Abstract
Infection with one of the four serotypes of dengue virus (DENV1-4) can result in a range of clinical manifestations in humans, from dengue fever to the more serious dengue hemorrhagic fever/dengue shock syndrome. Although T cells have been implicated in the immunopathogenesis of secondary infections with heterologous DENV serotypes, the role of T cells in protection against DENV is unknown. In this study, we used a mouse-passaged DENV2 strain, S221, to investigate the role of CD8+ T cells in the immune response to primary DENV infection. S221 did not replicate well in wild-type mice, but did induce a CD8+ T cell response, whereas viral replication and a robust CD8+ T cell response were observed after infection of IFN-α/βR-/- mice. Depletion of CD8+ T cells from IFN-α/βR-/- mice before infection resulted in significantly higher viral loads compared with undepleted mice. Mapping the specificity of the CD8+ T cell response led to the identification of 12 epitopes derived from 6 of the 10 DENV proteins, with a similar immunodominance hierarchy observed in wild-type and IFN-α/βR-/- mice. DENV-specific CD8+ T cells produced IFN-γ, TNF-α, expressed cell surface CD107a, and exhibited cytotoxic activity in vivo. Finally, immunization with four of the immunodominant CD8+ T cell epitopes enhanced viral clearance. Collectively, our results reveal an important role for CD8+ T cells in the host defense against DENV and demonstrate that the anti-DENV CD8+ T cell response can be enhanced by immunization, providing rationale for designing DENV-specific vaccines that induce cell-mediated immunity.
Dengue virus (DENV),3 a member of the Flaviviridae family, is transmitted by Aedes aegypti and Aedes albopictus mosquitoes. DENV is now endemic in more than 100 countries in the Americas, Africa, the Eastern Mediterranean, Southeast Asia, and the Western Pacific, and the number of dengue cases is increasing each year (1). It is estimated that DENV infections result in tens of millions of cases of dengue fever (DF) and 500,000 cases of dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS) worldwide per year (2). DF is a self-limiting yet debilitating febrile illness, whereas DHF and DSS are life-threatening and characterized by increased vascular permeability, thrombocytopenia, hemorrhagic manifestations, and in the case of DSS, shock. Currently, there is no approved vaccine or antiviral treatment available.
Infection with one of the four DENV serotypes presumably leads to protective immunity against that serotype but not the others. In fact, DHF/DSS is most often observed in individuals experiencing a secondary infection with a heterologous serotype (3, 4), and it has been postulated that serotype cross-reactive Abs and memory T cells are involved in the pathogenesis (5, 6). Thus, studies examining the T cell response to DENV have primarily focused on the pathogenic role of T cells during secondary DENV infections. Due to the 65-70% sequence homology between the four serotypes (7), there is a high potential for cross-reactivity during secondary heterologous DENV infections. Studies using patient samples have found that serotype-cross-reactive T cells are preferentially activated during secondary infection in a phenomenon termed “original antigenic sin” (8). These cross-reactive T cells have been shown to exhibit suboptimal degranulation (9) and altered cytokine production and cytolytic activity (10, 11). Aberrant cytokine production by T cells could contribute to severe disease, as higher levels of certain proinflammatory mediators are suspected to cause endothelial cell dysfunction or damage, leading to plasma leakage, a hallmark of DHF/DSS. Collectively, studies to date suggest that altered, suboptimal T cell responses during secondary infections with heterologous serotypes contribute to the immunopathogenesis of DHF/DSS. In contrast, little is known about the role of T cells in protection against either primary or secondary DENV infection. CD8+ T cells are activated in DENV-infected humans (12, 13) and both serotype-specific and serotype-cross-reactive CD8+ T cells that are cytolytic and/or produce IFN-γ and TNF-α have been detected in DENV-immune individuals (8, 9, 11, 14, 15). Similarly, CD8+ T cells are activated and produce IFN-γ in DENV-infected mice (16, 17), and DENV serotype-specific and serotype-cross-reactive murine CD8+ T cell clones have been described (18). However, whether CD8+ T cells protect against DENV infection has yet to be determined.
A better understanding of the role of CD8+ T cells in DENV infection is critical in the development of a vaccine, yet the lack of a good mouse model has hampered mechanistic investigations. The difficulty in developing a mouse model for DENV infections is largely due to the inability of human clinical isolates to replicate well in mice. However, we recently isolated a DENV2 strain, D2S10, which is more virulent in mice than the clinical isolate parental strain (19). S221, a triple-plaque-purified clone from the D2S10 population, was used in the present study to determine whether CD8+ T cells contribute to a protective host response against DENV. The specificity of the CD8+ T cell response in mice was mapped using a predictive algorithm to identify MHC class I-binding peptides from the entire proteome of DENV, which is approximately 3390 aa and encodes three structural (core (C), envelope (E), and membrane (M)) and seven nonstructural (NS) (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins. A broad anti-DENV CD8+ T cell response was observed, with the recognition of 12 epitopes derived from 6 DENV proteins in H-2b mice. We found that depletion of CD8+ T cells resulted in increased DENV replication, DENV-specific CD8+ T cells demonstrated in vivo cytotoxic activity, and immunization with dominant epitopes led to enhanced viral clearance, thereby revealing an important contribution of CD8+ T cells to the anti-DENV-immune response.
Materials and Methods
Mice and infections
C57BL/6 (H-2b) mice were obtained from The Jackson Laboratory and subsequently bred at the animal facility at the La Jolla Institute for Allergy and Immunology (LIAI). IFN-α/βR-/- mice on the C57BL/6 background were obtained from Dr. W. Yokoyama (Washington University, St. Louis, MO) via Dr. C. Ware (LIAI). B6.SJL mice were purchased from The Jackson Laboratory or Taconic Farms. Mice were used between 5 and 8 wk of age. Mice were infected i.v. in the lateral tail vein with 200 μl of S221 in 5% FBS/PBS. Blood was obtained from anesthetized mice by retro-orbital puncture. All mouse experiments were approved by the Animal Care Committee at LIAI.
Cell culture and viral stocks
The hybridoma clones SFR3 and 2.43, which produce rat anti-human HLA DR5 and anti-mouse CD8 IgG2b Ab, respectively, were from the American Type Culture Collection and were grown in protein-free hybridoma medium supplemented with penicillin, streptomycin, HEPES, GlutaMAX, and 2-ME (all from Invitrogen) at 37°C/5% CO2. C6/36, an Aedes albopictus cell line, was cultured in Leibovitz's L-15 medium (Invitrogen) supplemented with 10% FBS (Gemini Bio-Products), penicillin, streptomycin, and HEPES at 28°C in the absence of CO2. Alternate passaging of PL046 (a DENV2 clinical isolate obtained from Dr. H.-Y. Lei (National Cheng Kung University, Taiwan) (20)) through the sera of IFN-α/βR-/-, IFN-γR-/- mice, and C6/36 cells 10 times resulted in a novel DENV2 strain, termed D2S10, that causes more severe disease in those mice (19). S221, which differs from PL046 at E124, E128, and NS1228, is a triple-plaque-purified clone from the D2S10 population. Baby hamster kidney cells (BHK-21) were used to perform standard plaque assays, as described previously (21). Plaque-purified virus was amplified by two passages through C6/36 cells as described previously (21). Briefly, C6/36 monolayers were infected with DENV and cultured for 7 days. Viral supernatant was concentrated by ultracentrifugation at 50,000 × g for 90 min, resuspended in 20% FBS/PBS, and stored at -80°C. Infectious doses were determined based on genomic equivalents (GE), which were quantified by real-time RT-PCR. There are ~104 GE/PFU for S221.
