H, Binkley S, Siopes T, et al. Melatonin rhythms in the eyes, pineal bodies, and blood of Japanese quail (Coturnix coturnix japonica) Gen Comp Endocrinol. 1984;56:70–81. doi: 10.1016/0016-6480(84)90063-7. [DOI] [PubMed] [Google Scholar]
- 5.Ebihara S, Kawamura H. The role of pineal melatonin and the suprachiasmatic nucleus in the control of circadian locomotor rhythms in the Java sparrow, Padda orizivora. J Comp Physiol A. 1981;131:207–214. [Google Scholar]
- 6.Takahashi JS, Menaker M. Role of the suprachiasmatic nuclei in the circadian system of the house sparrow, Passer domesticus. J Neurosci. 1982;2:815–828. doi: 10.1523/JNEUROSCI.02-06-00815.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassone VM, Forsyth AM, Woodlee GL. Hypothalamic regulation of circadian noradrenergic input to the chick pineal gland. J Comp Physiol A. 1990;167:187–192. doi: 10.1007/BF00188110. [DOI] [PubMed] [Google Scholar]
- 8.Gaston S, Menaker M. Pineal function: the biological clock in the sparrow? Science. 1968;160:1125–1127. doi: 10.1126/science.160.3832.1125. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman NH, Menaker M. The pineal gland: a pacemaker within the circadian system of the house sparrow. Proc Natl Acad Sci USA. 1979;76:999–1003. doi: 10.1073/pnas.76.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein DC, Coon SL, Roseboom PH, et al. The melatonin rhythm-generating enzyme: molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res. 1997;52:307–358. [PubMed] [Google Scholar]
- 11.Hamm HE, Menaker M. Retinal rhythms in chicks: circadian variation in melatonin and serotonin N-acetyltransferase activity. Proc Natl Acad Sci USA. 1980;77:4998–5002. doi: 10.1073/pnas.77.8.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adachi A, Hasegawa M, Ebihara S. Measurement of circadian rhythms of ocular melatonin in the pigeon by in vivo microdialysis. Neuroreport. 1995;7:286–288. [PubMed] [Google Scholar]
- 13.Reppert SM, Weaver DR, Cassone VM, et al. Melatonin receptors are for the birds: molecular analysis of two receptor subtypes differentially expressed in chick brain. Neuron. 1995;15:1003–1015. doi: 10.1016/0896-6273(95)90090-x. [DOI] [PubMed] [Google Scholar]
- 14.Adachi A, Natesan AK, Whitfield-Rucker MG, et al. Functional melatonin receptors and metabolic coupling in cultured chick astrocytes. Glia. 2002;39:268–278. doi: 10.1002/glia.10109. [DOI] [PubMed] [Google Scholar]
- 15.Natesan AK, Cassone VM. Melatonin receptor mRNA localization and rhythmicity in the retina of the domestic chick, Gallus domesticus. Vis Neurosci. 2002;19:265–274. doi: 10.1017/s0952523802192042. [DOI] [PubMed] [Google Scholar]
- 16.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 17.Bell-Pedersen D, Cassone VM, Earnest DJ, et al. Circadian rhythms from multiple oscillators: lessons from diverse organism. Nat Rev Gen. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shearman LP, Sriram S, Weaver DR, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimura T, Suzuki Y, Makino E, et al. Molecular analysis of avian circadian clock genes. Brain Res Mol Brain Res. 2000;78:207–215. doi: 10.1016/s0169-328x(00)00091-7. [DOI] [PubMed] [Google Scholar]
- 20.Bailey MJ, Chong NW, Xiong J, et al. Chickens’ Cry2: molecular analysis of an avian cryptochrome in retinal and pineal photoreceptors. FEBS Lett. 2002;513:169–174. doi: 10.1016/s0014-5793(02)02276-7. [DOI] [PubMed] [Google Scholar]
- 21.Abraham U, Albrecht U, Brandstatter R. Hypothalamic circadian organization in birds. II. Clock gene expression. Chronobiol Int. 2003;20:657–669. doi: 10.1081/cbi-120022414. [DOI] [PubMed] [Google Scholar]
- 22.Bailey MJ, Beremand PD, Hammer R, et al. Transcriptional profiling of the chick pineal gland, a photoreceptive circadian oscillator and pacemaker. Mol Endocrinol. 2003;17:2084–2095. doi: 10.1210/me.2003-0121. [DOI] [PubMed] [Google Scholar]
- 23.Bailey MJ, Cassone VM. Opsin photoisomerases in the chick retina and pineal gland: characterization, localization, and circadian regulation. Invest Ophthalmol Vis Sci. 2004;45:769–775. doi: 10.1167/iovs.03-1125. [DOI] [PubMed] [Google Scholar]
