Abstract
Raman scattering provides valuable biochemical and molecular markers for studying bone tissue composition with use in predicting fracture risk in osteoporosis. Raman tomography can image through a few centimeters of tissue but is limited by low spatial resolution. X-ray computed tomography (CT) imaging can provide high-resolution image-guidance of the Raman spectroscopic characterization, which enhances the quantitative recovery of the Raman signals, and this technique provides additional information to standard imaging methods. This hypothesis was tested in data measured from Teflon® tissue phantoms and from a canine limb. Image-guided Raman spectroscopy (IG-RS) of the canine limb using CT images of the tissue to guide the recovery recovered a contrast of 145:1 between the cortical bone and background. Considerably less contrast was found without the CT image to guide recovery. This study presents the first known IG-RS results from tissue and indicates that intrinsically high contrasts (on the order of a hundred fold) are available.
1. Introduction
In many situations in medicine, structural imaging of tissue does not provide sufficient evidence for effective clinical assessment, and the addition of biochemical information would be beneficial. The ideal situation would be to design hybrid tomography systems which provide good spatial resolution together with high molecular contrast information. Optical spectroscopy can provide inherently high levels of intrinsic molecular contrast, including Raman measurements arising from functional and molecular changes [1]. Combining optical techniques with anatomical information from other low-contrast, high-resolution imaging modalities has been demonstrated with Magnetic resonance imaging [2], X ray [3] and ultrasound [4]. In these hybrid systems, the spatial structure of tissues is embedded into the optical image reconstruction to provide accurate and high-resolution molecular characterization. Work by Davis et al. [5] showed that the use of spatial priors is essential for recovering fluorophore distributions in complex tissue volumes [6, 7]. The proposed method is extended here to combine Raman spectroscopy with x-ray CT to allow image-guided Raman spectroscopy (IG-RS) using anatomical structures from CT as a priori knowledge. We present here, to the best of our knowledge, the first known IG-RS estimates using an excised canine limb as a model.
Raman spectroscopy is based on the principle of inelastically scattered light arising from vibrational energy states of molecular bonds, and it provides valuable biochemical markers to study tissue composition. Raman spectroscopy has been studied in various clinical applications [8–13]. Our interest is in exploring the use of Raman scattering for studying bone chemical composition and predicting the risk of bone fracture with age and disease, such as osteoporosis. Reports indicate that collagen, the primary organic component of bone extracellular matrix, undergoes changes with osteoporosis [14–17]. However, the current gold standard for bone health, dual-energy X-ray absorptiometry (DXA), measures only the mineral component of bone. With Raman spectroscopy both the mineral and organic phases can be measured. Results ex vivo showed that Raman spectroscopy could distinguish between normal and unhealthy tissue non-invasively [13] using time-resolved measurements in mouse genotypes. Also, Schulmerich et al. [18] have accurately recovered Raman bone spectra through 5 mm of tissue in a canine excised limb using a reflectance-based fiber optic probe. In addition, compositional differences observed in bone excised from women with and without hip fractures illustrated a higher carbonate-phosphate ratio from trabecular bone in women with fracture [19]. Because of the molecular specificity and quantitative potential of these measurements, deep tissue Raman measurements would provide valuable biomedical information if Raman scatter could be well localized to the region of origin and then used to tomographically recover the contrast (i.e. boundaries) between the desired target and background.
Diffuse optical spectroscopic measurements can be used to overlay molecular information onto imaging systems, but a transmission geometry limits the accuracy of imaging [20–22]. Preliminary studies on Raman tomographic imaging using a transmission geometry allowed three-dimensional visualization of spatial changes in biochemical composition [23], however, the localization of the origin of Raman signal was limited due to the diffuse nature of light propagation. Here we present the use of high-resolution CT images to guide the diffuse modeling of light in order to achieve a high-contrast molecular and structural characterization of tissue with accurate localization of signal origins.