Bioinformatic analyses
Experimental data of peptides binding to H-2b molecules previously generated in our laboratory were used to develop binding predictions. The predictions were performed in June 2006, and the data set available at the time comprised 521 eight-mer peptides binding to Kb and 319 nine-mer peptides binding to Db. In addition, combinatorial peptide libraries described previously (22) were available for 8-mer peptides binding to Kb and 9-mer peptides binding to Db. These two sources of data were combined to calculate scoring matrices that quantify the contribution of each residue in a fixed-length peptide to binding to a MHC molecule, as described previously (23). The entire DENV2 polyprotein PL046 was then scanned using these matrices for peptides binding to either MHC molecule with a predicted affinity of IC50 <500 nM. This approach selected 55 eight-mer peptides predicted to bind H-2Kb and 51 nine-mer peptides predicted to bind H-2Db.
Peptide synthesis
Peptides used in initial screening experiments were synthesized as crude material by Pepscan as described elsewhere (24). A total of 106 eight- and 9-mer peptides were made and combined into 10 pools. Peptides used in the binding assays and immunizations studies were synthesized by A and A Labs and purified to ≥95% homogeneity by reverse-phase HPLC. Purity of these peptides was determined using analytical reverse-phase HPLC and amino acid analysis, sequencing, and/or mass spectrometry. Peptides were radiolabeled with the chloramine-T method, as previously described (25).
MHC peptide-binding assays
MHC purification and quantitative assays to measure the binding affinity of peptides to purified H-2Kb and H-2Db molecules were performed as previously described (25, 26). Briefly, 0.1-1 nM radiolabeled peptide was coincubated at room temperature with 1 μM to 1 nM purified MHC in the presence of 1-3 μM human β2-microglobulin (Scripps Laboratories) and a mixture of protease inhibitors. After a 2-day incubation, binding of the radiolabeled peptide to the corresponding MHC class I molecule was determined by capturing MHC-peptide complexes on Lumitrac 600 microplates (Greiner Bio-One) coated with either the Y3 (anti-H-2Kb) or 28-14-8S (anti-H-2Db, Ld, and Dq) Ab, followed by measurement of bound cpm using the TopCount microscintillation counter (Packard).
IFN-γ ELISPOT assay
CD8+ T cells were isolated by magnetic bead positive selection (Miltenyi Biotec) from the spleens of C57BL/6 mice 7 days after infection with 1011 GE S221. CD8+ T cells (105) were stimulated with 7.5 × 105 uninfected splenocytes as APCs and 1 μg of individual DENV peptides in 96-well flat-bottom plates (Immobilon-P; Millipore) coated with anti-IFN-γ mAb (Mabtech). Following a 20-h incubation at 37°C, the wells were washed with PBS/0.05% Tween 20 and then incubated with biotinylated IFN-γ mAb (Mabtech) for 3 h. The spots were developed using Vectastain ABC peroxidase (Vector Laboratories) and 3-amino-9-ethylcarbazole (Sigma-Aldrich) and counted by computer-assisted image analysis (Zeiss KS ELISPOT reader). The response against an irrelevant, vaccinia virus-derived control peptide (VACV-WR B6R108-116, Kb restricted) was subtracted from the responses to each individual DENV peptide to obtain the net number of spots. The criteria for positivity were net spot-forming cells (SFC) per 106 cells of ≥20, a stimulation index of ≥2.0, and p < 0.05 in a t test comparing replicates with those from the irrelevant peptide.
Flow cytometric analyses
All Ab were purchased from eBioscience with the exception of anti-CD8α-PerCP and anti-CD107a-FITC which were obtained from BD Biosciences. RBC in blood were lysed using RBC lysis buffer (eBioscience). For surface staining, splenocytes or blood cells were washed, incubated with supernatant from 2.4G2-producing hybridoma cells to block Fc receptors, and labeled with anti-CD8α-PerCP, anti-CD44-allophycocyanin, and anti-CD62L-PE. For intracellular cytokine staining, 106 splenocytes or 105 blood cells were plated in 96-well U-bottom plates and stimulated with individual DENV peptides (0.1 μg/ml) in the presence of brefeldin A (GolgiPlug; BD Biosciences) for 5 h. For CD107a staining, CD107a-FITC (or rat IgG2a-FITC) was added to the wells at the same time as the peptide. Cells were washed, incubated with 2.4G2 supernatant, and labeled with anti-CD8α-PerCP. The cells were then fixed and permeabilized using a BD Biosciences Cytofix/Cytoperm Kit and stained with anti-IFN-γ-allophycocyanin and anti-TNF-α-PE. Samples were read on a FACSCaliber (BD Biosciences) and analyzed using FlowJo software (Tree Star).
CD8 depletions
Hybridoma supernatants were clarified by centrifugation, concentrated with Amicon filters (Millipore), and protein G purified (Pierce). The Ab preparations were quantified by spectrophotometry. IFN-α/βR-/- mice were injected i.p. with 250 μg of SFR3 or 2.43 in PBS (250-μl total volume) 3 days and 1 day before infection. At day 6 after infection, 85-95% of CD8+ T cells had been depleted.
Peptide immunizations
IFN-α/βR-/- mice were immunized s.c. at the base of the tail with 50 μg of each DENV peptide plus 100 μg of helper peptide (I-Ab-restricted hepatitis B virus core 128-140) emulsified in IFA (Difco). Mock-immunized mice received helper peptide and DMSO emulsified in IFA. Mice were infected 12-13 days after immunization.
In vivo cytotoxicity assay
IFN-α/βR-/- mice (recipients) were infected with 1010 GE S221. Splenocytes and lymph node cells (targets) were harvested from donor B6.SJL-congenic mice (CD45.1) 7 days later. RBC were lysed, and the target cells were pulsed with 0.5 μg/ml of the irrelevant vaccinia virus (VACV) peptide, C51-59, NS2A8-15, NS4B99-107, NS5237-245, or a pool of C51-59, NS2A8-15, NS4B99-107, and NS5237-245 for 1 h at 37°C. The cells were then washed and labeled with CFSE (Invitrogen) in PBS/0.1% BSA for 10 min at 37°C. DENV peptide-pulsed cells were labeled with 1 μM CFSE (CFSEhigh) and the irrelevant peptide-pulsed cells with 100 nM CFSE (CFSElow). After washing, the two cell populations were mixed at a 1:1 ratio and 5 × 106 cells from each population were injected i.v. into naive or infected recipient mice. After 4 h, the mice were sacrificed and splenocytes were stained with CD45.1-allophycocyanin and analyzed by flow cytometry, gating on CD45.1 cells. The percentage killing was calculated as follows: 100 - ((percentage DENV peptide-pulsed in infected mice/percentage irrelevant peptide-pulsed in infected mice)/(percentage DENV peptide-pulsed in naive mice/percentage irrelevant peptide-pulsed in naive mice)) × 100.
