Abstract
The Na+,K+-ATPase transforms the energy of ATP to the maintenance of steep electrochemical gradients for sodium and potassium across the plasma membrane. This activity is tissue specific, in particular due to variations in the expressions of the alpha subunit isoforms one through four. Several mutations in alpha2 and 3 have been identified that link the specific function of the Na+,K+-ATPase to the pathophysiology of neurological diseases such as rapid-onset dystonia parkinsonism and familial hemiplegic migraine type 2. We show a mapping of the isoform differences and the disease-related mutations on the recently determined crystal structure of the pig renal Na+,K+-ATPase and a structural comparison to Ca2+-ATPase. Furthermore, we present new experimental data that address the role of a stretch of three conserved arginines near the C-terminus of the alpha subunit (Arg1003–Arg1005).
Keywords: sodium pump; Na+,K+-ATPase; ATP1A2; ATP1A3; familial hemiplegic migraine; rapid-onset dystonia parkinsonism
1. Introduction
The Na+,K+-ATPase maintains the sodium and potassium gradients across the basolateral plasma membrane in virtually all animal cell types (Jorgensen et al. 2003). These gradients are of fundamental importance for signalling and secondary transport, control of cell volume and osmotic activity, and energization of the resting potential. The Na+,K+-ATPase was one of the earliest membrane protein enzymes discovered (Libet 1948; Skou 1957), and it founded the P-type ATPase family that encompasses not only the sodium, proton and calcium pumps, but also heavy-metal pumps and lipid flippases (Axelsen & Palmgren 2001).
During the catalytic cycle of the Na+,K+-ATPase, three sodium ions are transported out of the cell and two potassium ions are countertransported in. Sodium binding and ATP yield a high-energy intermediate state with phosphorylation of a conserved aspartic acid residue, the so-called E1P state, which is unique to the P-type ATPases. A conformational change of E1P to the E2P state leads to the active extrusion of bound sodium, and upon potassium binding the dephosphorylation of E2P is stimulated. Dissociation of the liberated phosphate along with ATP binding (Jorgensen & Pedersen 2001) stimulates potassium release, changing the enzyme from the E2 state to E1, thus completing the catalytic cycle. The complete functional cycle of the sarcoplasmic reticulum Ca2+-ATPase has been depicted through a range of crystal structures (Toyoshima et al. 2000, 2004; Toyoshima & Nomura 2002; Olesen et al. 2004, 2007; Sorensen et al. 2004; Toyoshima & Mizutani 2004; Jensen et al. 2006), and recently we have reported the first crystal structure of the pig renal Na+,K+-ATPase (Morth et al. 2007) in the form corresponding to the potassium-occluded [K2]E2·Pi state—i.e. a trapped product complex of the dephosphorylation state. The crystal structure revealed a near-complete structure of the alpha subunit and of the transmembrane regions of the beta and gamma subunits.
The minimal functional Na+,K+-ATPase consists of two subunits: the alpha subunit that performs ATP hydrolysis and cation binding, and the beta subunit that is important for stability and trafficking of the sodium pump. Four different isoforms of the alpha subunit have been found in humans (Lingrel et al. 2007). Alpha1 (ATP1A1 gene) is expressed in all tissues; it is the exclusive isoform in the outer medulla of the kidney and the predominant isoform in most other tissue types. Isoform 2 (ATP1A2 gene) is the predominant isoform in skeletal muscle, but is also found in the brain (astrocytes), heart, eyes and adipose tissue. The alpha3 isoform (ATP1A3 gene) is primarily found in the brain (neurons), and isoform 4 (ATP1A4 gene) is only expressed in testis (Woo et al. 2000). Three different isoforms of the beta subunit have been found with moderate differential effects on Na+,K+-ATPase activity (Geering 2001). Furthermore, the Na+,K+-ATPase is often associated with a tissue-specific regulatory subunit of the FXYD protein family (reviewed in Geering (2005) and Garty & Karlish (2006)).
Two rare human diseases are associated with mutations in the ATP1A2 and ATP1A3 genes, both autosomal dominant disorders with variable penetrance. Alpha2 haploinsufficiency can cause familial hemiplegic migraine type 2 (FHM2) and is implicated in sporadic hemiplegic migraine (SHM); both are migraines with aura where patients can experience hemiparesis, ataxia, coma or epileptic seizures (De Fusco et al. 2003; Estevez & Gardner 2004; de Vries et al. 2007). Alpha3 haploinsufficiency is associated with rapid-onset dystonia parkinsonism (RDP), which is characterized by rapid onset of involuntary movements and bulbar symptoms where dopaminergic medication has no effect (de Carvalho Aguiar et al. 2004).
In this paper, we present a further analysis of the Na+,K+-ATPase structure with particular focus on the comparison to the Ca2+-ATPase and the mapping of alpha subunit isoform differences and disease-related mutations, and we assess a novel functional role of a group of the FHM2/SHM mutations. Furthermore, we show that the proposed voltage sensing stretch of arginine residues near the C-terminus of the alpha subunit is unlikely to function independently within the pump.
2. Material and methods
(a) Generation of cRNAs encoding the Na+,K+-ATPase subunits
Plasmids encoding human alpha1, 2 and 3 and beta1 of the Na+,K+-ATPase were purchased from Origene (www.origene.com). Mutations Q118R and N129D (human alpha1 numbering) were introduced into all of the alpha isoforms to reduce their ouabain sensitivity (Price & Lingrel 1988). The sequences encoding Na+,K+-ATPase subunits were subcloned into the pXOON vector (Jespersen et al. 2002) using NotI (alpha1) or EcoR1/NotI (alpha2 and alpha3). Mutations 1010RRR-AAA in alpha1, 1009Y-N in alpha2 and 1002N-Y in alpha3 were introduced by PCR. All constructs were verified by sequencing. From NheI digested plasmids, cRNAs were transcribed with the mMESSAGE ULTRA kit (Ambion).
(b) Protein expression and electrophysiological measurements in Xenopus oocytes
Oocytes from Xenopus laevis were isolated and defollicated. Stage V–VI oocytes were coinjected with 1 ng of beta1 and 10 ng of one of the six alpha subunit cRNAs. After 1–3 days at 19°C, oocytes were loaded with sodium by incubation for at least 2 h in a potassium free solution with 110 mM NaCl, 2.5 mM Na citrate, 1 mM CaCl2, 10 mM Tris/MOPS, pH 7.4 as well as 10 μM ouabain to inhibit the endogenous oocyte Na+,K+-ATPase (Horisberger et al. 2004). Electrophysiological measurements were performed using the two-electrode voltage-clamp technique with an OC-725C voltage-clamp apparatus (Warner Instrument Corp.) in a buffer with 100 mM NaCl, 0.82 MgCl2, 0.41 CaCl2, 10 mM TEACl, 5 mM BaCl2, 5 mM Tris/MOPS, pH 7.4. Current–voltage curves were determined from the currents activated by replacing 10 mM NaCl with 10 mM KCl and running a series of 200 ms voltage steps every 20 mV between −130 and 30 mV. Data were recorded and analysed using pClamp 9.2 (Axon Instruments).
