Abstract
Background
Phylogenetic analyses of angiosperm relationships have used only a small percentage of available sequence data, but phylogenetic data matrices often can be augmented with existing data, especially if one allows missing characters. We explore the effects on phylogenetic analyses of adding 378 matK sequences and 240 26S rDNA sequences to the complete 3-gene, 567-taxon angiosperm phylogenetic matrix of Soltis et al.
Results
We performed maximum likelihood bootstrap analyses of the complete, 3-gene 567-taxon data matrix and the incomplete, 5-gene 567-taxon data matrix. Although the 5-gene matrix has more missing data (27.5%) than the 3-gene data matrix (2.9%), the 5-gene analysis resulted in higher levels of bootstrap support. Within the 567-taxon tree, the increase in support is most evident for relationships among the 170 taxa for which both matK and 26S rDNA sequences were added, and there is little gain in support for relationships among the 119 taxa having neither matK nor 26S rDNA sequences. The 5-gene analysis also places the enigmatic Hydrostachys in Lamiales (BS = 97%) rather than in Cornales (BS = 100% in 3-gene analysis). The placement of Hydrostachys in Lamiales is unprecedented in molecular analyses, but it is consistent with embryological and morphological data.
Conclusion
Adding available, and often incomplete, sets of sequences to existing data sets can be a fast and inexpensive way to increase support for phylogenetic relationships and produce novel and credible new phylogenetic hypotheses.
Background
Molecular data have had an enormous impact on angiosperm phylogenetic hypotheses (e.g. [1-5]), and the abundance of new sequence data provides the potential for further resolving angiosperm relationships. Still, molecular phylogenetic studies across all angiosperms have utilized only a small fraction of the available sequence data. While GenBank currently contains over 1.7 million core nucleotide sequences from angiosperms, with over 160,000 of these being from often phylogenetically useful plastid loci [6], few phylogenetic analyses of angiosperms have included more than a thousand sequences. We examine whether augmenting existing plant data matrices with incomplete data assembled from publicly available sources can enhance the understanding of the backbone phylogenetic relationships across angiosperms.
The sampling strategies of phylogenetic studies across angiosperms demonstrate a tradeoff between taxonomic sampling and the number of gene sequences per taxon. On one extreme, phylogenetic analyses using single genes such as rbcL [7,8], 18S rDNA [9], and matK [10] have sampled hundreds, or even thousands [8], of taxa. In some cases, analyses of single genes have provided strong support for many angiosperm relationships (e.g. [10]), but other parts of the single-gene trees have been unresolved, in disagreement with other single gene trees (albeit often without strong support), or simply anomalous (e.g. [7]). On the other extreme, the advent of chloroplast genome sequencing has led to phylogenetic analyses of angiosperms using sequences from up to 81 genes, although often with limited taxon sampling [11-17]. The limited taxon sampling can have undesirable effects on phylogenetic inferences from complete chloroplast genome sequences, as well as other large molecular data sets (see [13,14,18,19]). Other studies have used sampling strategies that attempt to compromise between taxon and gene sampling. For example, Qiu et al. [20,21] used 5 genes and 105 taxa, and later studies have used 9 genes and 100 taxa [22] or 8 genes and 162 taxa [23]. These studies generally have focused on the basal angiosperm splits, and more comprehensive taxon sampling is necessary to address backbone relationships throughout the angiosperms. The most comprehensive taxon sampling across angiosperms using multiple genes includes a study using atpB and rbcL [24] and a 567-taxon study with 18S rDNA, atpB, and rbcL [1-3].
While more data are needed to further resolve backbone relationships in angiosperms, it is not clear what the most efficient sampling strategies for adding new data would be. Previous analyses of molecular data across all angiosperms mostly used complete or nearly complete data matrices (but see [25,26]), in which all taxa have sequences from all, or nearly all, genes. Often it is possible to increase the number of taxa or genes in a phylogenetic matrix greatly using existing data by allowing small amounts of missing data (e.g. [27]). Furthermore, recent simulation studies have demonstrated that in some cases the total amount of data in a phylogenetic matrix may determine the performance of a phylogenetic analysis more than the percentage of missing data [28]. This suggests that adding sequences from genes with incomplete taxonomic coverage to existing complete data matrices can, at least in some cases, improve the phylogenetic inference. Generating large, complete data matrices can be both logistically daunting (e.g., assembling DNA samples from all taxa) and prohibitively expensive. In contrast, there is a wealth of publicly available sequence data that can be readily assembled into phylogenetic data matrices, providing a relatively fast and inexpensive way to augment existing data sets and possibly improve our phylogenetic inferences.
We explore the effect of adding existing plastid matK and nuclear 26S rDNA sequences to the 3-gene (18S rDNA, atpB, and rbcL) 567-taxon matrix of P. Soltis et al. ([1], see also [2,3]). Both matK and the nuclear 26S rDNA have been informative in large-scale angiosperm phylogenetic studies (e. g., matK: [10,22,23]; 26S rDNA: [29-32]), and consequently, sequence data from both genes are available for many angiosperm taxa. We assembled the available 26S rDNA and matK sequences from taxa represented in the 3-gene matrix and compared results from maximum likelihood phylogenetic analyses of the 3-gene 567-taxon complete matrix with the 5-gene 567-taxon incomplete matrix. Specifically, we examine if and how augmenting complete data sets with incomplete data affects our view of angiosperm phylogeny.
Results
Data set
We performed analyses on a 3-gene and a 5-gene data matrix that have the same set of 567-taxa (Fig. 1). In the 3-gene (18S rDNA, atpB, and rbcL), matrix, all taxa have sequences from all genes. The matrix is 4592 characters long and it contains 2.9% missing data, representing indels or small sections of missing gene sequences. The 5-gene matrix comprises the available matK and 26S rDNA sequences concatenated to the 3-gene matrix. The accession numbers for the matK and 26S rDNA sequences, as well as the 5-gene character matrix, are included as additional files (see Additional files 1, 2 and 3). In the 5-gene matrix, 170 (29.9%) taxa have sequences from all 5 genes, and 119 (20.9%) taxa have data from only the original 3-genes (18S rDNA, atpB, and rbcL; Fig. 1). In total 47.0% of the matK and 26S rDNA alignments are missing data, largely due to missing whole gene sequences, and overall, 27.5% of the cells in the 5-gene 567-taxon matrix are missing data.