Quantitation of DENV burden in mice
Mice were euthanized by isoflurane inhalation and blood was collected via cardiac puncture. Serum was separated from whole blood by centrifugation in serum separator tubes (Starsted). Small tissue pieces were immediately placed into RNAlater (Qiagen) and subsequently homogenized for 3 min at 4°C in 1 ml of tissue lysis buffer (Qiagen Buffer RLT) using a Mini-Beadbeater-8 (BioSpec Products). Immediately after homogenization, all tissues were centrifuged (5 min, 4°C, 16,000 × g) to pellet debris and RNA was isolated using the RNeasy Mini Kit (Qiagen). Serum RNA was isolated using a QIAamp Viral RNA Mini Kit (Qiagen). After elution, viral RNA was snap frozen in liquid N2 and stored at -80°C. Quantitative RT-PCR was performed according to a published protocol (27), except a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad) with an iScript One-Step RT-PCR Kit for Probes (Bio-Rad) were used. The DENV standard curve was generated with serial dilutions of a known concentration of DENV genomic RNA which was in vitro transcribed (MAXIscript Kit; Ambion) from a plasmid containing the cDNA template of S221 3′ untranslated region. After transcription, DNA was digested with DNase I and RNA was purified using a RNeasy Mini Kit and quantified by spectrophotometry. To control for RNA quality and quantity when measuring DENV in tissues, the level of 18S rRNA was measured using an 18S rRNA Control Kit (Eurogentec) in parallel real-time RT-PCR. A relative 18S standard curve was made from total splenic RNA.
Statistical analyses
Data were analyzed with Prism software version 5.0 (GraphPad Software). Statistical significance for CD8+ T cell numbers and viral load were determined using the unpaired t test with Welch's correction; for the ELISPOT, the unpaired t test was used.
Results
DENV induces CD8+ T cell activation in wild-type and IFN-α/βR-/- C57BL/6 mice
The protective and immunopathogenic roles of CD8+ T cells in DENV infection have not been thoroughly investigated, largely due to the lack of a suitable small animal model. We recently isolated a novel DENV2 strain, D2S10, which is more virulent in mice genetically deficient in both the IFN-α/βR and IFN-γR than the parental strain PL046 (19). A biological clone of D2S10, termed S221, was isolated by triple plaque purification. Wild-type C57BL/6 mice infected with S221 did not manifest any symptoms of disease, and the virus was undetectable by real-time RT-PCR in the serum 40 h after infection (Fig. 1A). To study the contribution of CD8+ T cells in protection against DENV infection, we sought to develop a model in which the virus replicates at detectable levels. Given the importance of the IFN system in most antiviral responses (28, 29), including the anti-DENV response (30, 31), we tested the susceptibility of IFN-α/βR-/- C57BL/6 mice to the S221 virus. Infection of IFN-α/βR-/- mice with S221 resulted in a productive infection, and the mice showed symptoms of disease, including hunched posture and ruffled fur at the peak of viremia, between 2 and 4 days after infection (Fig. 1A and our unpublished observations). The mice subsequently recovered and cleared the virus from the blood by approximately day 6.
FIGURE 1.
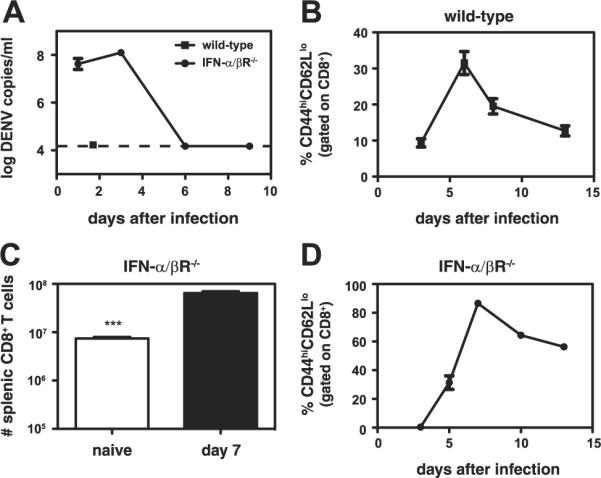
DENV infection results in a CD8+ T cell response in wild-type and IFN-α/βR-/- mice, but detectable levels of viremia only in IFN-α/βR-/- mice. A, Wild-type mice (n = 3) infected with 1011 GE of S221 and IFN-α/βR-/- mice (n = 6) infected with 1010 GE were bled and the DENV RNA levels in the serum were measured by real-time RT-PCR. The dashed line indicates the limit of detection. B, Blood lymphocytes were obtained from wild-type mice (n = 4) on days 3, 6, 8, and 13 after infection with 1011 GE of S221. The percentage of CD44highCD62Llow cells (gated on CD8+ T cells) is indicated. C, The numbers of splenic CD8+ T cells in naive IFN-α/βR-/- mice (n = 4) and IFN-α/βR-/- mice infected with 1010 GE of S221 (n = 7) are shown. ***, p < 0.0001 for naive vs infected mice. D, Blood lymphocytes were obtained from IFN-α/βR-/- mice (n = 3) on days 3, 5, 7, 10, and 13 after infection with 1010 GE of S221. The percentage of CD44highCD62Llow cells (gated on CD8+ T cells) is shown.
To investigate the CD8+ T cell response to S221, the number and activation state of splenic and blood CD8+ T cells in wild-type and IFN-α/βR-/- mice were examined. In wild-type mice, infection with S221 did not lead to an increase in splenic CD8+ T cell numbers (data not shown); however, the CD8+ T cells were activated, as measured by up-regulation of CD44 and down-regulation of CD62L on CD8+ T cells. In the spleen, 13% of CD8+ T cells were CD44highCD62Llow on day 7 after infection (13 ± 3%, mean ± SEM, n = 11). In the blood, activation peaked at day 6, with ~30% of the CD8+ T cells activated (Fig. 1B). UV-inactivated S221 did not induce CD8+ T cell activation, indicating that DENV replication or at least input strand translation is required for T cell activation (data not shown).
In S221-infected IFN-α/βR-/- mice, a 9-fold increase in splenic CD8+ T cell numbers was observed (Fig. 1C). Most of the splenic CD8+ T cells were CD44highCD62Llow on day 7 (76 ± 3%, n = 7). Activation of circulating CD8+ T cells peaked at approximately day 7, with 87% of CD8+ T cells activated (Fig. 1D). Taken together, these data demonstrate that DENV induces CD8+ T cell activation in both wild-type and IFN-α/βR-/- mice. As expected, a greater percentage of CD8+ T cells was activated in the IFN-α/βR-/- mice than in wild-type mice and CD8+ T cell proliferation was observed in the IFN-α/βR-/- mice but not wild-type mice, likely due to increased DENV replication.
CD8 depletion impairs DENV clearance
Because IFN-α/βR-/- mice mount a robust CD8+ T cell response following DENV infection, we sought to define the contribution of these cells to controlling infection. Therefore, IFN-α/βR-/- mice were CD8+ T cell depleted before infection with S221, and virus levels were measured in the serum, spleen, liver, and brain by real-time RT-PCR 3 and 6 days after infection. On day 3 after infection, all of the mice appeared hunched and ruffled, and DENV levels in the serum, spleen, liver, and brain were similar in control and CD8-depleted mice (data not shown). In contrast, by day 6 after infection, the control mice appeared healthy, whereas the CD8-depleted mice were still hunched and ruffled and some ex-hibited hind limb weakness (our unpublished observations). CD8-depleted mice had significantly higher DENV RNA levels than control mice in the serum, spleen, and brain, (30-, 460-, and 350- fold, respectively) but no differences were observed in the liver (Fig. 2). DENV RNA levels in the control mice declined from days 3 to 6, whereas viral RNA levels in the brain of CD8-depleted mice increased from days 3 to 6 (data not shown). These results dem- onstrate that CD8+ T cells control DENV infection in IFN-α/βR-/- mice.
FIGURE 2.