(c) Enzymatic characterization of wild-type and mutant expressed in COS cells
Introduction of the RRR–AAA triple mutation in the rat alpha1 isoform of Na+,K+-ATPase was carried out by PCR followed by expression in COS cells, and the previously described assays for phosphorylation and ATPase activity were used for the functional characterization (Toustrup-Jensen & Vilsen 2002; Rodacker et al. 2006). The Na+ dependence of phosphorylation (figure 3f(i)) was determined in the presence of 2 μM [γ-32P]ATP in the absence of K+. The K+ dependence of Na+,K+-ATPase activity (figure 3f(ii)) was determined in the presence of 40 mM Na+ and 3 mM ATP. The ATP and vanadate dependencies of the Na+,K+-ATPase activity (figure 3f(iii)(iv)) were determined in the presence of 130 mM Na+ and 20 mM K+.
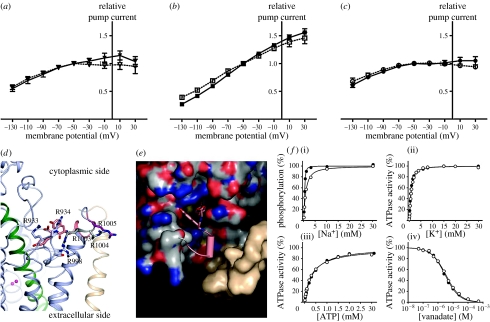
Figure 3.
A C-terminal arginine patch in Na+,K+-ATPase is not critical for function. (a–c) Oocytes were injected with cRNAs encoding human beta1 plus wild-type (closed symbols) or mutant (open symbols) alpha. (a) Solid line with filled triangle, alpha3; dashed line with open triangle, alpha3 N-Y; (b) Solid line with filled square, alpha2; dashed line with open square, alpha2 Y-N; (c) Solid line with filled circle, alpha1; dashed line with open circle, alpha1 RRR–AAA. The K+-activated currents determined at different membrane potentials were normalized to the value of the current at −50 mV. Data are the mean of 5–9 oocytes; error bars represent the standard deviations. The average current at −50 mV was 84 nA, standard deviation 62 nA. (d) Side view of the arginine patch presumably located at the cytosol–membrane interface. The arginine coloured purple (R1005, pig alpha1 numbering) is the only position that is not conserved between the three isoforms, being a tyrosine in alpha2 and an asparagine in alpha3. (e) Top view of the c-terminal binding pocket. The C-terminal is represented as a cartoon except for the two terminal tyrosine residues given as sticks. The binding pocket is represented as spacefilled: oxygens (red), nitrogens (blue), sulphurs (yellow) and carbons (grey); the beta subunit is shown in beige. (f(i)–(iv)) Functional analysis of the triple mutant in rat alpha1 (open symbols) compared to wild-type rat alpha1 (closed symbols) with the apparent affinities for Na+,K+, ATP and vanadate given as K0.5 values. (i) Filled circles, K0.5=0.430 mM; open circles, K0.5=0.912 mM; (ii) filled circles, K0.5=0.669 mM; open circles, K0.5=0.843 mM; (iii) filled circles, K0.5=0.350 mM; open circles, K0.5=0.312 mM; (iv) filled circles, K0.5=2.98 μM; open circles, K0.5=3.35 μM.
3. Structural comparison of the Na+,K+- and Ca2+-ATPases
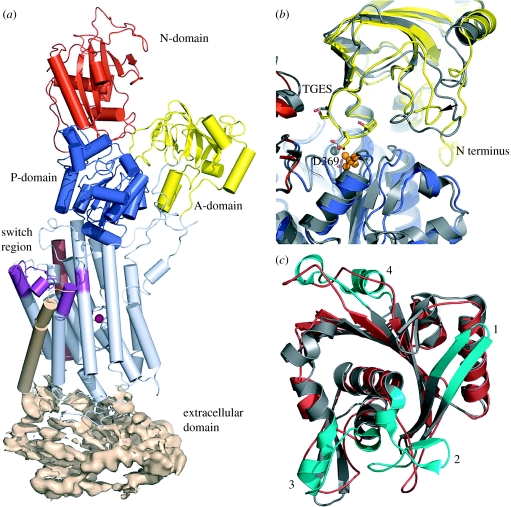
We have compared the Na+,K+-ATPase structure to the structure of the equivalent form of the Ca2+-ATPase in the absence of calcium and in the presence of either aluminium fluoride (Olesen et al. 2004) or magnesium fluoride (Toyoshima et al. 2004). The comparison reveals that the overall arrangements of domains and transmembrane helices are nearly identical (figure 1a), except for a slight shift of the nucleotide binding domain (N-domain). This finding supports the suggested mechanism of the Ca2+-ATPase that dephosphorylation is coupled to occlusion of the countertransported protons (Olesen et al. 2004) since rubidium/potassium in this structural state of Na+,K+-ATPase is occluded (Morth et al. 2007). The conserved residues of the cytoplasmic domains forming the phosphorylation site, the ATP binding pocket and the Mg2+ coordination, as well as residues assisting in catalysis of phosphorylation and dephosphorylation, are found in nearly the same positions (with Na+,K+-ATPase (pig)/Ca2+-ATPase (rabbit) numbering); e.g. Asp 369/351, Thr 371/353, Thr 610/625, Gly 611/626, Lys 691/684, Asp 710/703 and Asn 713/706 in the phosphorylation domain (P-domain), Phe 475/487 and Arg 544/560 in the N-domain, and Glu 214/183 in the TGES motif of the actuator domain (A-domain). The architectures of the A- and P-domains are similar in the two pumps, and the magnesium fluoride bound at the phosphorylation site is coordinated by residues from both domains. As in the and structures of the Ca2+-ATPase (Olesen et al. 2004; Toyoshima et al. 2004), the invariant TGES loop of the A-domain (figure 1b, residues 212–215 in the Na+,K+-ATPase) is inserted into the catalytic site, thus reflecting its role in the catalysis of E2P dephosphorylation.