2. Materials and methods
2.1 Instrumentation
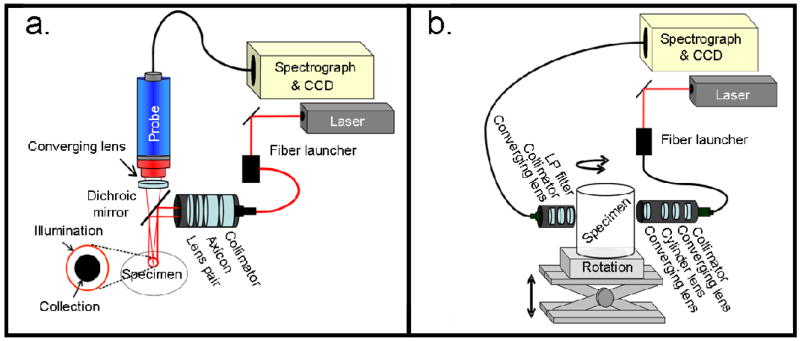
Data were collected using a Raman system operated in two modes: reflectance mode and transmission mode. A comparison of the two imaging systems is shown in Figure 1. The system has been detailed elsewhere [24–26]. Briefly, this system comprised a 400-mW, 785-nm external cavity diode laser (Invictus, Kaiser Optical Systems, Inc., Ann Arbor, MI) and a near infra-red -optimized imaging spectrograph (HoloSpec, f/1.8, Kaiser Optical Systems, Inc.) fitted with a 50-μm slit to provide 6–8 cm−1 resolution. The detector was a thermoelectrically-cooled, deep-depletion 1024×256 pixel CCD (Model DU420-BR-DD, Andor Technology, Belfast, Northern Ireland) operated at −75°C with no binning. The dispersion axis of the spectrograph was calibrated against the neon discharge lamp of a Raman Calibration Accessory (Kaiser Optical Systems, Inc.). The diffused quartz-halogen light of the Calibration Accessory was used to flat-field the CCD.
Fig. 1.
(a) Reflectance mode instrumentation for Raman measurements is shown using a ring/disk geometry. The ring diameters can be varied to sample multiple depths for tomographic reconstruction. (b) Transmission mode configuration is shown using a rectangular array of collection fibers at 180 degrees from the line source illumination. Multiple projections across the tissue can be obtained by rotating the sample with respect to the source-collection fiber setup.
Reflectance geometry
When operated in reflectance mode, as for measurements on the tissue phantom and canine tissue, the sampling system was a filtered fiber optic probe with a bundle of fifty 100-μm core collection fibers arranged in a close-packed circle (PhAT probe, Kaiser Optical Systems, Inc., Ann Arbor, MI). At the spectrograph end, the collection fibers were arranged in a line for coupling into the spectrograph. For the tissue phantom measurements, the collection probe was fitted with a 500-mm focal length fused silica lens that focused to a 14-mm diameter collection field of view. For the canine measurements, a 250-mm focal length fused silica lens was used to provide a collection field of view that was 7 mm in diameter. The laser power available at the sample/specimen was attenuated to 200 mW. To generate the illumination ring, the laser light was launched into a 200-μm NIR optical fiber (Multimode Fiber Optics, Inc., Hackettstown, NJ). The light was collimated (F810FC-780, Thorlabs, Inc., Newton, NJ) and directed through a 175° axicon (Delmar Ventures, San Diego, CA). Telephoto optics (positive/negative lens pair with variable spacing) were placed after the axicon and used to vary the ring diameter. A dichroic mirror (Chroma Technology Corp., Rockingham, VT) reflected the 785-nm light to the sample and transmitted the Raman signal for collection.