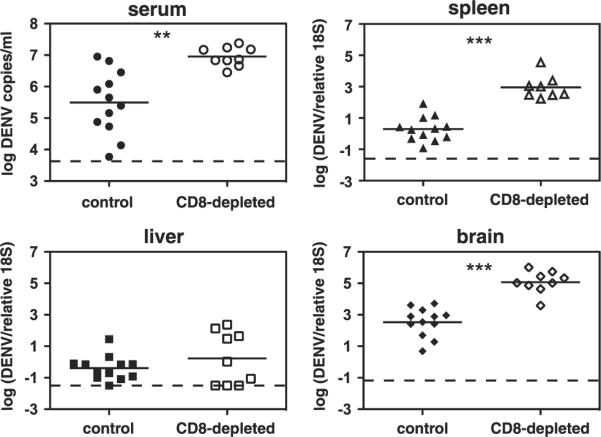
Depletion of CD8+ T cells before DENV infection results in increased viral loads. IFN-α/βR-/- mice were depleted of CD8+ T cells by administration of an anti-CD8 Ab (or given an isotype control Ab) 3 days and 1 day before infection with 1011 GE of S221. Mice were sacrificed 6 days later, and DENV RNA levels in the serum, spleen, liver, and brain were quantified by real-time RT-PCR. Data are expressed as DENV copies per ml of sera or DENV units normalized to 18S rRNA levels for the spleen, liver, and brain. Each symbol represents one mouse, the bar represents the geometric mean, and the dashed line is the limit of detection. *, p < 0.001 for serum; ***, p < 0.0001 for spleen and brain; and p = 0.39 for viral load in the liver of CD8-depleted mice compared with control mice.
Identification of DENV CD8+ T cell epitopes
To study the anti-DENV CD8+ T cell response in more detail, the specificity of the MHC class I-restricted CD8+ T cell response to DENV was mapped using a bioinformatics prediction method that has been previously used to define the total CD8+ T cell response to lymphocytic choriomeningitis virus (LCMV) in C57BL/6 mice (32). Briefly, the proteome of PL046 was inspected for the presence of peptides predicted to bind H-2b class I molecules (Kb and Db) with high affinity. A total of 106 potential H-2b-binding peptides were identified. These peptides were combined into 10 pools and tested in IFN-γ ELISPOT assays using CD8+ T cells from S221-infected wild-type mice. The criteria for positivity were net SFC per 106 cells ≥20, a stimulation index of ≥2.0, and p < 0.05 when compared with an irrelevant control peptide.
Positive pools were deconvoluted by testing the peptides individually and 12 positive peptide epitopes were identified (Fig. 3). The identified epitopes are derived from 6 of the 10 DENV proteins, including all three structural DENV proteins (C, M, and E) and three of the seven nonstructural proteins (NS2A, NS4B, and NS5). The highest responses were observed against C51-59, E-2-6, E451-458, NS2A8-15, NS4B99-107, and NS5237-245. No peptides were identified from the remaining proteins, despite the fact that numerous candidate epitopes were predicted and tested.
FIGURE 3.
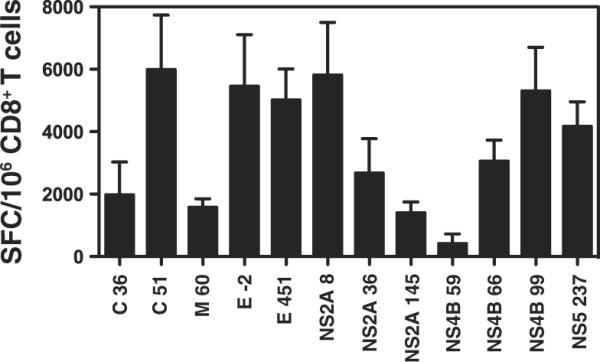
Identification of DENV-derived epitopes recognized by CD8+ T cells. IFN-γ ELISPOT was performed using CD8+ T cells isolated from wild-type mice 7 days after infection with 1011 GE of S221. The data are expressed as the mean number of net SFC per 106 CD8+ T cells. Three independent experiments performed in triplicate were averaged and the error bars represent the SEM. The criteria for positivity were net SFC per 106 cells of ≥20, a stimulation index of ≥2.0, and p < 0.05 when compared with an irrelevant control peptide. The 12 positive peptides identified are shown.
Further characterization of the epitopes identified
To confirm the MHC class I restriction of the 12 identified epitopes, we measured their MHC-binding capacity using purified Kb and Db molecules in an in vitro-binding assay. The results are shown in Table I. Seven of the 12 epitopes tested bound the predicted allele with either high or intermediate affinity, as indicated by an IC50 of < 50 or < 500 nM, respectively. The five remaining epitopes still bound with biologically relevant affinities in the 500-2000 nM range.
Table I.
DENV-derived CD8+ T cell epitopes
| Binding Affinity (IC50, nM) |
||||
|---|---|---|---|---|
| Epitope | Sequence | Restriction Size | H-2Db | H-2Kb |
| C36-44 | GMLQGRGPL | Db 9-mer | 820 | 1336 |
| C51-59 | VAFLRFLTI | Kb 9-mer | 1379 | 20 |
| M60-67 | RALIFILL | Kb 8-mer | ― | 22 |
| E-2-6 | MTMRCIGI | Kb 8-mer | ― | 1085 |
| E451-458 | VSWTMKIL | Kb 8-mer | ― | 458 |
| NS2A8-15 | FSLGVLGM | Kb 8-mer | 1803 | ― |
| NS2A36-44 | VAVSFVTLI | Db 9-mer | 997 | 12 |
| NS2A145-153 | LAVTIMAIL | Db 9-mer | 644 | ― |
| NS4B59-66 | SSVNVSLT | Kb 8-mer | 1451 | ― |
| NS4B66-74 | TAIANQATV | Db 9-mer | 1.3 | ― |
| NS4B99-107 | YSQVNPITL | Db 9-mer | 1.3 | 1893 |
| NS5237-245 | RMLINRFTM | Db 9-mer | 21 | ― |
―, An IC50 <2000 nM.
To verify that the IFN-γ response to these MHC class I-binding DENV peptides was mediated by CD8+ T cells, we performed intracellular cytokine staining (ICS) on CD8+ T cells isolated from S221-infected wild-type mice. The number of CD8+ T cells producing IFN-γ in response to the seven dominant epitopes is shown (Fig. 4A). The responses ranged from an average of 3.3 × 104 CD8+ T cells recognizing E451-458 to 6.3 × 105 cells specific for NS4B99-107 (which corresponds to 0.6 and 5.9% of splenic CD8+ T cells, respectively). Responses against the remaining five epitopes were not reproducibly above background levels, which is consistent with the greater sensitivity of the ELISPOT assay compared with ICS (33). Thus, we have identified a set of 12 H-2b-restricted CD8+ T cell epitopes derived from DENV, seven of which induce responses detectable by ICS.
FIGURE 4.