Figure 1.
Diagram representing the distribution of data in the total 5-gene data matrix. All taxa in the matrix contain sequences from the first 3 genes (18S rDNA, atpB, and rbcL), 378 taxa have matK sequences, and 240 taxa have 26S rDNA sequences. Only 170 taxa have sequences from both matK and 26S rDNA, and 119 taxa have no matK or 26S rDNA sequences.
Phylogenetic Analyses
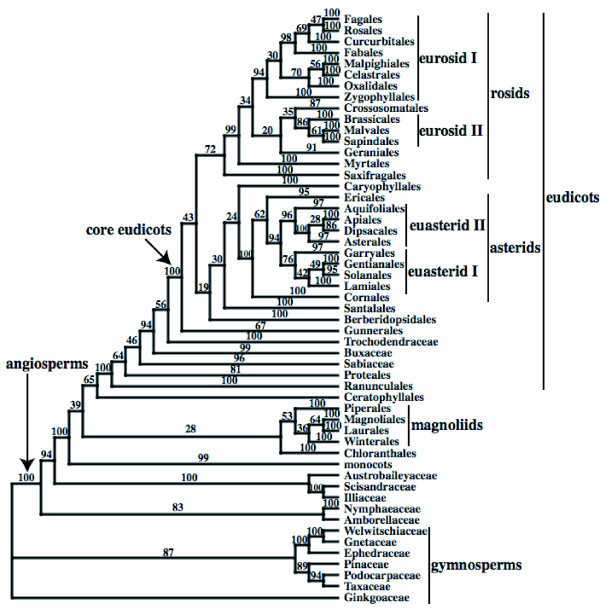
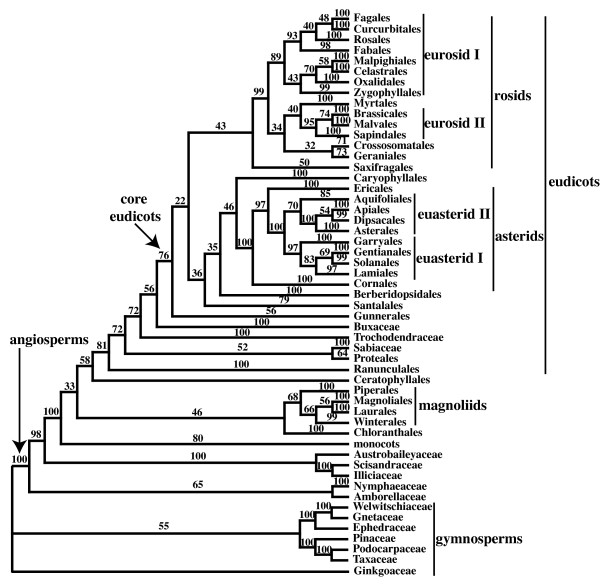
The bootstrap trees from the 5-gene and 3-gene analyses are included as additional data (see Additional files 4 and 5). The 5-gene ML bootstrap analysis produced higher levels of support than the 3-gene ML bootstrap analysis. First, the analysis of the 5-gene matrix produced more clades with high bootstrap support score (Table 1, Figs. 2 and 3; Additional files 6 and 7). Also, the average quartet similarity among bootstrap trees was higher in the 5-gene analysis than the 3-gene analysis (0.963 vs. 0.934; Table 1). Still, bootstrap support for some clades, decreased. For example, bootstrap support for monocot, eudicot, and core eudicot clades decreased from 99% or 100% in the 3-gene analysis to between 76% and 81% the 5-gene analyses (Figs. 2 and 3; Additional files 6 and 7). There are few major differences in the topologies of the 3-gene and 5-gene ML bootstrap 50% majority rule consensus trees, and these topologies are largely consistent with results from previous 3-gene parsimony [1,2] and Bayesian analyses [3]. Therefore, we include the full bootstrap consensus trees as additional files (see Additional files 6 and 7).
Table 1.
Summary of support for 3-gene and 5-gene 567-taxon maximum likelihood bootstrap analyses.
| Clades with Bootstrap Support | ||||||
| 100% | ≥95% | ≥90% | ≥70% | ≥50% | Ave. Quartet Similarity | |
| 5-gene ML | 223 | 290 | 318 | 409 | 487 | 0.963 |
| 3-gene ML | 190 | 273 | 298 | 378 | 461 | 0.934 |
The first 5 columns show the number of clades (out of a possible 564) that have the specified level of bootstrap support. The last column shows the average percent of quartets (4-taxon unrooted trees) that are identical among bootstrap trees.
Figure 2.
Summary of the majority rule consensus from the 3-gene (18S rDNA, atpB, and rbcL) ML analysis. Names of the orders and informal names follow APG II [4] and Soltis et al. [2,3], with Hydrostachys in Cornales. Numbers above the branches are bootstrap percentages. This tree was rooted using all gymnosperm taxa as outgroups.
Figure 3.
Summary of the majority rule consensus from the 5-gene (18S rDNA, atpB, rbcL, matK, and 26S rDNA) ML analysis. Names of the orders and informal names follow APG II [4] and Soltis et al. [2,3], with Hydrostachys in Lamiales. Numbers above the branches are bootstrap percentages. This tree was rooted using all gymnosperm taxa as outgroups.