Figure 1.
Structural comparison of Na+,K+-ATPase with Ca2+-ATPase. (a) A domain overview of Na+,K+ ATPase. The N-domain is in red, P-domain in blue, A-domain in yellow. The transmembrane region is coloured light blue, the β subunit light brown and the C-terminal switch region purple. (b) Na+,K+-ATPase (blue, PDB 3B8E, Rb2E2 : complex) and Ca2+-ATPase (yellow, PDB 1WPG, H2-3E2 : Thapsigargin : complex) were structurally aligned on the basis of the A- and P-domains. Comparison of the interactions at the interface between the A-domain (upper part) and P-domain (lower part). The invariant TGES loop is represented with sticks and labelled. In both pumps, it comes into close contact with the phosphate analogue (orange) and the Mg2+ ion (grey) in the catalytic site. The loop region corresponding to residues 225–235 in Na+,K+-ATPase is shorter than the corresponding region of Ca2+-ATPase indicated by the arrow. Also shown is the traceable part of the N-terminus of Na+,K+-ATPase, starting with a short helical fragment at residue 21. (c) N-domain insertions unique to the Ca2+-ATPase are coloured cyan, the most prominent differences are indicated with numbers.
A notable difference between the A-domains of the Na+,K+-ATPase and the Ca2+-ATPase is in another loop critical for E2P dephosphorylation in the Ca2+-ATPase (residues 189–202; Kato et al. 2003), which is a few residues shorter in the Na+,K+-ATPase (residues 225–235; figure 1a). In the Ca2+-ATPase, Arg 198 (exposed to tryptic cleavage, ‘T2 site’) and Val 200 probably mediate a contact to the P-domain at Glu 660, thereby stabilizing the A-domain in the correct position for dephosphorylation (Toyoshima et al. 2004), but these residues are missing in the Na+,K+-ATPase, which could in part explain its very slow E2P dephosphorylation kinetics in the absence of potassium (Post et al. 1972; Glynn 1993; Einholm et al. 2005).
The N-domain is rather loosely associated with the rest of the molecule and is rotated approximately 30° away from the A-domain relative to the position in the structure of the Ca2+-ATPase. There is no nucleotide present in the crystallization buffer to lock the domain, and the relative position between the N-domain and the A- and P-domains is inherently flexible and may be influenced by crystal contacts. The Na+,K+-ATPase N-domain is smaller than that of the Ca2+-ATPase, with the major differences in exposed loop regions (see figure 1c, 1). Thus, a β-hairpin at the start of the N-domain is approximately 10 residues shorter in the Na+,K+-ATPase: the loop starting at Glu 454 lacks a small α-helical region compared with the Ca2+-ATPase (figure 1c, 2), and the loop starting at Phe 552 does not have the two short helices seen in the Ca2+-ATPase. These insertions in the Ca2+-ATPase are all located on the surface of the N-domain at some distance from the nucleotide binding site.
Superimposition of the previously determined structures of isolated N-domains from human alpha2 (Hakansson 2003) and rat alpha1 (Hilge et al. 2003) shows a general agreement except at a loop region near the ATP binding site (Thr 396–Ser 403), which could not be modelled in the human alpha2 crystal structure (PDB ID 1Q3I) (Hakansson 2003) and which appears significantly different in the rat alpha1 NMR structure (PDB ID 1MO8, Hilge et al. 2003).
4. Mapping of alpha subunit isoform differences
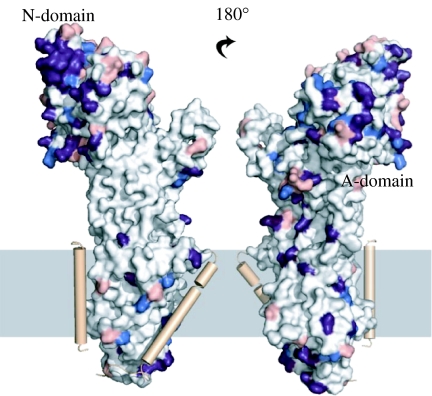
The differences in expression profiles of the Na+,K+-ATPase alpha subunits together with specific characteristics in affinities for sodium, potassium and cardiotonic steroids, and in voltage sensitivity, indicate varied physiological roles of the different isoforms (Pressley et al. 2005). To visualize the differences, the three major human isoforms alpha1–3 were modelled with MODELLER (Sali & Blundell 1993). Figure 2 shows that the isoforms mapped on to the model of human alpha1 with differences between alpha1 and 2 in blue, differences between alpha1 and 3 in light red and differences between alpha1 and both 2 and 3 in purple.
Figure 2.
Isoform differences mapped on the alpha1 surface. The alpha subunit is represented as a molecular surface. The beta and gamma subunits are represented as single helices in brown. The white regions are those in which alpha1 is identical to alpha2 and 3. Red marks differences between alpha1 and 2. Blue marks differences between alpha1 and 3. Purple marks regions where both alpha2 and 3 differ from alpha1. The models were built with MODELLER (Sali & Blundell 1993) using the human alpha sequences on the pig alpha1.
From figure 2, it is evident that there are very few isoform differences in the transmembrane region and in the regions interacting with the transmembrane segments of the beta and gamma chains, suggesting that these areas are conserved and functionally important for the basic transport mechanism and stability. By contrast, large clusters of isoform differences map at surface-exposed regions of the A- and N-domains, possibly reflecting isoform specific interactions with other cellular components. A recent example of an isoform specific interaction is in trafficking, where adaptor protein 1 was shown to recruit alpha1 to the membrane via Tyr 255 (Efendiev et al. 2008), a residue not conserved in alpha2 and alpha3.
5. A voltage modulator of the alpha subunit?
The electrogenic character of the Na+,K+-ATPase and the tremendous importance it has in the energization of the plasma membrane is consistent with the observation that the membrane potential influences the activity of the sodium pump. Previous studies have shown that the alpha subunit isoforms display large differences in their voltage sensitivities (Crambert et al. 2000), and a cluster of five arginines near the C-terminal part of alpha1 (figure 3d), presumably located at the membrane–cytosol interface, was proposed to act as a voltage sensitive modulatory patch (Morth et al. 2007). Arginines at the membrane interface have previously been demonstrated to form a voltage sensing paddle in voltage-gated potassium channels (Jiang et al. 2003).