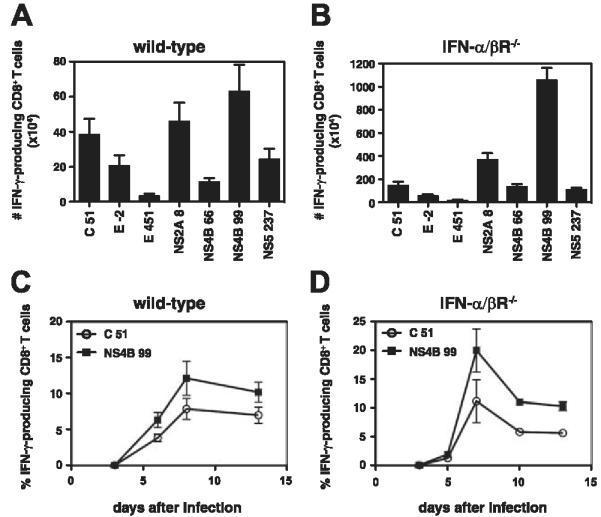
Confirmation of DENV-derived CD8+ T cell epitope identification in wild-type and IFN-α/βR-/- mice by ICS. A and B, Splenocytes harvested from wild-type (A) or IFN-α/βR-/- (B) mice 7 days after infection with 1011 or 1010 GE of S221, respectively, were restimulated in vitro with individual DENV peptides or an irrelevant peptide. Cells were then stained for surface CD8 and intracellular IFN-γ and analyzed by flow cytometry. The response to the irrelevant peptide was subtracted from the response to each DENV peptide, and the number of CD8+ T cells producing IFN-γ is indicated. Results are expressed as the mean ± SEM of 13 wild-type and 7 IFN-α/βR-/- mice tested in at least three independent experiments. C and D, Kinetics of the DENV-specific CD8+ T cell response. Wild-type mice (C, n = 4) and IFN-α/βR-/- mice (D, n = 3) were infected with 1011 or 1010 GE of S221, respectively, and blood lymphocytes were isolated at various time points. Stimulation and ICS were performed as in A and B and the percentage of CD8+ T cells producing IFN-γ is shown.
Epitopes recognized in IFN-α/βR-/- mice
The initial epitope identification experiments were performed in wild-type mice. Because IFN-α/βR-/- mice were used to investigate the contribution of CD8+ T cells to controlling DENV infection, we next examined the specificity of the CD8+ T cell response in these mice. The responses to the seven dominant DENV epitopes identified in wild-type mice were examined in IFN-α/βR-/- mice by ICS (Fig. 4B). The responses ranged from an average of 1.6 × 105 CD8+ T cells specific for E451-458 to 1.1 × 107 recognizing NS4B99-107 (0.3 and 16.5% of splenic CD8+ T cells, respectively). The specificity of the response was similar in the IFN-α/βR-/- and wild-type mice, with C51-59, NS2A8-15, and NS4B99-107 dominating. However, the immunodominance hierarchies were not identical; for example, the response to NS4B66-74 was higher than the response to E-2-6 in the IFN-α/βR-/- but not wild-type mice. In addition, the magnitude of the response differed, with a greater number of CD8+ T cells specific for each epitope in the IFN-α/βR-/- mice. Finally, a kinetic analysis revealed that the DENV-specific response of circulating CD8+ T cells peaked at approximately day 7/8 in both the wild-type and IFN-α/βR-/- mice (Fig. 4, C and D).
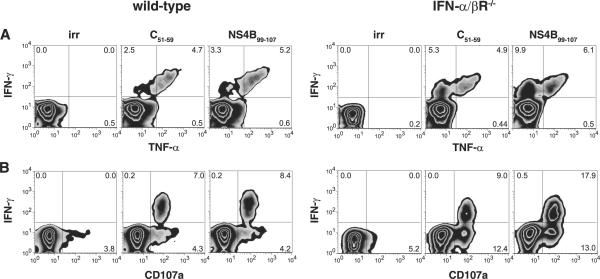
Because polyfunctional CD8+ T cell responses are associated with protection (34), we examined the phenotype of the DENV-specific CD8+ T cells in wild-type and IFN-α/βR-/- mice in more detail by measuring CD107a expression (CD107a, or LAMP-1, is expressed on the cell surface upon degranulation) and TNF-α production for two of the DENV-specific CD8+ T cell populations. The CD8+ T cells recognizing C51-59 and NS4B99-107 produced TNF-α (Fig. 5A) and expressed CD107a on the surface (Fig. 5B). Not all of the IFN-γ+ cells also produced TNF-α, but all of the IFN-γ+ cells were CD107a+. The CD107a+ cells were IFN-γ-TNF-α-, IFN-γ+TNF-α-, or IFN-γ+TNF-α+ (data not shown). Thus, the specificity of the CD8+ T cell response is similar in wild-type and IFN-α/βR-/- mice, with DENV-specific CD8+ T cells producing IFN-γ, TNF-α, and degranulating. Together with the results shown in Fig. 1, our data reveal that the CD8+ T cell response to DENV is polyfunctional and that the response, including proliferation, degranulation, and cytokine production, can develop in the absence of IFN-α/β signaling.
FIGURE 5.
DENV-specific CD8+ T cells have a polyfunctional phenotype. Splenocytes harvested from wild-type or IFN-α/βR-/- mice 7 days after infection with 1011 or 1010 GE of S221, respectively, were restimulated in vitro with C51-59, NS4B99-107, or an irrelevant peptide. An anti-CD107a Ab was added for the duration of the stimulation. Cells were then stained for surface CD8, intracellular IFN-γ and TNF-α, and analyzed by flow cytometry. Representative density plots are shown.
In vivo killing of DENV peptide-pulsed target cells
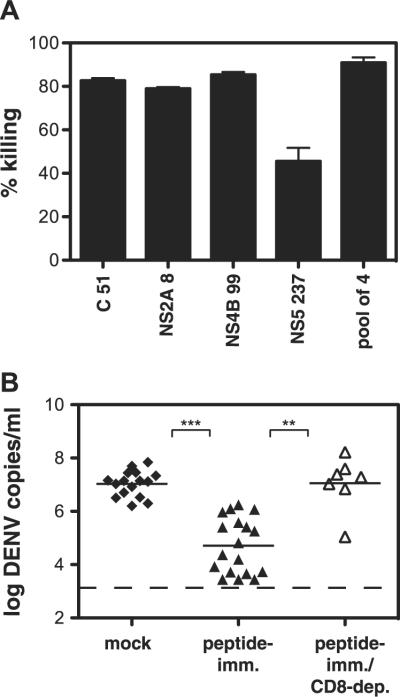
Having found that DENV-specific CD8+ T cells express a degranulation marker, we sought to analyze their cytotoxic activity using an in vivo cytotoxicity assay. Briefly, CFSE-labeled splenocytes pulsed with individual immunodominant DENV peptides (C51-59, NS2A8-15, NS4B99-107, NS5237-245) or a pool of the four peptides were transferred into DENV-immune IFN-α/βR-/- mice, and the percentage of target cell killing was calculated after 4 h. Target cells pulsed with the pool of four peptides were efficiently killed (91% killing), and killing of targets pulsed with the individual peptides ranged from 46% for NS5237-245 to 85% for NS4B99-107 (Fig. 6A). These results show that DENV-specific CD8+ T cells mediate cytotoxicity in DENV-infected IFN-α/βR-/- mice.
FIGURE 6.
DENV-specific CD8+ T cells are cytotoxic and protective in vivo. A, In vivo killing of DENV peptide-pulsed cells. IFN-α/βR-/- mice infected 7 days previously with 1010 GE of S221 were injected i.v. with CFSE-labeled target cells pulsed with C51-9, NS2A8-15, NS4B99-107, NS5237-245 or a pool of the four peptides (n = 3-6 mice/group). After 4 h, splenocytes were harvested, analyzed by flow cytometry, and the percentage killing was calculated. B, Peptide immunization results in enhanced DENV clearance. IFN-α/βR-/- mice were immunized s.c. with 50 μg each of four DENV peptides (C51-59, NS2A8-15, NS4B99-107, NS5237-245) and 100 μg of helper peptide in IFA or mock immunized with 100 μgof helper peptide and DMSO in IFA. Mice were infected with 1011 GE of S221 virus 12-13 days later and then sacrificed 4 days after infection. A separate group of peptide-immunized mice were depleted of CD8+ T cells before infection. DENV RNA levels in the serum were quantified by realtime RT-PCR. Each symbol represents one mouse; the bar represents the geometric mean, and the dashed line the limit of detection. ***, p < 0.0001 comparing peptide-immunized with mock-immunized mice; **, p < 0.001 comparing peptide-immunized with peptide-immunized/CD8-depleted mice.