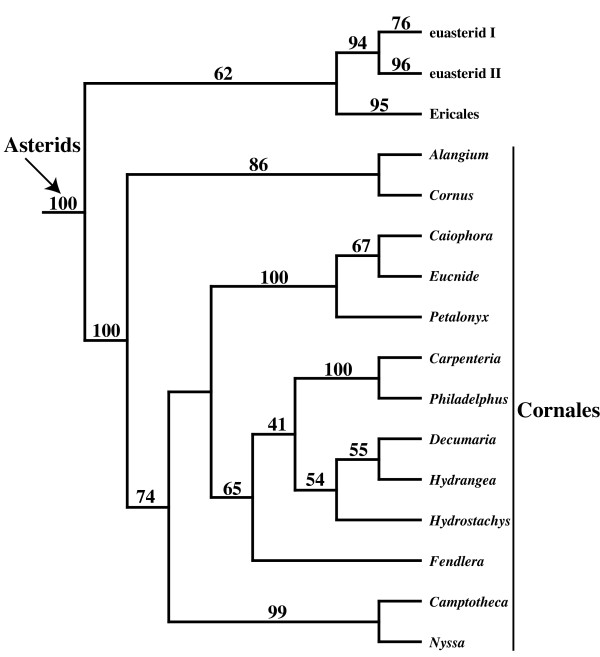
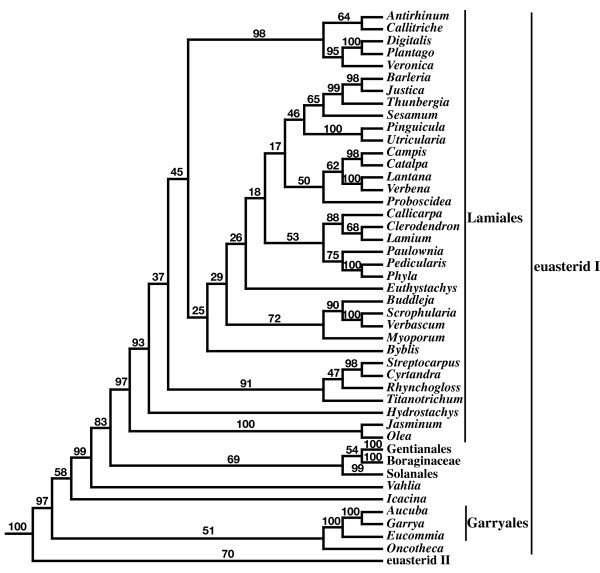
Perhaps the biggest single change in topology between the 5-gene and 3-gene ML analyses was the position of Hydrostachys (Figs. 4 and 5), which has a matK but no 26S rDNA sequence in the 5-gene matrix. In the 3-gene ML analysis, there was 100% bootstrap support for placing Hydrostachys within Cornales (Fig. 4), which is sister to all remaining asterids (the Ericales + euasterid I and II clade) (Figs. 2 and 4). In contrast, the 5-gene ML analysis placed Hydrostachys in Lamiales (Fig. 5), within the euasterid I clade (Figs. 3 and 5). Although there was 97% bootstrap support for the Lamilaes clade containing Hydrostachys in the 5-gene analysis, the placement of Hydrostachys within Lamiales was largely unresolved (Fig. 5). Within Lamilaes, there was 93% support for a clade of all taxa (including Hydrostachys) except Olea and Jasminum (Fig. 5).
Figure 4.
Detail of the position of Hydrostachys within Cornales in the majority rule consensus from the 3-gene ML analysis.
Figure 5.
Detail of the position of Hydrostachys within Lamiales in the majority rule consensus from the 5-gene ML analysis.
The average taxon quartet similarity, the average quartet similarity between all pairs of bootstrap trees for only the quartets that contain the taxon, was higher for every taxon in the 5-gene analysis than the 3-gene analysis (not shown). In the 5-gene bootstrap analyses, the taxa with the lowest average taxon quartet similarity values, or whose position were least supported (or vary most) among bootstrap trees, are largely from early diverging eudicot clades (e.g., Santalales, Dilleniaceae, and Berberidopsidales), and the core eudicots (Gunnerales) (Table 2).
Table 2.
Taxa with the least support in the 5-gene ML analysis.
| Taxa | Ave. Taxon Quartet Similarity | |
| 1 | Dendrophthora | 0.779 |
| 2 | Cercidiphyllum | 0.824 |
| 3 | Tetracera | 0.827 |
| 4 | Dillenia | 0.828 |
| 5 | Myrothamnus | 0.863 |
| 6 | Berberidopsis | 0.869 |
| 7 | Aextoxicon | 0.875 |
| 8 | Schoepfia | 0.889 |
| 9 | Eubrachion | 0.889 |
| 10 | Santalum | 0.889 |
The average taxon quartet similarity is the average percentage of all quartets (4-taxon unrooted trees) that include the taxon that are identical among bootstrap trees.
Effects of adding loci with incomplete taxonomic coverage
We further examined the effects of adding incomplete data sets by comparing the support within the 3-gene and 5-gene, 567-taxon analyses for relationships among the 170 taxa having all five gene sequences (the original three genes plus both 26S rDNA and matK sequences) and the 119 taxa having neither 26S rDNA nor matK sequences (Fig. 1). To do this, we examined reduced consensus bootstrap trees (e.g. [33]) that included just the 170 taxa with data from all 5 genes and also that included only the 119 taxa with data from only 3 genes (missing both matK and 26S rDNA; Figure 1). The reduced consensus trees were made by pruning all the taxa except the specified sets of 170 or 119 from the 567-taxon bootstrap trees and then making a majority rule consensus from the pruned bootstrap trees. Adding both matK and 26S rDNA resulted in greater increases in support among relationships between the 170 taxa with 5 genes than among the 119 taxa with 3 genes (Table 3). There were increases in the number of highly supported clades in the reduced consensus tree containing the 170 taxa with 5 genes, and the average quartet similarity among the 170-taxon bootstrap trees increases by 0.044 with the addition of matK and 26S rDNA (Table 3). In contrast, the number of clades with 100%, ≥95%, and ≥90% bootstrap support in the 119-taxon reduced consensus is lower in the 5-gene than in the 3-gene analysis, and the average quartet similarity among the 119-taxon bootstrap trees increases by only 0.010 with the addition of matK and 26S rDNA (Table 3).
Table 3.