To test if the arginine cluster is a site of voltage-dependent modulation, we first examined whether the most C-terminal of the five arginines (R1005) could be responsible for the different voltage sensitivities seen for the alpha subunit isoforms. R1005 is a variable and is being exchanged for a tyrosine in alpha2 and an asparagine in alpha3, while the four other arginines are conserved between alpha1, 2 and 3 (figure 3d). It was previously reported that alpha2 shows the steepest voltage dependence, whereas alpha3 is rather insensitive to voltage (Crambert et al. 2000; Horisberger & Kharoubi-Hess 2002), so we examined whether interchanging the asparagine and tyrosine residues (alpha2 Y-N and alpha3 N-Y, figure 3a,b) would alter their voltage profiles. We confirmed that the wild-type alpha2 form shows much steeper voltage dependence than alpha3, but mutation of the variable position in the arginine patch did not change the current–voltage curves. Therefore, we also tested a radical mutation where the three C-terminal arginines in the patch were simultaneously replaced by alanines in alpha1, but again this did not influence the voltage profile of the pump (figure 3c).
Consistent with these findings, functional analysis of the rat alpha1 RRR-to-AAA mutant expressed in a mammalian COS cell line demonstrated that the ATP- and vanadate dependencies were unaffected by the arginine cluster mutation, indicating that it does not alter the E1–E2 conformational equilibrium. Moreover, the mutation was without influence on the potassium affinity and only reduced the sodium affinity approximately twofold (figure 3f).
These results imply that the third arginine position in the patch cannot account for the differences between the voltage profiles of the isoforms, and that voltage regulation of the Na+,K+-ATPase involves residues different from or additional to the arginine cluster.
6. Mapping disease-related mutations on the Na+,K+-ATPase structure
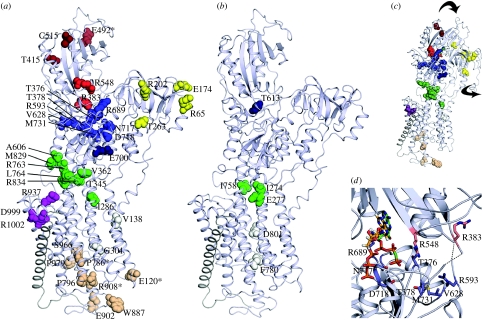
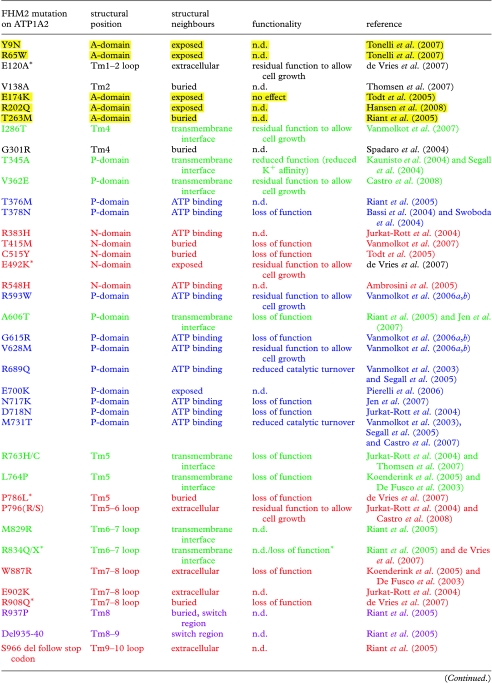
With the recent discoveries that mutations in the ATP1A2 gene contribute to FHM2 and SHM, and mutations in the ATP1A3 gene lead to RDP, the positioning of these mutations represents excellent leads in the analysis of structure and function of the Na+,K+-ATPase. The FHM2 and SHM mutations were plotted on the human alpha2 homology model (figure 4a; model coordinates available at http://www.pumpkin.au.dk), and the RDP mutations were plotted on the human alpha3 homology model (figure 4b). We find the mutations to cluster in six regions that are summarized in table 1 with colour coding referring to figure 4.
Figure 4.
Mutations causing FHM2 and RDP plotted on alpha2 and alpha3. Cartoon representations of the alpha2 and alpha3 isoforms modelled as described in figure 2. (a) The alpha2 isoform with mutations causing FHM2 plotted in sphere representation. The colour coding is region specific: mutations in the N-domain are red (near ATP binding light red), in the A-domain yellow, in the P-domain blue (near ATP binding light blue), at the interface between the transmembrane region and the P-domain green, at the C-terminus magenta, in the transmembrane region white and extracellular mutations are coloured beige. Mutations specific to SHM are marked with asterisks. (b) The alpha3 isoform with RDP causing mutations coloured as above. (c) A model of the alpha2 isoform docked onto the E1 state of the Ca2+-ATPase (PDB 1T5S, Sorensen et al. 2004) with the major structural rearrangements indicated by arrows. (d) A close view of the N- and P-domain interface in the E1·AMPCP state. The mutations causing FHM2 are shown in stick representation. The AMPPCP described in the Ca2+-ATPase is coloured green, and the ATP described in the rat alpha1 NMR structure (Hilge et al. 2003) is coloured yellow.
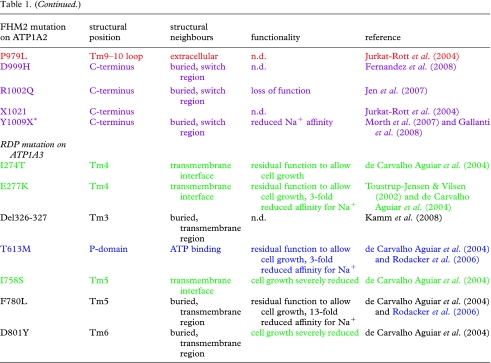
Table 1.
Compilation of mutations implicated in FHM2, SHM and RDP. (Mutations found in FHM2, SHM and RDP are summarized; the colour coding is consistent with that used in the structural representation (figure 4a,b). The structural position points to the structural domain or secondary structure element; the structural neighbours describe possible interaction partners. Mutations marked with asterisks belong to the SHM group.)
 |
 |
Mutations in the N-domain are indicated in red. Three of the five mutations have been characterized, the T415M and C515Y mutants show loss of function (Todt et al. 2005) and E492K shows partial function (de Vries et al. 2007). These effects are likely due to impaired nucleotide binding.
Five mutations are located in the A-domain (yellow). Only the E174K mutation has been characterized (Todt et al. 2005), but showed no difference from the wild-type alpha2, suggesting a more subtle effect on protein function than revealed by the Xenopus oocyte activity assay used.