Vaccination with DENV CD8+ T cell epitopes controls viral load
Since depleting CD8+ T cells resulted in increased viral loads and DENV-specific CD8+ T cells demonstrated in vivo cytotoxic activity, we examined whether enhancing the anti-DENV CD8+ T cell response through peptide immunization would contribute to protection against a subsequent DENV challenge. Specifically, the effect of peptide vaccination on viremia was determined by immunizing IFN-α/βR-/- mice with DENV peptides before infection with S221. Mice were immunized with four dominant DENV epitopes in an attempt to induce a multispecific T cell response, which is desirable to prevent possible viral escape through mutation (35). At day 4 after infection, viremia in the serum was measured by real-time RT-PCR. Peptide immunization resulted in enhanced control of DENV infection, with 350-fold lower serum DENV RNA levels in peptide-immunized mice than mock-immunized mice (Fig. 6B). To confirm that the protection was mediated by CD8+ T cells, we depleted CD8+ T cells from a group of peptide-immunized mice before infection and found that this abrogated the protective effect. Thus, our data demonstrate that a preexisting DENV-specific CD8+ T cell response induced by peptide vaccination enhances viral clearance.
Discussion
The investigation of the cellular immune response to DENV is of considerable interest, as DHF/DSS are presumed to result from immunopathology possibly caused in part by serotype-cross-reactive T cell responses during secondary heterologous infection. Lack of an adequate animal model has hindered studies on the role of T cells in protection against DENV and the mechanisms by which cross-reactive T cells may contribute to DHF/DSS. In this study, we have characterized the CD8+ T cell response to DENV using a mouse model of DENV infection. Specifically, our data reveal that a CD8+ T cell response to DENV can develop in the absence of IFN-α/β signaling and plays an important role in controlling DENV infection. Additionally, our data show that the Agspecific CD8+ T cell response simultaneously targets multiple determinants encoded by the virus and is polyfunctional, with DENV-specific CD8+ T cells producing IFN-γ, TNF-α, and killing target cells in vivo. Finally, a CD8+ T cell response elicited via peptide vaccination can limit DENV infection, as judged by a dramatic reduction in viral load in the serum.
We used a novel murine model that allows investigations of CD8+ T cell responses and DENV replication. Our laboratory previously reported the isolation of the DENV2 strain D2S10, which induces an early, TNF-α-mediated death with vascular leakage, a hallmark of DHF/DSS, in double-deficient IFN-α/βR-/-, IFN-γR-/- mice (19). The S221-infected IFN-α/βR-/- mouse model described here better resembles DF, which constitutes the majority of symptomatic dengue cases (2). DF is a self-limiting febrile illness, in which viremia peaks at or soon after the onset of illness and lasts for 2-12 days, with fever observed during the viremic period (36). In our model, mice exhibited signs of illness (hunched posture, ruffled fur) at the peak of viremia (approximately day 3) and then cleared the virus around day 6.
A caveat to using the IFN-α/βR-/- mice is that IFN-α/β cytokines have been reported to directly affect CD8+ T cell expansion and memory formation after LCMV infection (37). However, it was also reported that during LCMV infection, increased IL-12 can compensate for the lack of IFN-α/β signaling in mediating CD8+ T cell proliferation and IFN-γ production (38). Moreover, dependence on IFN-α/β signaling appears to be pathogen specific, as the lack of the IFN-α/βR did not significantly impair CD8+ T cell expansion in response to VACV, vesicular stomatitis virus, or Listeria monocytogenes (39). Similarly, IFN-α/βR-/- mice generate efficient primary and memory CD8+ T cell responses following Sendai virus infection (40). Our results indicate that the CD8+ T cell response to primary DENV infection does not require signaling through the IFN-α/βR. CD8+ T cell expansion, activation, and Ag-specific IFN-γ production (based on CD8+ T cell number) were enhanced in IFN-α/βR-/- mice compared with wild-type mice, likely a result of increased viral replication. In addition, the percentages of TNF-α+ or CD107a+ CD8+ T cells were similar or greater in the IFN-α/βR-/- mice. Thus, in terms of proliferation, cytokine production, and degranulation, the CD8+ T cell response to DENV does not require type I IFNs. It is probable that IL-2 and IL-12 compensate for the lack of IFN-α/β signaling in inducing CD8+ T cell proliferation and IFN-γ production, respectively.
Our data also show that the specificity of the CD8+ T cell response was similar in the wild-type and IFN-α/βR-/- mice. All of the epitopes identified in wild-type mice were also positive in IFNα/βR-/- mice and the immunodominance hierarchy of the seven dominant epitopes was similar. A study of LCMV found the responses to two dominant LCMV epitopes were altered in IFN-α/βR-/- mice compared with wild-type mice, although only responses to those two epitopes were measured (38). Similar to our findings, the IFN-α/βR-/- mice had a higher viral load, which has been shown to affect the immunodominance of the anti-LCMV CD8+ T cell response (41). However, the difference in DENV replication between wild-type and IFN-α/βR-/- mice did not significantly alter the hierarchy of the seven dominant epitopes. Overall, our results suggest that the lack of IFN-α/β signaling does not appreciably affect the immunodominance of the CD8+ T cell response to DENV. This finding, combined with the high viremia and CD8+ T cell activation observed in S221-infected IFN-α/βR-/- mice lends credence to the use of this model to study the role of CD8+ T cells during DENV infection.
Although it is well known that CD8+ T cells are important in the host response to many viral pathogens, including the related flavivirus, West Nile virus (42), their protective role in DENV infection has not been clearly demonstrated, as studies to date have mainly focused on the potentially pathogenic role of T cells. Determining the contribution of CD8+ T cells to protection is imperative in designing a DENV vaccine and, to our knowledge, our data are the first to demonstrate that CD8+ T cells contribute to protection during primary DENV infection. We found that depletion of CD8+ T cells resulted in an impaired ability of the mice to clear DENV and significantly higher viral loads in the serum, spleen, and brain. Nervous system involvement in human DENV infection is rare (43), but it is a major target in mouse models of DENV infection (20, 31). Whether CD8+ T cells play a role in preventing DENV dissemination to the brain or limiting viral replication in the brain is as yet unknown. Although no difference in DENV levels in the liver between control and depleted mice was observed, the viral load was low in both groups, and the liver therefore does not appear to be a major target of DENV in this mouse model. Altogether, our data show that in the absence of IFN-α/β signaling, CD8+ T cells contribute to controlling DENV infection.
In general, the antiviral activity of CD8+ T cells is mediated by the production of cytokines, including IFN-γ and TNF-α, and direct killing of infected cells. We found that DENV-specific CD8+ T cells produce IFN-γ and TNF-α and demonstrate potent cytotoxic activity in vivo. As DENV-specific CD8+ T cells degranulate, it is likely that perforin/granzyme-mediated killing contributes to viral clearance.
Most dengue infections are asymptomatic or classified as DF, whereas DHF/DSS accounts for a small percentage of dengue cases, indicating that in most infections the host immune response is protective. Our data suggest that CD8+ T cells contribute to protection during primary infection by reducing viral load. As mentioned above, however, cross-reactive memory T cells have been postulated to contribute to immunopathology during heterologous infections, and the relative contribution of CD8+ T cells to protection vs pathogenesis during secondary infections remains to be determined.