Transmission geometry
When operated in transmission mode, as for transmission Raman measurements from excised canine limb, the sampling system was a rectangular fiber optic array of 100-μm core fibers arranged in a 5×10 close-packed rectangle (FiberTech Optica Inc., Kitchener, ON, Canada). At the other end of the bundle, these collection fibers were arranged in a line for coupling into the spectrograph. An 800-nm long-pass filter (HQ800LP, Chroma Technologies Corp., VT) and a 250-mm focal length fused silica lens were placed in front of the rectangular bundle to reject ambient and Raleigh scattered light and to focus the fibers to a 9 mm × 5 mm collection field of view at the specimen. For illumination, laser light was launched diametrically opposite (at 180 degrees) to the collection fibers into a 200-μm NIR optical fiber (Multimode Fiber Optics, Inc., Hackettstown, NJ). The light was collimated (F810FC-780, Thorlabs, Inc., Newton, NJ) and directed through a 50-mm focal length cylinder lens that was positioned between a telescope comprising two 50-mm focal length fused silica lenses. The optical elements were translated to project an 8 mm × 1.5 mm illumination line onto the specimen. Power at the specimen was attenuated to 200 mW.
2.2 Tissue specimens
Tissue phantom
A cylindrical tissue phantom made with agar and Intralipid® was used for testing IG-RS in an experimental setup. Details of the phantom preparation have been described elsewhere [27]. Briefly, this phantom (diameter 59 mm, height 45 mm) contained a single Teflon® inclusion of diameter 3/8″ (~9 mm) embedded 10 mm below the top surface of the phantom. Two concentrations of the background Intralipid®, 0.5% and 1%, were used to obtain two phantom measurement sets. Nine acquisitions were taken for each data set using the ring/disk probe in reflectance mode, with the illuminations rings incremented from 4.0 to 13.5 mm in diameter. The acquisition time for each measurement was 10 seconds.
Dog tibia
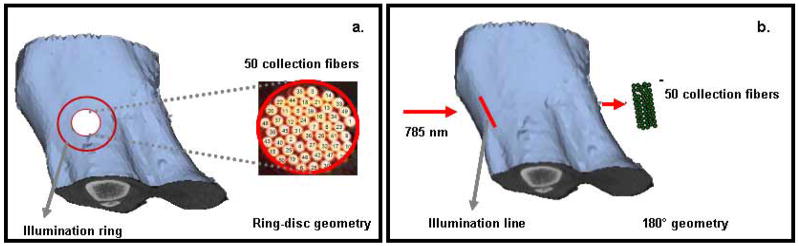
An excised canine tibia was used to obtain Raman measurements in reflectance and transmission modes. The limb was harvested from an animal that was euthanized in approved (University Committee on Use and Care of Animals) studies at the University of Michigan Medical School. CT images of the limb were obtained using a micro-CT scanner (eXplore Locus RS, GE Healthcare, Ontario, Canada). The canine tibia was prepared using glycerol for optical clearing, and reflectance measurements were acquired using the ring/disk configuration. The ring dimensions were changed for ten different acquisitions, from 6 to 16 mm, and the data acquisition time was 1 minute per ring. The thickness of the canine limb ranged from 24 to 45 cm. After all transcutaneous measurements were taken, the overlying tissue was removed with a scalpel, and an exposed bone measurement was taken for validation.
To obtain transmission measurements from the canine limb, the tibia was clamped onto a 360° rotation stage, and 13 projections were obtained at 8° intervals around the limb. The acquisition time was 5 minutes per projection. The source illumination was incident on the anterior side of the tibia, and the collection fibers were positioned diametrically opposite, focused on the limb’s posterior side. Figure 2 shows a representation of the reflectance and transmission measurement geometries for the canine limb.
Fig. 2.
(a) Schematic representation of the ring/disk measurements on the canine limb is shown using surface rendering from CT images. The ring diameter was varied between 6 and 16 mm for 10 different separations between source ring and collection fibers. (b) Representation of the transmission measurements from the canine tibia is shown. A total of 13 projections were obtained at 8° intervals around the limb.