The mutations in the P-domain cluster into two distinct fractions, one coloured in light blue close to the ATP binding site, and the other coloured green at the interface of the P-domain and the transmembrane region. Only FHM2/SHM mutations have been identified in the A- and N-domains, but in the P-domain, four RDP mutations are also seen. Most mutations in the ATP binding region cause loss or reduction of function, as expected if ATP binding is compromised. An alpha1 equivalent of the alpha3 mutant T613M has been characterized biochemically, showing a displacement of the E1–E2 conformational equilibrium in favour of E2 (Rodacker et al. 2006). Similar investigations of the RDP E277K mutation at the interface between the P-domain and the transmembrane region showed significantly reduced Na+ affinity (Toustrup-Jensen & Vilsen 2002), and generally, the investigated mutations at the interface show reduced or even lost function, likely because the coupling between the phosphorylation site and the transmembrane domain is impaired.
Within the transmembrane region, two FHM2 and two RDP mutations are found (white). Like the P-domain RDP mutants, an alpha1 equivalent of the alpha3 F780L showed reduced Na+ affinity (Rodacker et al. 2006), and the mutation reduced the ability to support cell growth (de Carvalho Aguiar et al. 2004). The RDP mutant D801Y was unable to support cell growth (de Carvalho Aguiar et al. 2004), and D801 is an essential part of the ion-binding pocket, probably donating two oxygen ligands to K+ binding (Morth et al. 2007). Thus, an obvious speculation is that the transmembrane mutations interfere with ion transport.
Eight FHM2/SHM mutations cluster close to the extracellular side (orange). The two characterized mutants (P796S and W887R) have reduced or lost function (Jurkat-Rott et al. 2004; De Fusco et al. 2003). These mutations are likely to interfere with the extracellular entry/exit pathway of the transported ions.
Finally, a cluster of mutations was found around the C-terminus (purple) that has been recently discovered to be a potential switch region in the Na+,K+-ATPase (Morth et al. 2007). The only characterized mutation (R1002Q) abolished the ability of the Na+,K+-ATPase to support growth of transfected HeLa cells (Jen et al. 2007).
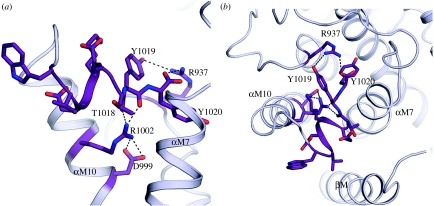
Deletion in the C-terminus was shown to have profound effects on sodium affinity (Morth et al. 2007). We also find that a C-terminal deletion alters the voltage dependence substantially (H. Poulsen, J. P. Morth & P. Nissen 2008, unpublished results). Naturally, it would cause large perturbations to extend (X1021R) or delete (Y1009X) the C-terminus, and from the structure, it is apparent that the other four identified FHM2 mutations in this region may also cause destabilization of the C-terminus (figure 5). Indeed, while writing this manuscript, a study of the X1021R mutation was published and the authors find a strong voltage dependency of the Na+,K+-ATPase associated with this particular FHM2 mutation (Tavraz et al. 2008). The loop region between transmembrane helices M8 and M9 would be affected by either deleting residues 935–940 or by changing arginine 937 to a proline (figure 5b). Arg937 probably adopts another mode of interaction, but whether it is loss of the arginine or a disruption of the structural motif is difficult to deduce from the structure. A proline mutation would cause the loop region between transmembrane helices M8 and M9 to become more rigid (figure 5b). Arg1002 points towards the end of the helical motif at Thr1018, an interaction likely to break the α-helix, and Asp999, which is positioned right below Arg1002, could be part of a binding motif that allows the functionally important C-terminal tyrosines to adopt the desired conformation for binding in the pocket (figure 5). We therefore propose that this group of mutations describe a novel functionally important motif of the sodium pump, and we will determine their effects on the alpha2 voltage profile in future studies.
Figure 5.
Structure model of C-terminal FHM2 mutations. The C-terminal residues W1013–Y1020 and three FHM2 causing mutations in alpha2 are shown as sticks. The structural motif formed by R1002, D999 and the carbonyls of T1018 and Y1018 are shown as dashed lines below the helical fragment formed by residues W1013–T1014. The possible interaction between R937 and Y1019 and the terminal carboxyl group of Y1020 is also shown with dashed lines. Side view (a), and top view, facing the cytoplasmic side (b), of the C-terminal region.
7. Discussion and conclusion
Hopefully, future biochemical investigations and further structural characterization of the Na+,K+-ATPase, in particular of the sodium-bound form, will allow us to distinguish between subtle and determining factors on the function of this key enzyme of the cell. Mutational screening, structural analysis and perhaps also advanced interaction network models of Na+,K+-ATPase in the tissue-specific context of channels, transporters and pumps for ionic flow and steady state will be important sources of new, testable hypotheses on the pathophysiology of neurological diseases linked to Na+,K+-ATPase mutations.
Comparing figures 2 and 4, it is interesting to note that there is a general lack of overlap between isoform differences and disease-causing mutations, underscoring that FHM2 and RDP are generally due to severe impairment of pump function, i.e. these mutations hit vital residues, while the variations between the isoforms are probably selected to fine-tune the basic machinery for different specific functions, which are likely to be determined by interplay with other cellular factors.
Very significantly, we find further support for the importance of the C-terminus by a most recent study indicating that a deletion of the terminal 11 residues of the alpha2 isoform is linked to SHM (Gallanti et al. 2008). Thus as predicted (Morth et al. 2007), the C-terminus may represent a sensitive determinant of Na+,K+-ATPase activity, and it will be of key importance to reveal the underlying mechanisms of this element in regulation of sodium levels in the cell, and how it may be targeted by pharmacological intervention.
Footnotes
One contribution of 16 to a Discussion Meeting Issue ‘Membrane transport in flux: the ambiguous interface between channels and pumps’.