- 24.Bernard M, Iuvone PM, Cassone VM, et al. Avian melatonin synthesis: photic and circadian regulation of serotonin N-acetyltransferase mRNA in the chicken pineal gland and retina. J Neurochem. 1997;68:213–224. doi: 10.1046/j.1471-4159.1997.68010213.x. [DOI] [PubMed] [Google Scholar]
- 25.Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zool Sci. 2004;21:359–368. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- 26.Tanoue S, Krishnan P, Krishnan B, et al. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Yoo SH, Yamazaki S, Lowrey PL, et al. Period2::luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen G, Rappe J, Earnest DJ, et al. Oscillating on borrowed time: diffusible signals from immortalized suprachiasmatic nucleus cells regulate circadian rhythmicity in cultured fibroblasts. J Neurosci. 2001;21:7937–7943. doi: 10.1523/JNEUROSCI.21-20-07937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen GC, Farnell Y, Bell-Pedersen D, et al. Effects of altered Clock gene expression on the pacemaker properties of SCN2.2 cells and oscillatory properties of NIH/3T3 cells. Neuroscience. 2004;127:989–999. doi: 10.1016/j.neuroscience.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Peters JL, Cassone VM, Zoran MJ. Melatonin modulates intercellular communication among cultured chick astrocytes. Brain Res. 2005;1031:10–19. doi: 10.1016/j.brainres.2004.09.064. [DOI] [PubMed] [Google Scholar]
- 31.Zatz M, Mullen DA, Moskal JR. Photoendocrine transduction in cultured chick pineal cells: effects of light, dark, and potassium on the melatonin rhythm. Brain Res. 1988;438:199–215. doi: 10.1016/0006-8993(88)91339-x. [DOI] [PubMed] [Google Scholar]
- 32.Fraser S, Cowen P, Franklin M, et al. Direct radioimmunoassay for melatonin in plasma. Clin Chem. 1983;29:396–397. [PubMed] [Google Scholar]
- 33.Cassone VM. Melatonin’s role in vertebrate circadian rhythms. Chronobiol Int. 1998;15:457–473. doi: 10.3109/07420529808998702. [DOI] [PubMed] [Google Scholar]
- 34.Yasuo S, Yoshimura T, Bartell PA, et al. Effect of melatonin administration on qPer2, qPer3, and qClock gene expression In the suprachiasmatic nucleus of Japanese quail. Eur J Neurosci. 2002;16:1541–1546. doi: 10.1046/j.1460-9568.2002.02222.x. [DOI] [PubMed] [Google Scholar]
- 35.Lu J, Cassone VM. Pineal regulation of circadian rhythms of 2-deoxy[14C]glucose uptake and 2[125I]iodomelatonin binding in the visual system of the house sparrow, Passer domesticus. J Comp Physiol A. 1993;173:765–774. [Google Scholar]
- 36.Cassone VM, Chesworth MJ, Armstrong SM. Dose-dependent entrainment of rat circadian rhythms by daily injection of melatonin. J Biol Rhythms. 1986;1:219–229. doi: 10.1177/074873048600100304. [DOI] [PubMed] [Google Scholar]
- 37.Cassone VM, Roberts MH, Moore RY. Effects of melatonin on 2-deoxy-[1-14C]glucose uptake within rat suprachiasmatic nucleus. Am J Physiol. 1988;255:R332–R337. doi: 10.1152/ajpregu.1988.255.2.R332. [DOI] [PubMed] [Google Scholar]
- 38.McArthur AJ, Gillette MU, Prosser RA. Melatonin directly resets the rat suprachiasmatic circadian clock in vitro. Brain Res. 1991;565:158–161. doi: 10.1016/0006-8993(91)91748-p. [DOI] [PubMed] [Google Scholar]
- 39.Bae K, Jin X, Maywood ES, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 40.Ebisawa T, Uchiyama M, Kajimura N, et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–346. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereira DS, Tufik S, Louzada FM, et al. Association of the length polymorphism in the human Per3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep. 2005;28:29–32. [PubMed] [Google Scholar]
- 42.Brown AM. Brain glycogen re-awakened. J Neurochem. 2004;89:537–552. doi: 10.1111/j.1471-4159.2004.02421.x. [DOI] [PubMed] [Google Scholar]
- 43.Lu J, Cassone VM. Daily melatonin administration synchronizes circadian patterns of brain metabolism and behavior in pinealectomized house sparrows, Passer domesticus. J Comp Physiol A. 1993;173:775–782. [Google Scholar]
- 44.Cassone VM, Brooks DS. The sites of melatonin action in the house sparrow brain. J Exp Zool. 1991;260:302–309. [Google Scholar]