Summary of the support for relationships among the 170 taxa with sequences from 5 genes and the 119 taxa with sequences from 3 genes.
| Clades With Bootstrap Support | ||||||
| 100% | ≥95% | ≥90% | ≥70% | ≥50% | Ave. Quartet Similarity | |
| 5-Gene 170-taxa | 89 | 103 | 109 | 128 | 146 | 0.946 |
| 3-Gene 170-taxa | 70 | 93 | 99 | 119 | 137 | 0.902 |
| Change | 19*** | 10 | 10 | 9 | 9 | 0.044*** |
| 5-Gene 119-taxa | 39 | 51 | 56 | 82 | 94 | 0.952 |
| 3-Gene 119-taxa | 49 | 54 | 61 | 77 | 87 | 0.942 |
| Change | -10*** | -3* | -5** | 5 | 7 | 0.010*** |
The support values were obtained from reduced consensus trees, which were made by pruning all but either the selected 170 or 119 taxa from the bootstrap trees and making a consensus from the pruned trees. The first 5 columns show the number of clades in the reduced consensus trees that have the specified level of bootstrap support. The last column shows the average percent of quartets (4-taxon unrooted trees) that are identical between the pruned bootstrap trees. We tested if the observed changes in the bootstrap support or average quartet similarity are greater in the 170 taxa with 5-genes than they would be from a random sample of 170 taxa. Similarly, we tested if the changes in support are less in the 119 taxa with 3 genes than they would be from a random sample of 119 taxa. Significant results are noted with "*" symbols (* p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.005).
We next tested the hypothesis that the observed changes in bootstrap and average quartet similarity scores for relationships among the 170 taxa with 5 genes were greater than we would expect from a random sample of 170 taxa. We did this by comparing the observed changes in bootstrap and quartet similarity scores from the 170 taxa with 5 genes to changes in support for relationships among 200 sets of 170 randomly selected taxa. We found significantly greater than expected increases in the number of clades with 100% bootstrap support and in the average quartet similarity (p ≤ 0.005; Table 3). Similarly, we tested to see if the changes in support for relationships among the 119 taxa with 3 genes were less than we would expect from a random sample of 119 taxa. We examined changes in support from 200 sets of 119 randomly selected taxa and found significantly greater than expected decreases in the number of clades with 100%, ≥95%, and ≥90% bootstrap support (p ≤ 0.005, 0.05, and 0.01 respectively; Table 3) and in the average quartet similarity (p ≤ 0.005; Table 3).
Finally, we compared changes in the average taxon quartet similarity among bootstrap trees from the 3-gene to the 5-gene analysis for all taxa, just the 170 taxa with data from 5 genes, and just the 119 taxa with data from 3 genes. The mean change in the average taxon quartet similarity for the 170 5-gene taxa was greater than the mean across all taxa, and the mean change for the 119 3-gene taxa was less than that across all taxa (Table 4). The mean change in the average taxon similarity for the 170 5-gene taxa is greater than we would expect from a random sample of 170 taxa (p ≤ 0.001; Table 4), and the mean change for the 119 3-gene taxa is less than we would expect from a random sample of 119 taxa (p ≤ 0.001; Table 4).
Table 4.
Summary of average taxon quartet similarity scores across bootstrap trees.
| Mean Ave. Taxon Quartet Similarity | |||
| 3-Gene ML | 5-Gene ML | Ave. Change | |
| All taxa | 0.937 | 0.958 | 0.021 |
| 170-taxa | 0.928 | 0.954 | 0.026*** |
| 119-taxa | 0.941 | 0.957 | 0.016*** |
The average taxon quartet similarity is the average proportion of quartets (4-taxon unrooted trees) that include a particular taxon that have the same topology among bootstrap trees. The "170 taxa" are the taxa with sequences from all 5 genes, while the "119 taxa" are those with sequences from only the original three genes (18S rDNA, atpB, rbcL). "Ave. Change" shows the difference in the mean of the average taxon quartet similarity between the 3-gene and 5-gene ML bootstrap analyses. We tested if the average change among the 170 taxa is significantly greater than would be expected by randomly selecting without replacement 1000 sets of 170 taxa. Similarly, we tested if the average change among the 119 taxa with 3 genes is significantly less than would be expected by randomly selecting without replacement 1000 sets of 119 taxa. Both tests were highly significant (*** = p ≤ 0.001).
Discussion
Augmenting existing data sets with other available sequence data can enhance our understanding of angiosperm phylogeny, even if the new sequence data are incomplete. Although adding available matK and 26S rDNA sequences to the complete 3-gene angiosperm data set increases the overall percentage of missing data in the character matrix from 2.9% to 27.5%, analyses with the new sequences provide higher overall levels of support than analyses of the original 3 genes alone (Figs. 2 and 3, Table 1). The gains in support are especially evident in the relationships among the 170 taxa with both matK and 26S rDNA sequences (Tables 3 and 4). In contrast, there is little increase in support for relationships among the 119 taxa that do not have matK or 26S rDNA sequences (Tables 3 and 4). This relationship between the amount of data and support suggests that, although incorporating available sequence data can increase support for much of the tree, a complete understanding of the relationships among all taxa likely will require more complete sampling. Specifically, among angiosperms, the average taxon quartet scores indicate that more data is especially needed to resolve the early diverging branches within eudicots (Table 2).
The gains in bootstrap and quartet similarity support resulting from the new matK and 26S rDNA sequences mostly are incremental, and they do not notably alter our view of most angiosperm relationships. One major exception is the placement of Hydrostachys, an aquatic genus from Madagascar and southern Africa, which is in Cornales in the 3-gene ML analyses and the Lamiales in the 5-gene ML analyses (Fig. 4). The placement of Hydrostachys within Lamiales also is strongly supported by maximum parsimony bootstrap analyses of the 5-gene matrix (Burleigh, unpublished). This new result appears to be driven by the matK sequences, and an ML analysis of just our matK data also places Hydrostachys in Lamiales (not shown). The new Hydrostachys matK sequence was originally included as an outgroup for an analysis of Loasaceae [34], and it is from not only the same species but the same vouchered specimen that was used to obtain the data for the other three genes [Schatz et al. 3413 (MO)].