References
- Ambrosini A., et al. Familial basilar migraine associated with a new mutation in the ATP1A2 gene. Neurology. 2005;65:1826–1828. doi: 10.1212/01.wnl.0000187072.71931.c0. doi:10.1212/01.wnl.0000187072.71931.c0 [DOI] [PubMed] [Google Scholar]
- Axelsen K.B., Palmgren M.G. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 2001;126:696–706. doi: 10.1104/pp.126.2.696. doi:10.1104/pp.126.2.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi M.T., et al. A novel mutation in the ATP1A2 gene causes alternating hemiplegia of childhood. J. Med. Genet. 2004;41:621–628. doi: 10.1136/jmg.2003.017863. doi:10.1136/jmg.2003.017863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro M.J., et al. Recurrent ATP1A2 mutations in Portuguese families with familial hemiplegic migraine. J. Hum. Genet. 2007;52:990–998. doi: 10.1007/s10038-007-0205-7. doi:10.1007/s10038-007-0205-7 [DOI] [PubMed] [Google Scholar]
- Castro M.J., et al. Two novel functional mutations in the Na+,K+-ATPase α2-subunit ATP1A2 gene in patients with familial hemiplegic migraine and associated neurological phenotypes. Clin. Genet. 2008;73:37–43. doi: 10.1111/j.1399-0004.2007.00918.x. doi:10.1111/j.1399-0004.2007.00918.x [DOI] [PubMed] [Google Scholar]
- Crambert G., Hasler U., Beggah A.T., Yu C., Modyanov N.N., Horisberger J.D., Lelievre L., Geering K. Transport and pharmacological properties of nine different human Na,K-ATPase isozymes. J. Biol. Chem. 2000;275:1976–1986. doi: 10.1074/jbc.275.3.1976. doi:10.1074/jbc.275.3.1976 [DOI] [PubMed] [Google Scholar]
- de Carvalho Aguiar P., et al. Mutations in the Na+/K+-ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron. 2004;43:169–175. doi: 10.1016/j.neuron.2004.06.028. doi:10.1016/j.neuron.2004.06.028 [DOI] [PubMed] [Google Scholar]
- De Fusco M., Marconi R., Silvestri L., Atorino L., Rampoldi L., Morgante L., Ballabio A., Aridon P., Casari G. Haploinsufficiency of ATP1A2 encoding the Na+/K+ pump alpha2 subunit associated with familial hemiplegic migraine type 2. Nat. Genet. 2003;33:192–196. doi: 10.1038/ng1081. doi:10.1038/ng1081 [DOI] [PubMed] [Google Scholar]
- de Vries B., et al. Systematic analysis of three FHM genes in 39 sporadic patients with hemiplegic migraine. Neurology. 2007;69:2170–2176. doi: 10.1212/01.wnl.0000295670.01629.5a. doi:10.1212/01.wnl.0000295670.01629.5a [DOI] [PubMed] [Google Scholar]
- Efendiev R., Budu C.E., Bertorello A.M., Pedemonte C.H. G-protein-coupled receptor-mediated traffic of Na,K-ATPase to the plasma membrane requires the binding of adaptor protein1 to a Tyr-255 based sequence in the α-subunit. J. Biol. Chem. 2008;283:17 561–17 567. doi: 10.1074/jbc.M709260200. doi:10.1074/jbc.m709260200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einholm A.P., Toustrup-Jensen M., Andersen J.P., Vilsen B. Mutation of Gly-94 in transmembrane segment M1 of Na+,K+-ATPase interferes with Na+ and K+ binding in E2P conformation. Proc. Natl Acad. Sci. USA. 2005;102:11 254–11 259. doi: 10.1073/pnas.0501201102. doi:10.1073/pnas.0501201102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M., Gardner K.L. Update on the genetics of migraine. Hum. Genet. 2004;114:225–235. doi: 10.1007/s00439-003-1055-9. doi:10.1007/s00439-003-1055-9 [DOI] [PubMed] [Google Scholar]
- Fernandez D.M., Hand C.K., Sweeney B.J., Parfrey N.A. A novel ATP1A2 gene mutation in an Irish familial hemiplegic migraine kindred. Headache. 2008;48:101–108. doi: 10.1111/j.1526-4610.2007.00848.x. doi:10.1111/j.1526-4610.2008.00856.x [DOI] [PubMed] [Google Scholar]
- Gallanti A., Tonelli A., Cardin V., Bussone G., Bresolin N., Bassi M.T. A novel de novo nonsense mutation in ATP1A2 associated with sporadic hemiplegic migraine and epileptic seizures. J. Neurol. Sci. 2008;273:123–126. doi: 10.1016/j.jns.2008.06.006. doi:10.1016/j.jns.2008.06.006 [DOI] [PubMed] [Google Scholar]
- Garty H., Karlish S.J. Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 2006;68:431–459. doi: 10.1146/annurev.physiol.68.040104.131852. doi:10.1146/annurev.physiol.68.040104.131852 [DOI] [PubMed] [Google Scholar]
- Geering K. The functional role of β subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 2001;33:425–438. doi: 10.1023/a:1010623724749. doi:10.1023/A:1010623724749 [DOI] [PubMed] [Google Scholar]
- Geering K. Function of FXYD proteins, regulators of Na,K-ATPase. J. Bioenerg. Biomembr. 2005;37:387–392. doi: 10.1007/s10863-005-9476-x. doi:10.1007/s10863-005-9476-x [DOI] [PubMed] [Google Scholar]
- Glynn I.M. Annual review prize lecture. ‘All hands to the sodium pump’. J. Physiol. 1993;462:1–30. doi: 10.1113/jphysiol.1993.sp019540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson K.O. The crystallographic structure of Na,K-ATPase N-domain at 2.6 Å resolution. J. Mol. Biol. 2003;332:1175–1182. doi: 10.1016/j.jmb.2003.07.012. doi:10.1016/j.jmb.2003.07.012 [DOI] [PubMed] [Google Scholar]
- Hansen J.M., Thomsen L.L., Marconi R., Casari G., Olesen J., Ashina M. Familial hemiplegic migraine type 2 does not share hypersensitivity to nitric oxide with common types of migraine. Cephalalgia: Int. J. Headache. 2008;28:367–375. doi: 10.1111/j.1468-2982.2008.01542.x. doi:10.1111/j.1468-2982.2008.01542.x [DOI] [PubMed] [Google Scholar]
- Hilge M., Siegal G., Vuister G.W., Guntert P., Gloor S.M., Abrahams J.P. ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase. Nat. Struct. Biol. 2003;10:468–474. doi: 10.1038/nsb924. doi:10.1038/nsb924 [DOI] [PubMed] [Google Scholar]