- 45.Cantwell EL, Cassone VM. Daily and circadian fluctuation in 2-deoxy[14C]-glucose uptake in circadian and visual system structures of the chick brain: effects of exogenous melatonin. Brain Res Bull. 2002;57:603–611. doi: 10.1016/s0361-9230(01)00753-5. [DOI] [PubMed] [Google Scholar]
- 46.Magistretti PJ. Cellular bases of functional brain imaging: insights from neuron-glia metabolic coupling. Brain Res. 2000;886:108–112. doi: 10.1016/s0006-8993(00)02945-0. [DOI] [PubMed] [Google Scholar]
- 47.Travnickova-Bendova Z, Cermakian N, Reppert SM, et al. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci USA. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlberg C. Gene regulation by melatonin. Ann NY Acad Sci. 2000;917:387–396. doi: 10.1111/j.1749-6632.2000.tb05403.x. [DOI] [PubMed] [Google Scholar]
- 49.Sato TK, Panda S, Miraglia LJ, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Guldner FH. Numbers of neurons and astroglial cells in the suprachiasmatic nucleus of male and female rats. Exp Brain Res. 1983;50:373–376. doi: 10.1007/BF00239203. [DOI] [PubMed] [Google Scholar]
- 51.Van den Pol AN. The hypothalamic suprachiasmatic nucleus of rat: intrinsic anatomy. J Comp Neurol. 1980;191:661–702. doi: 10.1002/cne.901910410. [DOI] [PubMed] [Google Scholar]
- 52.Peters JL, Earnest BJ, Tjalkens RB, et al. Modulation of intercellular calcium signaling by melatonin in avian and mammalian astrocytes is brain region-specific. J Comp Neurol. 2005;493:370–380. doi: 10.1002/cne.20779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juknat AA, Mendez Mdel V, Quaglino A, et al. Melatonin prevents hydrogen peroxide-induced Bax expression in cultured rat astrocytes. J Pineal Res. 2005;38:84–92. doi: 10.1111/j.1600-079X.2004.00166.x. [DOI] [PubMed] [Google Scholar]
- 54.Martin V, Sainz RM, Antolin I, et al. Several antioxidant pathways are involved in astrocyte protection by melatonin. J Pineal Res. 2002;33:204–212. doi: 10.1034/j.1600-079x.2002.02113.x. 3. [DOI] [PubMed] [Google Scholar]
- 55.Poirel VJ, Boggio V, Dardente H, et al. Contrary to other non-photic cues, acute melatonin injection does not induce immediate changes of clock gene mRNA expression in the rat suprachiasmatic nuclei. Neurosci. 2003;120:745–755. doi: 10.1016/s0306-4522(03)00344-0. [DOI] [PubMed] [Google Scholar]
- 56.Warren WS, Hodges DB, Cassone VM. Pinealectomized rats entrain and phase-shift to melatonin injections in a dose-dependent manner. J Biol Rhythms. 1993;8:233–245. doi: 10.1177/074873049300800306. [DOI] [PubMed] [Google Scholar]
- 57.Blask DE, Dauchy RT, Sauer LA, et al. Melatonin uptake and growth prevention in rat hepatoma 7288 CTC in response to dietary melatonin: melatonin receptor-mediated inhibition of tumor linoleic acid metabolism to the growth signaling molecule 13-hydroxydecadienoic acid and the potential role of phytomelatonin. Carcinogenesis. 2004;25:951–960. doi: 10.1093/carcin/bgh090. [DOI] [PubMed] [Google Scholar]
- 58.Cini G, Neri B, Pacini A, et al. Antiproliferative activity of melatonin by transcriptional inhibition of cyclin D1 expression: a molecular basis for melatonin-induced oncostatic effects. J Pineal Res. 2005;39:12–20. doi: 10.1111/j.1600-079X.2004.00206.x. [DOI] [PubMed] [Google Scholar]
- 59.Jung B, Ahmad N. Melatonin in cancer management. Cancer Res. 2006;66:9789–9793. doi: 10.1158/0008-5472.CAN-06-1776. [DOI] [PubMed] [Google Scholar]
- 60.Niles LP, Armstrong KJ, Rincón Castro LM, et al. Neural stem cells express melatonin receptors and neurotrophic factors: colocalization of the MT1 receptor with neuronal and glial markers. BMC Neurosci. 2004;5:41–49. doi: 10.1186/1471-2202-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sainz RM, Mayo JC, Tan DX, et al. Antioxidant activity of melatonin in Chinese hamster ovarian cells: changes in cellular proliferation and differentiation. Biochem Biophys Res Comm. 2003;302:625–634. doi: 10.1016/s0006-291x(03)00230-4. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez-Osorio N, Kim IJ, Wang H, et al. Melatonin increases cleavage rate of porcine preimplantation embryos in vitro. J Pineal Res. 2007;43:283–288. doi: 10.1111/j.1600-079X.2007.00475.x. [DOI] [PubMed] [Google Scholar]