While the 5-gene analysis provides a new perspective on the placement of Hydrostachys, its placement has always been enigmatic. Xiang et al. [35] and Fan and Xiang [36] placed Hydrostachys among the earlier diverging Cornales lineages, but these studies did not sample extensively outside Cornales. A 3-gene analysis by Albach et al. [37] also placed Hydrostachys within in Cornales [38], but the authors also noted that Hydrostachys had long molecular branches and few morphological characters to support its placement in Cornales [38]. In contrast, placement of Hydrostachys within Lamiales is consistent with embryological [37,39] and floral morphological [40] data. Consequently, it was classified within Lamiales by Dahlgren [41], Cronquist [42], and Takhtajan [43]. Still, while the placement of Hydrostachys within Lamiales is intriguing and credible, we urge caution in interpreting this result. Additional molecular data, as well as analyses to examine the causes for the various molecular results and sources of phylogenetic error, are necessary to confirm the position of Hydrostachys.
Adding matK and 26S rDNA sequences with incomplete taxonomic sampling appears to be beneficial for the inference of the angiosperm phylogeny, but there are still troubling aspects of the 5-gene analysis. Perhaps foremost is the reduction of bootstrap support for a few well-accepted clades such as monocots, eudicots, and core eudicots (Fig. 3). In simulation, adding characters with missing data can decrease the probability of resolving the true phyogeny [44,45], and this is consistent with a reduction in bootstrap support. Furthermore, adding characters with missing data also can have a similar effect as reducing taxon sampling; that is, they can effectively increase the length of branches from sampled taxa and the proportion of characters that support erroneous topologies [44]. Taxonomic sampling can drastically affect the results of phylogenetic analyses (e.g. [46-48]), and sufficient taxonomic sampling is an especially relevant concern in analyses of angiosperm relationships [13,14,17]. Still, there is no obvious evidence of strong taxon sampling-like error in the 5-gene analysis. Each of the added genes had sequences from at least 240 taxa, so no parts of the character matrix had especially poor taxon sampling. Furthermore, bootstrap support for monocots, eudicots, and core eudicots are still at least 78% (Fig. 3), and, besides the placement of Hydrostachys, there are few strongly supported, major differences in the 5-gene and 3-gene results.
Our results suggest other directions for future phylogenetic research. In our study, we increased sampling by taking a fixed set of taxa and adding genes that had many sequences from these taxa. We might also expand our sampling by adding new taxa that have sequences from most or all of the 5-genes (e.g. [25]). This may ameliorate any potential problems associated with inadequate taxon sampling. We also note that conventional nonparametric bootstrapping methods (e.g. [49]) do not explicitly account for missing data, and it may be profitable to explore bootstrapping methods explicitly designed for incomplete data sets (e.g. [50]).
Conclusion
Although there has been much recent progress in elucidating angiosperm phylogeny, there are still many unresolved relationships that are critical to understanding the angiosperm evolution. New data are needed; yet assembling new, complete data sets across all angiosperms is both extremely time-consuming and expensive. While most major analyses of angiosperm relationships have used complete or nearly complete data sets, this study demonstrates that exploring new ways to exploit existing angiosperm data can be a fast, cost-effective, and informative complement to more conventional systematic efforts to sequence new genes.
Methods
Taxon sampling and data sets
The original 3-gene (nuclear 18S rDNA, plastid atpB and rbcL) matrix includes 567 taxa, and the alignment is 4592 characters in length. The set of excluded characters, and thus the total length of the alignment, differs slightly from previous analyses [[1-3]; see Additional file 3]. Each terminal "taxon" in the 567-taxon matrix represents a single genus, and in some cases, gene sequences from congeneric species were combined (see [2]). (The original matrix contains two species from the genus "Saxifraga", but Saxifraga integrifolia is now Micranthes integrifolia). Thus, we also added data from some congeneric species for matK and 26S rDNA (see Additional files 1 and 2). We first searched GenBank for matK sequences from genera that were included in the original 567-taxon matrix. If multiple matK sequences were found from a particular genus, we chose the longest one. Consequently, some of the sequences include sections of the trnK intron regions that flank matK. Additionally, we added a new matK sequence from Gunnera. In total, we had at least partial matK sequences from 378 of the 567 terminal taxa (Fig. 1; Additional file 2). The matK sequences were aligned using on the protocol of Hilu et al. [10], and the total matK alignment was 2379 characters in length (Fig. 1; Additional file 3). The 26S rDNA sequence data were taken from previously analyzed data sets [[30-32]; Additional file 1]. They were aligned using Clustal W [51] and further edited manually, deleting a few small sections in which we could not confidently determine the homology of characters across taxa. In total, 240 out of the original 567 taxa have 26S rDNA sequences, and the 26S rDNA alignment was 3428 characters in length (Fig. 1; see Additional file 3). We concatenated the matK and 26S rDNA sequences to the original 3-locus complete data matrix to generate the 5-locus data matrix (Fig. 1; Additional file 3). All phylogenetic analyses were performed on both the 3-gene (18S rDNA, atpB, and rbcL) 567-taxon data set, which has no missing gene sequences, and the 5-gene 567-taxon matrix, which includes taxa with missing gene sequences.