- Horisberger J.D., Kharoubi-Hess S. Functional differences between α subunit isoforms of the rat Na,K-ATPase expressed in Xenopus oocytes. J. Physiol. 2002;539(Pt 3):669–680. doi: 10.1113/jphysiol.2001.013201. doi:10.1113/jphysiol.2001.013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horisberger J.D., Kharoubi-Hess S., Guennoun S., Michielin O. The fourth transmembrane segment of the Na,K-ATPase α subunit: a systematic mutagenesis study. J. Biol. Chem. 2004;279:29 542–29 550. doi: 10.1074/jbc.M400585200. doi:10.1074/jbc.M400585200 [DOI] [PubMed] [Google Scholar]
- Jen J.C., Klein A., Boltshauser E., Cartwright M.S., Roach E.S., Mamsa H., Baloh R.W. Prolonged hemiplegic episodes in children due to mutations in ATP1A2. J. Neurol. Neurosurg. Psychiatry. 2007;78:523–526. doi: 10.1136/jnnp.2006.103267. doi:10.1136/jnnp.2006.103267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen A.M., Sorensen T.L., Olesen C., Moller J.V., Nissen P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 2006;25:2305–2314. doi: 10.1038/sj.emboj.7601135. doi:10.1038/sj.emboj.7601135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen T., Grunnet M., Angelo K., Klaerke D.A., Olesen S.P. Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. Biotechniques. 2002;32:536–540. doi: 10.2144/02323st05. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Ruta V., Chen J., Lee A., MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. doi:10.1038/nature01581 [DOI] [PubMed] [Google Scholar]
- Jorgensen P.L., Pedersen P.A. Structure–function relationships of Na+,K+, ATP, or Mg2+ binding and energy transduction in Na,K-ATPase. Biochim. Biophys. Acta. 2001;1505:57–74. doi: 10.1016/s0005-2728(00)00277-2. doi:10.1016/S0005-2728(00)00277-2 [DOI] [PubMed] [Google Scholar]
- Jorgensen P.L., Hakansson K.O., Karlish S.J.D. Structure and mechanism of Na,K-ATPase: functional sites and their interactions. Annu. Rev. Physiol. 2003;65:817–849. doi: 10.1146/annurev.physiol.65.092101.142558. doi:10.1146/annurev.physiol.65.092101.142558 [DOI] [PubMed] [Google Scholar]
- Jurkat-Rott K., et al. Variability of familial hemiplegic migraine with novel A1A2 Na+/K+-ATPase variants. Neurology. 2004;62:1857–1861. doi: 10.1212/01.wnl.0000127310.11526.fd. [DOI] [PubMed] [Google Scholar]
- Kamm C., Fogel W., Wachter T., Schweitzer K., Berg D., Kruger R., Freudenstein D., Gasser T. Novel ATP1A3 mutation in a sporadic RDP patient with minimal benefit from deep brain stimulation. Neurology. 2008;70(Pt 2):1501–1503. doi: 10.1212/01.wnl.0000310431.41036.e0. doi:10.1212/01.wnl.0000310431.41036.e0 [DOI] [PubMed] [Google Scholar]
- Kato S., Kamidochi M., Daiho T., Yamasaki K., Gouli W., Suzuki H. Val200 residue in Lys189–Lys205 outermost loop on the A domain of sarcoplasmic reticulum Ca2+-ATPase is critical for rapid processing of phosphoenzyme intermediate after loss of ADP sensitivity. J. Biol. Chem. 2003;278:9624–9629. doi: 10.1074/jbc.M208861200. doi:10.1074/jbc.M208861200 [DOI] [PubMed] [Google Scholar]
- Kaunisto M.A., et al. A novel missense ATP1A2 mutation in a Finnish family with familial hemiplegic migraine type 2. Neurogenetics. 2004;5:141–146. doi: 10.1007/s10048-004-0178-z. doi:10.1007/s10048-004-0178-z [DOI] [PubMed] [Google Scholar]
- Koenderink J.B., Zifarelli G., Qiu L.Y., Schwarz W., De Pont J.J., Bamberg E., Friedrich T. Na,K-ATPase mutations in familial hemiplegic migraine lead to functional inactivation. Biochim. Biophys. Acta. 2005;1669:61–68. doi: 10.1016/j.bbamem.2005.01.003. doi:10.1016/j.bbamem.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Libet B. Adenosinetriphosphatase (ATP-ASE) in nerve. Fed. Proc. 1948;7:72. [PubMed] [Google Scholar]
- Lingrel J.B., Williams M.T., Vorhees C.V., Moseley A.E. Na,K-ATPase and the role of alpha isoforms in behavior. J. Bioenerg. Biomembr. 2007;39:385–389. doi: 10.1007/s10863-007-9107-9. doi:10.1007/s10863-007-9107-9 [DOI] [PubMed] [Google Scholar]
- Morth J.P., Pedersen B.P., Toustrup-Jensen M.S., Sorensen T.L., Petersen J., Andersen J.P., Vilsen B., Nissen P. Crystal structure of the sodium–potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. doi:10.1038/nature06419 [DOI] [PubMed] [Google Scholar]
- Olesen C., Sorensen T.L., Nielsen R.C., Moller J.V., Nissen P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science (NY) 2004;306:2251–2255. doi: 10.1126/science.1106289. doi:10.1126/science.1106289 [DOI] [PubMed] [Google Scholar]
- Olesen C., Picard M., Winther A.M., Gyrup C., Morth J.P., Oxvig C., Moller J.V., Nissen P. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–1042. doi: 10.1038/nature06418. doi:10.1038/nature06418 [DOI] [PubMed] [Google Scholar]
- Pierelli F., et al. A novel ATP1A2 mutation in a family with FHM type II. Cephalalgia: Int. J. Headache. 2006;26:324–328. doi: 10.1111/j.1468-2982.2006.01002.x. doi:10.1111/j.1468-2982.2006.01002.x [DOI] [PubMed] [Google Scholar]
- Post R.L., Hegyvary C., Kume S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 1972;247:6530–6540. [PubMed] [Google Scholar]
- Pressley T.A., Duran M.J., Pierre S.V. Regions conferring isoform-specific function in the catalytic subunit of the Na,K-pump. Front. Biosci. J. Virtual Library. 2005;10:2018–2026. doi: 10.2741/1677. doi:10.2741/1677 [DOI] [PubMed] [Google Scholar]
- Price E.M., Lingrel J.B. Structure–function relationships in the sodium–potassium ATPase alpha subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry. 1988;27:8400–8408. doi: 10.1021/bi00422a016. doi:10.1021/bi00422a016 [DOI] [PubMed] [Google Scholar]
- Riant F., et al. ATP1A2 mutations in 11 families with familial hemiplegic migraine. Hum. Mutation. 2005;26:281. doi: 10.1002/humu.9361. doi:10.1002/humu.9361 [DOI] [PubMed] [Google Scholar]