Two DENV-derived CD8 + T cell epitopes (NS3298-306 and E331-339) have been identified in mice by isolating T cell clones from DENV-immunized BALB/c (H-2d) mice (18, 44). However, ours is the first study to use a predictive approach spanning the entire proteome to map the CD8+ T cell response to DENV. A total of 12 epitopes derived from 6 of the 10 DENV proteins were identified in wild-type C57BL/6 mice. In general, the positive peptides exhibited intermediate or high binding affinity to MHC class I in vitro. As expected, the immunodominance hierarchy did not strictly correlate with in vitro binding affinities, as other factors, such as T cell precursor frequency and Ag processing during infection affect immunodominance (33). Considering the small size of the DENV genome, this CD8+ T cell response is relatively broad and is similar to the 16 CD8+ T cell epitopes identified in influenza A-infected C57BL/6 mice (45). Broad CD8+ T cell responses in C57BL/6 mice have been identified for LCMV (28 epitopes) (32) and for larger viruses such as murine CMV (24 epitopes) (46) and VACV (49 epitopes) (47).
A number of DENV-derived CD8+ T cell epitopes have been identified in humans, including epitopes from the E, NS3, NS4A, NS4B, and NS5 proteins (8, 11, 48-51). Based on the isolation of a number of NS3-specific CD8+ T cell clones, this protein has been postulated as a major target of the T cell response (52). However, we did not identify any immunogenic NS3 epitopes, even though seven NS3 peptides were predicted and tested by IFN-γ ELISPOT, which could be due to species-specific differences in the CD8+ T cell responses.
Finally, we found that immunization with four dominant epitopes before infection resulted in enhanced DENV clearance, and this protection was mediated by CD8+ T cells. These results have implications for vaccine development, they provide a proofof-principle that vaccination with T cell epitopes can reduce viremia. Current DENV vaccines under development attempt to elicit a neutralizing Ab response and need to protect against all four serotypes so as to avoid Ab-dependent enhancement. Vaccine research thus far has focused on live, attenuated tetravalent vaccines, but subunit and DNA vaccines are in development (53). Our results indicate that CD8+ T cells are an important component to a protective immune response. A desirable attribute of a DENV vaccine would be the induction of T cell responses to highly conserved epitopes that will protect against all four serotypes (54). Based on the original antigenic sin hypothesis, vaccination with variable CD8+ T cell epitopes could lead to pathology by activating cross-reactive memory T cells that respond aberrantly and should therefore be avoided. The nonstructural proteins of DENV, and NS3, NS4B, and NS5 in particular, are highly conserved across serotypes and likely contain good vaccine candidates.
In conclusion, our results reveal a critical role for CD8+ T cells in the immune response to an important human pathogen and provide a rationale for the inclusion of CD8+ T cell epitopes in DENV vaccines. Furthermore, identification of the CD8+ T cell epitopes recognized during DENV infection in combination with our new mouse model have laid the foundation for elucidating the protective vs pathogenic role of CD8+ T cells during secondary infections.
Acknowledgments
We thank Sarala Joshi, Steven Lada, and Carla Oseroff for technical assistance.
This study was supported by National Institutes of Health Grants AI060989 and AI077099-01 (to S.S.), by a developmental project award from the Pacific Southwest Regional Center of Excellence (U54 AI065359 to S.S.), and by fellowships from the Center for Infectious Diseases and the Diabetes & Immune Disease National Research Institute (to L.E.Y.). This is Kirin publication no. 1027.
Footnotes
Abbreviations used in this paper: DENV, dengue virus; DF, dengue fever; DHF, dengue hemorrhagic fever; DSS, dengue shock syndrome; GE, genomic equivalent; SFC, spot-forming cell; LCMV, lymphocytic choriomeningitis virus; ICS, intracellular cytokine staining; VACV, vaccinia virus.
Disclosures The authors have no financial conflict of interest.
References
- 1.WHO . Dengue haemorrhagic fever: diagnosis, treatment prevention and control. 2nd Ed. WHO; Geneva: 1997. [Google Scholar]
- 2.Kyle JL, Harris E. Global spread and persistence of dengue. Annu. Rev. Microbiol. 2008 doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 3.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead SB. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand: I. The 1980 outbreak. Am. J. Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 4.Guzman MG, Kouri G, Valdes L, Bravo J, Alvarez M, Vazques S, Delgado I, Halstead SB. Epidemiologic studies on dengue in Santiago de Cuba, 1997. Am. J. Epidemiol. 2000;152:793–799. doi: 10.1093/aje/152.9.793. discussion 804. [DOI] [PubMed] [Google Scholar]
- 5.Green S, Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr. Opin. Infect. Dis. 2006;19:429–436. doi: 10.1097/01.qco.0000244047.31135.fa. [DOI] [PubMed] [Google Scholar]
- 6.Halstead SB. Dengue. Lancet. 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 7.Fu J, Tan BH, Yap EH, Chan YC, Tan YH. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90) Virology. 1992;188:953–958. doi: 10.1016/0042-6822(92)90560-c. [DOI] [PubMed] [Google Scholar]
- 8.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 9.Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, Khemnu N, Tangthawornchaikul N, Chotiyarnwong P, Sae-Jang K, et al. T cell responses in dengue hemorrhagic fever: are crossreactive T cells suboptimal? J. Immunol. 2006;176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 10.Mangada MM, Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J. Immunol. 2005;175:2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- 11.Imrie A, Meeks J, Gurary A, Sukhbataar M, Kitsutani P, Effler P, Zhao Z. Differential functional avidity of dengue virus-specific T-cell clones for variant peptides representing heterologous and previously encountered serotypes. J. Virol. 2007;81:10081–10091. doi: 10.1128/JVI.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurane I, Innis BL, Nimmannitya S, Nisalak A, Meager A, Janus J, Ennis FA. Activation of T lymphocytes in dengue virus infections: high levels of soluble interleukin 2 receptor, soluble CD4, soluble CD8, interleukin 2, and interferon-γ in sera of children with dengue. J. Clin. Invest. 1991;88:1473–1480. doi: 10.1172/JCI115457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green S, Pichyangkul S, Vaughn DW, Kalayanarooj S, Nimmannitya S, Nisalak A, Kurane I, Rothman AL, Ennis FA. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J. Infect. Dis. 1999;180:1429–1435. doi: 10.1086/315072. [DOI] [PubMed] [Google Scholar]
- 14.Livingston PG, Kurane I, Dai LC, Okamoto Y, Lai CJ, Men R, Karaki S, Takiguchi M, Ennis FA. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones: recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J. Immunol. 1995;154:1287–1295. [PubMed] [Google Scholar]
- 15.Mathew A, Kurane I, Rothman AL, Zeng LL, Brinton MA, Ennis FA. Dominant recognition by human CD8+ cytotoxic T lymphocytes of dengue virus nonstructural proteins NS3 and NS1.2a. J. Clin. Invest. 1996;98:1684–1691. doi: 10.1172/JCI118964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen HC, Lai SY, Sung JM, Lee SH, Lin YC, Wang WK, Chen YC, Kao CL, King CC, Wu-Hsieh BA. Lymphocyte activation and hepatic cellular infiltration in immunocompetent mice infected by dengue virus. J. Med. Virol. 2004;73:419–431. doi: 10.1002/jmv.20108. [DOI] [PubMed] [Google Scholar]