Phylogenetic analyses
For both the 3-gene and 5-gene data matrices, we performed 100 maximum likelihood (ML) bootstrap replicates using GARLI v. 0.951 [52], which implements a genetic heuristic algorithm for the tree search. We relied on bootstrap support because 1) we are interested in directly assessing how adding characters with missing data affects sampling variance and 2) the 567-taxon data sets appear to have much trouble reaching stationarity in Bayesian analyses [3]. All tree searches started from a neighbor-joining topology [53] and otherwise used the default settings from GARLI. The likelihood function incorporated the general time reversible substitution model (GTR; [54]), which allows different substitution rates for each type of nucleotide substitution, with rate variation among sites estimated using a discrete gamma distribution with four rate categories [55] and a separate parameter for the percentage of invariable sites. The bootstrapped data sets were generated (sampled with replacement from the original data set) using HyPhy [56]. We found that the ML estimates of 567-taxon topology using GARLI occasionally differed, suggesting that that the GARLI tree search may get trapped in local optima. Therefore, for each bootstrap replicate, we performed 5 runs of GARLI and selected a tree with the highest likelihood across the five runs. We also performed maximum parsimony bootstrap analyses on the 3-gene and 5-gene data sets using a parsimony ratchet search strategy [57] implemented in PAUP* [58]. The overall bootstrap support was slightly lower than in the ML analyses, but the results were very similar. Therefore, we just focus on ML results for this paper.
Comparison of tree topologies
We used two measures to compare levels of support from the 3-gene and 5-gene phylogenetic analyses. First, we simply compared the bootstrap support from the two analyses, specifically the number of partitions (clades in a rooted tree) that had bootstrap support of 100%, ≥95%, ≥90%, ≥70%, and ≥50%. However, the bootstrap support for each partition does not describe the support for relationships among all sets of taxa. Therefore, we also used a measure based on the quartet distance (e.g. [59]) to examine and compare the support in the 3-gene and 5-gene analyses in more detail.
A quartet is set of four taxa, and it represents the smallest unit of phylogenetic information in a tree. In a 567-taxon tree, there are over 4.26 billion possible quartets. The quartet similarity (or 1- quartet distance) is the percentage of all quartets with identical unrooted topologies in two trees. We first calculated the average quartet similarity between all pairs of bootstrap trees. We also quantified the phylogenetic support for each taxon in the bootstrap trees using quartet distances (e.g. [59]). To do this, we measured the average taxon quartet similarity, which, for a specified taxon, is the average quartet similarity between all pairs of bootstrap trees for only the quartets that contain the specified taxon. For example, in the 567-taxon tree, only just over 30 million of the 4.26 billion possible quartets involve any single taxon, like Amborella. The average taxon quartet similarity for Amborella measures the average quartet similarity among all pairs of bootstrap trees only for the ~30 million quartets that contain Amborella. Taxa whose positions vary among bootstrap trees will have lower average taxon quartet similarity scores than taxa with similar positions among bootstrap trees. For each set of bootstrap trees, there is a single average quartet similarity score, but there are 567 average taxon quartet similarity scores, one for each taxon. The quartet similarity scores were computed with QDist [60] and a series of perl scripts.
Authors' contributions
This study was conceived and designed by all authors. DES provided initial 26S rDNA sequence alignments and a new matK sequence. JGB assembled the remaining matK sequences, which KWH aligned. JGB designed and performed the computational analyses and wrote the manuscript with help from KWH and DES. All authors read and approved the final manuscript.
Supplementary Material
26S.accession. Accession table for 26SrDNA sequences used in this study.
matK.accession. Accession table for matK sequences used in this study.
5gene. Text file in nexus format with the sequence alignment used for analyses in this study.
3gene.AllMLBS. Text file containing the bootstrap trees from the 3-gene maximum likelihood bootstrap analysis.
5gene.AllMLBS. Text file containing the bootstrap trees from the 5-gene maximum likelihood bootstrap analysis.
3gene.MLBS. PDF file with a figure showing the full majority rule consensus tree from the 3-gene maximum likelihood bootstrap analysis.
5gene.MLBS. PDF file with a figure showing the full majority rule consensus tree from the 5-gene maximum likelihood bootstrap analysis.
Acknowledgments
Acknowledgements
Michael Moore provided a new matK sequence for Gunnera, and Anya Hinkle assisted with the matK alignment. This work was funded by the angiosperm tree of life grant (NSF EF 04-31266 to DES, PSS, KWH, et al.), and NESCent, NSF EF-0423641 (JGB).
Contributor Information
J Gordon Burleigh, Email: gburleigh@ufl.edu.
Khidir W Hilu, Email: hilukw@vt.edu.
Douglas E Soltis, Email: dsoltis@botany.ufl.edu.
References
- Soltis PS, Soltis DE, Chase MW. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature. 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, Axtell M, Swensen SM, Prince LM, Kress WJ, Nixon KC, Farris JS. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Biol J Linn Soc. 2000;133:381–461. [Google Scholar]
- Soltis DE, Gitzendanner MA, Soltis PS. A 567-taxon data set for angiosperms: the challenges posed by Bayesian analyses of large data sets. Int J Plant Sci. 2007;168:137–157. doi: 10.1086/509788. [DOI] [Google Scholar]