- Rodacker V., Toustrup-Jensen M., Vilsen B. Mutations Phe785Leu and Thr618Met in Na+,K+-ATPase, associated with familial rapid-onset dystonia parkinsonism, interfere with Na+ interaction by distinct mechanisms. J. Biol. Chem. 2006;281:18 539–18 548. doi: 10.1074/jbc.M601780200. doi:10.1074/jbc.M601780200 [DOI] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. doi:10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Segall L., Scanzano R., Kaunisto M.A., Wessman M., Palotie A., Gargus J.J., Blostein R. Kinetic alterations due to a missense mutation in the Na,K-ATPase α2 subunit cause familial hemiplegic migraine type 2. J. Biol. Chem. 2004;279:43 692–43 696. doi: 10.1074/jbc.M407471200. doi:10.1074/jbc.M407471200 [DOI] [PubMed] [Google Scholar]
- Segall L., Mezzetti A., Scanzano R., Gargus J.J., Purisima E., Blostein R. Alterations in the alpha2 isoform of Na,K-ATPase associated with familial hemiplegic migraine type 2. Proc. Natl Acad. Sci. USA. 2005;102:11 106–11 111. doi: 10.1073/pnas.0504323102. doi:10.1073/pnas.0504323102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skou J.C. The influence of some cation on an adenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta. 1957;23:394–401. doi: 10.1016/0006-3002(57)90343-8. doi:10.1016/0006-3002(57)90343-8 [DOI] [PubMed] [Google Scholar]
- Sorensen T.L., Moller J.V., Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science (NY) 2004;304:1672–1675. doi: 10.1126/science.1099366. doi:10.1126/science.1099366 [DOI] [PubMed] [Google Scholar]
- Spadaro M., Ursu S., Lehmann-Horn F., Veneziano L., Antonini G., Giunti P., Frontali M., Jurkat-Rott K. A G301R Na+/K+-ATPase mutation causes familial hemiplegic migraine type 2 with cerebellar signs. Neurogenetics. 2004;5:177–185. doi: 10.1007/s10048-004-0183-2. doi:10.1007/s10048-004-0183-2 [DOI] [PubMed] [Google Scholar]
- Swoboda K.J., Kanavakis E., Xaidara A., Johnson J.E., Leppert M.F., Schlesinger-Massart M.B., Ptacek L.J., Silver K., Youroukos S. Alternating hemiplegia of childhood or familial hemiplegic migraine? A novel ATP1A2 mutation. Ann. Neurol. 2004;55:884–887. doi: 10.1002/ana.20134. doi:10.1002/ana.20134 [DOI] [PubMed] [Google Scholar]
- Tavraz N.N., Friedrich T., Du¨rr K.L., Koenderink J.B., Bamberg E., Freilinger T., Dichgans M. Diverse functional consequences of mutations in the Na+,K+-ATPase α2-subunit causing familial hemiplegic migraine type 2. J. Biol. Chem. 2008;283:31 097–31 106. doi: 10.1074/jbc.M802771200. doi:10.1074/jbc.M802771200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen L.L., et al. The genetic spectrum of a population-based sample of familial hemiplegic migraine. Brain: J. Neurol. 2007;130(Pt 2):346–356. doi: 10.1093/brain/awl334. doi:10.1093/brain/awl334 [DOI] [PubMed] [Google Scholar]
- Todt U., et al. Rare missense variants in ATP1A2 in families with clustering of common forms of migraine. Hum. Mutation. 2005;26:315–321. doi: 10.1002/humu.20229. doi:10.1002/humu.20229 [DOI] [PubMed] [Google Scholar]
- Tonelli A., et al. Amino acid changes in the amino terminus of the Na,K-adenosine triphosphatase alpha-2 subunit associated to familial and sporadic hemiplegic migraine. Clin. Genet. 2007;72:517–523. doi: 10.1111/j.1399-0004.2007.00892.x. [DOI] [PubMed] [Google Scholar]
- Toustrup-Jensen M., Vilsen B. Importance of Glu282 in transmembrane segment M3 of the Na+,K+-ATPase for control of cation interaction and conformational changes. J. Biol. Chem. 2002;277:38 607–38 617. doi: 10.1074/jbc.M203665200. doi:10.1074/jbc.M203665200 [DOI] [PubMed] [Google Scholar]
- Toyoshima C., Mizutani T. Crystal structure of the calcium pump with a bound ATP analogue. Nature. 2004;430:529–535. doi: 10.1038/nature02680. doi:10.1038/nature02680 [DOI] [PubMed] [Google Scholar]
- Toyoshima C., Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. doi:10.1038/nature00944 [DOI] [PubMed] [Google Scholar]
- Toyoshima C., Nakasako M., Nomura H., Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. doi:10.1038/35015017 [DOI] [PubMed] [Google Scholar]
- Toyoshima C., Nomura H., Tsuda T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432:361–368. doi: 10.1038/nature02981. doi:10.1038/nature02981 [DOI] [PubMed] [Google Scholar]
- Vanmolkot K.R., et al. Novel mutations in the Na+,K+-ATPase pump gene ATP1A2 associated with familial hemiplegic migraine and benign familial infantile convulsions. Ann. Neurol. 2003;54:360–366. doi: 10.1002/ana.10674. doi:10.1002/ana.10674 [DOI] [PubMed] [Google Scholar]
- Vanmolkot K.R., et al. Two de novo mutations in the Na,K-ATPase gene ATP1A2 associated with pure familial hemiplegic migraine. Eur. J. Hum. Genet. 2006a;14:555–560. doi: 10.1038/sj.ejhg.5201607. doi:10.1038/sj.ejhg.5201607 [DOI] [PubMed] [Google Scholar]
- Vanmolkot K.R., et al. Severe episodic neurological deficits and permanent mental retardation in a child with a novel FHM2 ATP1A2 mutation. Ann. Neurol. 2006b;59:310–314. doi: 10.1002/ana.20760. doi:10.1002/ana.20760 [DOI] [PubMed] [Google Scholar]
- Vanmolkot K.R., et al. First case of compound heterozygosity in Na,K-ATPase gene ATP1A2 in familial hemiplegic migraine. Eur. J. Hum. Genet. 2007;15:884–888. doi: 10.1038/sj.ejhg.5201841. doi:10.1038/sj.ejhg.5201841 [DOI] [PubMed] [Google Scholar]
- Woo A.L., James P.F., Lingrel J.B. Sperm motility is dependent on a unique isoform of the Na,K-ATPase. J. Biol. Chem. 2000;275:20 693–20 699. doi: 10.1074/jbc.M002323200. doi:10.1074/jbc.M002323200 [DOI] [PubMed] [Google Scholar]