- 17.Beaumier CM, Mathew A, Bashyam HS, Rothman AL. Crossreactive memory CD8+ T cells alter the immune response to heterologous secondary dengue virus infections in mice in a sequence-specific manner. J. Infect. Dis. 2008;197:608–617. doi: 10.1086/526790. [DOI] [PubMed] [Google Scholar]
- 18.Rothman AL, Kurane I, Ennis FA. Multiple specificities in the murine CD4+ and CD8+ T-cell response to dengue virus. J. Virol. 1996;70:6540–6546. doi: 10.1128/jvi.70.10.6540-6546.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shresta S, Sharar KL, Prigozhin DM, Beatty PR, Harris E. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J. Virol. 2006;80:10208–10217. doi: 10.1128/JVI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin YL, Liao CL, Chen LK, Yeh CT, Liu CI, Ma SH, Huang YY, Huang YL, Kao CL, King CC. Study of dengue virus infection in SCID mice engrafted with human K562 cells. J. Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 2000;74:7814–7823. doi: 10.1128/jvi.74.17.7814-7823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udaka K, Wiesmuller KH, Kienle S, Jung G, Tamamura H, Yamagishi H, Okumura K, Walden P, Suto T, Kawasaki T. An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics. 2000;51:816–828. doi: 10.1007/s002510000217. [DOI] [PubMed] [Google Scholar]
- 23.Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidney J, Southwood S, Oseroff C, del Guercio MF, Sette A, Grey HM. Current Protocols of Immunology. 2001. Measurement of MHC/peptide interactions by gel filtration. Chap 18, Unit 18 13. [DOI] [PubMed] [Google Scholar]
- 26.Vitiello A, Yuan L, Chesnut RW, Sidney J, Southwood S, Farness P, Jackson MR, Peterson PA, Sette A. Immunodominance analysis of CTL responses to influenza PR8 virus reveals two new dominant and subdominant Kb-restricted epitopes. J. Immunol. 1996;157:5555–5562. [PubMed] [Google Scholar]
- 27.Houng HH, Hritz D, Kanesa-thasan N. Quantitative detection of dengue 2 virus using fluorogenic RT-PCR based on 3′-noncoding sequence. J. Virol. Methods. 2000;86:1–11. doi: 10.1016/s0166-0934(99)00166-4. [DOI] [PubMed] [Google Scholar]
- 28.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 29.van den Broek MF, Muller U, Huang S, Aguet M, Zinkernagel RM. Antiviral defense in mice lacking both α/β and γ interferon receptors. J. Virol. 1995;69:4792–4796. doi: 10.1128/jvi.69.8.4792-4796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diamond MS, Roberts TG, Edgil D, Lu B, Ernst J, Harris E. Modulation of dengue virus infection in human cells by α, β, and γ interferons. J. Virol. 2000;74:4957–4966. doi: 10.1128/jvi.74.11.4957-4966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson AJ, Roehrig JT. New mouse model for dengue virus vaccine testing. J. Virol. 1999;73:783–786. doi: 10.1128/jvi.73.1.783-786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotturi MF, Peters B, Buendia-Laysa F, Jr., Sidney J, Oseroff C, Botten J, Grey H, Buchmeier MJ, Sette A. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J. Virol. 2007;81:4928–4940. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol. Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- 35.Welsh RM, Fujinami RS. Pathogenic epitopes, heterologous immunity and vaccine design. Nat. Rev. Microbiol. 2007;5:555–563. doi: 10.1038/nrmicro1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cousens LP, Peterson R, Hsu S, Dorner A, Altman JD, Ahmed R, Biron CA. Two roads diverged: interferon α/β- and interleukin 12-mediated pathways in promoting T cell interferon γ responses during viral infection. J. Exp. Med. 1999;189:1315–1328. doi: 10.1084/jem.189.8.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson LJ, Kolumam GA, Thomas S, Murali-Krishna K. Innate inflammatory signals induced by various pathogens differentially dictate the IFN-I dependence of CD8 T cells for clonal expansion and memory formation. J. Immunol. 2006;177:1746–1754. doi: 10.4049/jimmunol.177.3.1746. [DOI] [PubMed] [Google Scholar]
- 40.Lopez CB, Yount JS, Hermesh T, Moran TM. Sendai virus infection induces efficient adaptive immunity independently of type I interferons. J. Virol. 2006;80:4538–4545. doi: 10.1128/JVI.80.9.4538-4545.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Probst HC, Tschannen K, Gallimore A, Martinic M, Basler M, Dumrese T, Jones E, van den Broek MF. Immunodominance of an antiviral cytotoxic T cell response is shaped by the kinetics of viral protein expression. J. Immunol. 2003;171:5415–5422. doi: 10.4049/jimmunol.171.10.5415. [DOI] [PubMed] [Google Scholar]
- 42.Shrestha B, Diamond MS. Role of CD8+ T cells in control of West Nile virus infection. J. Virol. 2004;78:8312–8321. doi: 10.1128/JVI.78.15.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patey O, Ollivaud L, Breuil J, Lafaix C. Unusual neurologic manifestations occurring during dengue fever infection. Am. J. Trop. Med. Hyg. 1993;48:793–802. doi: 10.4269/ajtmh.1993.48.793. [DOI] [PubMed] [Google Scholar]
- 44.Spaulding AC, Kurane I, Ennis FA, Rothman AL. Analysis of murine CD8+ T-cell clones specific for the dengue virus NS3 protein: flavivirus cross-reactivity and influence of infecting serotype. J. Virol. 1999;73:398–403. doi: 10.1128/jvi.73.1.398-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong W, Reche PA, Lai CC, Reinhold B, Reinherz EL. Genome-wide characterization of a viral cytotoxic T lymphocyte epitope repertoire. J. Biol. Chem. 2003;278:45135–45144. doi: 10.1074/jbc.M307417200. [DOI] [PubMed] [Google Scholar]
- 46.Munks MW, Gold MC, Zajac AL, Doom CM, Morello CS, Spector DH, Hill AB. Genome-wide analysis reveals a highly diverse CD8 T cell response to murine cytomegalovirus. J. Immunol. 2006;176:3760–3766. doi: 10.4049/jimmunol.176.6.3760. [DOI] [PubMed] [Google Scholar]
- 47.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 48.Bashyam HS, Green S, Rothman AL. Dengue virus-reactive CD8+ T cells display quantitative and qualitative differences in their response to variant epitopes of heterologous viral serotypes. J. Immunol. 2006;176:2817–2824. doi: 10.4049/jimmunol.176.5.2817. [DOI] [PubMed] [Google Scholar]
- 49.Simmons CP, Dong T, Chau NV, Dung NT, Chau TN, Thao le TT, Hien TT, Rowland-Jones S, Farrar J. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J. Virol. 2005;79:5665–5675. doi: 10.1128/JVI.79.9.5665-5675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zivny J, DeFronzo M, Jarry W, Jameson J, Cruz J, Ennis FA, Rothman AL. Partial agonist effect influences the CTL response to a heterologous dengue virus serotype. J. Immunol. 1999;163:2754–2760. [PubMed] [Google Scholar]
- 51.Mathew A, Kurane I, Green S, Stephens HA, Vaughn DW, Kalayanarooj S, Suntayakorn S, Chandanayingyong D, Ennis FA, Rothman AL. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J. Virol. 1998;72:3999–4004. doi: 10.1128/jvi.72.5.3999-4004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothman AL. Dengue: defining protective versus pathologic immunity. J. Clin. Invest. 2004;113:946–951. doi: 10.1172/JCI21512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat. Rev. Microbiol. 2007;5:518–528. doi: 10.1038/nrmicro1690. [DOI] [PubMed] [Google Scholar]
- 54.Khan AM, Miotto O, Heiny AT, Salmon J, Srinivasan KN, Nascimento EJ, Marques ET, Jr., Brusic V, Tan TW, August JT. A systematic bioinformatics approach for selection of epitope-based vaccine targets. Cell. Immunol. 2006;244:141–147. doi: 10.1016/j.cellimm.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]