- APG II (Angiosperm Phylogeny Group II) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants. Bot J Linn Soc. 2003;141:399–436. doi: 10.1046/j.1095-8339.2003.t01-1-00158.x. [DOI] [Google Scholar]
- Soltis DE, Soltis PS, Endress PK, Chase MW. Phylogeny and evolution of angiosperms. Sunderland, Massachusetts: Sinauer; 2005. [Google Scholar]
- GenBank http://www.ncbi.nlm.nih.gov/
- Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qiu Y-L, Kron KA, Rettig JH, Conti E, Palmer JD, Manhart JR, Sytsma KJ, Michael HJ, Kress WJ, Karol KA, Clark WD, Hedrén M, Gaut BS, Jansen RK, Kim KJ, Wimpee CF, Smith JF, Furnier GR, Strauss SH, Xiang QY, Plunkett GM, Soltis PS, Swensen SM, Williams SE, Gadek PA, Quinn CJ, Eguiarte LE, Golenberg E, Learn GH, Graham SW, Jr, Barrett SCH, Dayanandan S, Albert VA. Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Missouri Bot Gard. 1993;80:528–580. doi: 10.2307/2399846. [DOI] [Google Scholar]
- Källersjö M, Farris JS, Chase MW, Bremer B, Fay MF, Humphries CJ, Peterson G, Seberg O, Bremer K. Simultaneous parsimony jackknife analysis of 2538 rbcL DNA sequences reveals support for major clades of green plants, land plants, seed plants, and flowering plants. Plant Syst Evol. 1998;213:259–287. doi: 10.1007/BF00985205. [DOI] [Google Scholar]
- Soltis DE, Soltis PS, Nickrent DL, Johnson LA, Hahn WJ, Hoot SB, Sweere JA, Kuzoff RK, Kron KA, Chase MW, Swenson SM, Zimmer EA, Chaw SM, Gillespie LJ, Kress WJ, Sytsma KJ. Angiosperm phylogeny inferred from 18S ribosomal DNA sequences. Ann Missouri Bot Gard. 1997;84:1–49. doi: 10.2307/2399952. [DOI] [Google Scholar]
- Hilu KW, Brosch T, Müller K, Soltis DE, Soltis PS, Savolainen V, Chase MW, Powell MP, Alice LA, Evans R, Sauquet H, Neinhuis C, Slotta TAB, Rohwer JG, Campbell CS, Chatrou LW. Angiosperm phylogeny based on matK sequence information. Amer J Bot. 2003;90:1758–1776. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- Goremykin V, Hirsch-Ernst KI, Wölfl S, Hellwig FH. Analysis of Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol. 2003;20:1499–1505. doi: 10.1093/molbev/msg159. [DOI] [PubMed] [Google Scholar]
- Goremykin V, Hirsch-Ernst KI, Wölfl S, Hellwig FH. The chloroplast genome of Nymphaea alba, whole genome analysis and the problem of identifying the most basal angiosperm. Mol Biol Evol. 2004;21:1445–1454. doi: 10.1093/molbev/msh147. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Rice DW, Palmer JD. Long branch attraction, taxon sampling, and the earliest angiosperms: Amborella or monocots? BMC Evol Biol. 2004;4:35. doi: 10.1186/1471-2148-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack J, Raubeson LA, Cui L, Kuehl JV, Fourcade MH, Chumley TW, Boore JL, Jansen RK, dePamphilis CW. Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one's way out of the Felsenstein zone. Mol Biol Evol. 2005;22:1948–1963. doi: 10.1093/molbev/msi191. [DOI] [PubMed] [Google Scholar]
- Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, dePamphilis CW, Boore JL, Jansen RK. Complete plastid genome sequences of Drimys, Liriodedron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 2006;6:77. doi: 10.1186/1471-2148-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller K, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee S-B, Peery R, McNeal JR, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Amborella not a "basal angiosperm"? Not so fast. Am J Bot. 2004;91:997–1001. doi: 10.3732/ajb.91.6.997. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Savolainen V, Hilu K, Qiu Y-L, Chase MW, Farris JS, Stefanovic S, Rice DW, Palmer JD, Soltis PS. Genome-scale data, angiosperm relationships, and 'ending incongruence': a cautionary tale in phylogenetics. Trends Plant Sci. 2004;9:477–483. doi: 10.1016/j.tplants.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Qiu Y-L, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW. The earliest angiosperms: evidence from mitochondrial, plastid, and nuclear genomes. Nature. 1999;402:404–407. doi: 10.1038/46536. [DOI] [PubMed] [Google Scholar]
- Qiu Y-L, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW. Phylogeny of basal angiosperms: analyses of five genes from three genomes. Int J Plant Sci. 2000;161:S3–S27. doi: 10.1086/317584. [DOI] [Google Scholar]
- Qiu Y-L, Dombrovska O, Lee J, Li L, Whitlock BA, Bernasconi-Quadroni F, Rest JS, Davis CC, Borsch T, Hilu KW, Renner SS, Soltis DE, Soltis PS, Zanis MJ, Cannone JJ, Gutell RR, Powell M, Savolainen V, Chatrou LW, Chase MW. Phylogenetic analyses of basal angiosperms based on nine plastid, mitochondrial, and nuclear genes. Int J Plant Sci. 2005;166:815–842. doi: 10.1086/431800. [DOI] [Google Scholar]
- Qiu Y-L, Li L, Hendry TA, Li R, Taylor DW, Issa MJ, Ronen AJ, Vekaria ML, White AM. Reconstructing the basal angiosperm phylogeny: evaluating information content of mitochondrial genes. Taxon. 2006;55:837–856. [Google Scholar]
- Savolainen V, Chase MW, Hoot SB, Morton CM, Soltis DE, Bayer C, Fay MF, de Bruijn AY, Sullivan S, Qiu Y-L. Phylogenetic analysis of flowering plants based upon a combined analysis of plastid atpB and rbcL gene sequences. Syst Biol. 2000;49:306–362. doi: 10.1093/sysbio/49.2.306. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Darwin's abominable mystery: insights from a supertree of the angiosperms. Proc Natl Acad Sci USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell AC, Ané C, Burleigh JG, McMahon MM, O'Meara BC, Sanderson MJ. Prospects for building the tree of life from large sequence databases. Science. 2004;306:1172–1174. doi: 10.1126/science.1102036. [DOI] [PubMed] [Google Scholar]
- Yan C, Burleigh JG, Eulenstein O. A method for finding incomplete phylogenetic datasets from large sequence databases. Mol Phylogenet Evol. 2005;35:528–535. doi: 10.1016/j.ympev.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Wiens JJ. Missing data, incomplete taxa, and phylogenetic accuracy. Syst Biol. 2003;52:528–538. doi: 10.1080/10635150390218330. [DOI] [PubMed] [Google Scholar]
- Kuzoff RK, Sweere JA, Soltis DE, Soltis PS, Zimmer EA. The phylogenetic potential of entire 26S rDNA sequences in plants. Mol Biol Evol. 1998;15:251–263. doi: 10.1093/oxfordjournals.molbev.a025922. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Senters AE, Zanis M, Kim S, Thompson JD, Soltis PS, Ronse De Craene LP, Endress PK, Farris JS. Gunnerales are sister to other core eudicots: implications for the evolution of pentamery. Am J Bot. 2003;90:461–470. doi: 10.3732/ajb.90.3.461. [DOI] [PubMed] [Google Scholar]
- Zanis MJ, Soltis PS, Qiu Y-L, Zimmer EA, Soltis DE. Phylogenetic analysis and perianth evolution in basal angiosperms. Ann Missouri Bot Gard. 2003;90:129–150. doi: 10.2307/3298579. [DOI] [Google Scholar]
- Kim S, Soltis DE, Soltis PS, Zanis MJ, Suh Y. Phylogenetic relationships among early-diverging eudicots based on four genes: were the eudicots ancestrally woody? Mol Phylogenet Evol. 2004;31:16–30. doi: 10.1016/j.ympev.2003.07.017. [DOI] [PubMed] [Google Scholar]
- Wilkinson M. Majority-rule reduced consensus trees and their use in bootstrapping. Mol Biol Evol. 1996;13:437–444. doi: 10.1093/oxfordjournals.molbev.a025604. [DOI] [PubMed] [Google Scholar]
- Hufford L, McMahon MM, Sherwood AM, Reeves G, Chase MW. The major clades of Loasaceae: phylogenetic analysis using the plastid matK and trnL-trnF regions. Am J Bot. 2003;90:1215–1228. doi: 10.3732/ajb.90.8.1215. [DOI] [PubMed] [Google Scholar]
- Fan C, Xiang Q-Y. Phylogenetic analyses of cornales based on 26S rRNA and combined 26S rRNA-matK -rbcL sequence data. Amer J Bot. 2003;90:1357–1372. doi: 10.3732/ajb.90.9.1357. [DOI] [PubMed] [Google Scholar]
- Albach DC, Soltis DE, Chase MW, Soltis DE. Phylogenetic placement of the enigmatic angiosperm Hydrostachys. Taxon. 2001;50:781–805. doi: 10.2307/1223707. [DOI] [Google Scholar]
- Jäger-Zürn I. Zur Frage der systematischen Stellung der Hydrostachyaceae auf Grund ihrer Embryologie, Blüten- und Infloreszenzmorphologie. Österr Bot Z. 1965;112:621–639. doi: 10.1007/BF01373191. [DOI] [Google Scholar]
- Xiang Q-Y, Moody ML, Soltis DE, Fan CZ, Soltis PS. Relationships within Cornales and circumscription of Cornaceae- matK and rbcL sequence data and effects of outgroups and long branches. Mol Phylogenet Evol. 2002;24:35–57. doi: 10.1016/S1055-7903(02)00267-1. [DOI] [PubMed] [Google Scholar]
- Rauh W, Jäger-Zürn I. Le probleme de la position systematique des Hydrostachyacées. Adansonia. 1967;6:515–523. [Google Scholar]
- Leins P, Erbar C. Einige Bemerkungen zur Blütenentwicklung und systematischen Stellung der Wasserpflanzen Callitriche, Hippuris und Hydrostachys. Beitr Biol Pflanzen. 1988;63:157–178. [Google Scholar]
- Dahlgren RMT. A revised system of classification of the angiosperms. Bot J Linn Soc. 1980;80:91–124. doi: 10.1111/j.1095-8339.1980.tb01661.x. [DOI] [Google Scholar]
- Cronquist A. An integrated system of classification of flowering plants. Columbia University Press, New York; 1981. [Google Scholar]
- Takhtajan A. Diversity and classification of flowering plants. New York: Columbia University Press; 1997. [Google Scholar]
- Wiens JJ. Does adding characters with missing data increase or decrease phylogenetic accuracy? Syst Biol. 1998;47:625–640. doi: 10.1080/106351598260635. [DOI] [PubMed] [Google Scholar]
- Hartmann S, Vision TJ. Using ESTs for phylogenomics: can one accurately infer a phylogenetic tree from a gappy alignment? BMC Evol Biol. 2008;8:5. doi: 10.1186/1471-2148-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybeal A. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol. 1998;47:9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 2002;51:588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Pollock DD, McGuire JA, Zwickl DJ. Is sparse taxon sampling a problem for phylogenetic inference? Syst Biol. 2003;52:124–126. doi: 10.1080/10635150390132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Efron B. Missing data, imputation, and the bootstrap. J Am Stat Assoc. 1994;89:463–475. doi: 10.2307/2290846. [DOI] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. GARLI Genetic algorithm for rapid likelihood inference. 2006. https://www.nescent.org/wg_garli/Main_Page
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Tavaré S. Some probabilistic and statistical problems on the analysis of DNA sequences. In: Miura RM, editor. Lectures on mathematics in the life sciences, 5786. Providence, Rhode Island: American Mathematical Society; 1986. [Google Scholar]
- Yang Z. Maximizing likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost DW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Nixon KC. The parsimony ratchet: a new method for rapid parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Version 40 beta 10. Sinauer Assoc., Sunderland, Massachusetts; 2002. PAUP* Phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- Estabrook GF, McMorris FR, Meacham CA. Comparison of undirected phylogenetic trees based on subtrees of 4 evolutionary units. Syst Zool. 1985;34:193–200. doi: 10.2307/2413326. [DOI] [Google Scholar]
- Mailund T, Pedersen CNS. QDist – Quartet distance between evolutionary trees. Bioinformatics. 2004;20:1636–1637. doi: 10.1093/bioinformatics/bth097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
26S.accession. Accession table for 26SrDNA sequences used in this study.
matK.accession. Accession table for matK sequences used in this study.
5gene. Text file in nexus format with the sequence alignment used for analyses in this study.
3gene.AllMLBS. Text file containing the bootstrap trees from the 3-gene maximum likelihood bootstrap analysis.
5gene.AllMLBS. Text file containing the bootstrap trees from the 5-gene maximum likelihood bootstrap analysis.
3gene.MLBS. PDF file with a figure showing the full majority rule consensus tree from the 3-gene maximum likelihood bootstrap analysis.
5gene.MLBS. PDF file with a figure showing the full majority rule consensus tree from the 5-gene maximum likelihood bootstrap analysis.