2.3 Raman data reduction
Preprocessing included removing spikes due to cosmic rays and removing high-frequency components of the collected signal using a Blackman-Harris windowing function [28]. Each collection fiber was projected onto 5 adjacent rows of the CCD detector. Only signal from the central three rows was used because the outer rows included small amounts of signal from adjacent fibers. To generate a tomographic reconstruction of the buried target (i.e. Teflon® sphere or bone) we needed to determine a measure of the Raman signal generated by the buried target relative to signal originating from overlying layers. To do this, the intense Teflon® band at 732 cm−1 was used as a measure of Teflon® content, and this signal was normalized to the Intralipid® band at 842cm−1. All operations were performed in MATLAB (v.6.1, The MathWorks, Inc., Natick, MA). The normalized signal for each collection fiber was input into the reconstruction algorithm.
Additional preprocessing of the canine limb data included removing the auto-fluorescent background with a 5th-order polynomial using the algorithm of Lieber and Mahadevan-Jansen [29]. The data were truncated to an 833–1500 cm−1 region of interest, chosen to include Raman bands that are correlated with changes in the mechanical properties of bone [19, 30, 31]. The spectra were then normalized to the phosphate ν1 band, the most intense band in the bone spectrum. The transcutaneous measurements acquired with the probe in reflectance mode were used to recover the Raman bone spectrum non-invasively. The recovery protocols have been described elsewhere [24]. Briefly, three of the reflectance measurements were selected as having an optimal separation between the illumination ring and collection field of view. The Raman signal for the three measurements were combined into a single data set, and band target entropy minimization (BTEM) was used to target the phosphate ν1 band (930–990 cm−1). Five eigenvectors were necessary to recover the Raman bone spectrum. Bone scores were calculated from a regression of the signal detected by each of the collection fibers with respect to the recovered Raman bone spectrum. Bone scores were used as inputs to the tomographic reconstruction in both transmission and reflectance modes.
The detected Raman data was registered with the mesh by marking the sample/specimen at each illumination and collection point with a fine point indelible ink marker and then manually entering the coordinates of the illumination and collection fibers onto the mesh. The position of the collection fibers were registered by projecting a white-light source through the fibers onto the sample/specimen and noting the location of a feature on the fiber bundle relative to the orientation of the sample/specimen.
2.4 Computational modeling for IG-RS
A finite element model of the diffusion approximation, developed for fluorescence imaging, was used in this work. Since both fluorescence and Raman imaging involve emission of light by tissue, the same numerical models are applicable to both. A set of coupled diffusion equations was used to model both the propagation of laser light at the excitation wavelength and the propagation of emitted light that resulted from target site fluorescence at the emission wavelength [32]. The coupled equations can be written as a function of frequency ω and spatial position r as:
| (1) |
| (2) |
where Φx and Φm are the isotropic fluence at the excitation and emission wavelengths, respectively. Subscript x denotes excitation, and m denotes emission; μax is the absorption coefficient at the excitation wavelength, Dx is the diffusion coefficient at the excitation wavelength and Sx is the source term at the excitation wavelength. Similarly, μam, Dm and Sm are the absorption coefficient, diffusion coefficient and the source term at the emission wavelength. The source term at the emission wavelength is related to the fluence at the excitation wavelength as:
| (3) |
where ημaf is the fluorescence yield (Raman yield in this case) and τ is the Raman lifetime. The implementation details of this model for fluorescence were published previously [5].
The inverse problem involves recovering images of Raman yield given measurements of the emission field at the boundary of the imaging domain. A modified Newton’s method was used for this purpose, and, using a Taylor series approximation, the equation for update in the source term ∂γ can be given as:
| (4) |
where ∂Φ refers to the change in the emission field sampled at the detector locations on the boundary and the calculated field at the same locations using the forward solver. ℑ is the Jacobian, the matrix containing the sensitivity of the boundary data to a change in γ and
| (5) |
Using this procedure, 3D estimates of the Raman yield were reconstructed. To implement the spatial information from the CT images, a matrix transformation was applied to the Jacobian so that the recovered ∂γ then contained homogeneous updates for the specified tissue types from CT. Details of this transformation can be found in Dehghani et al. [7].
The intrinsic optical properties at the emission and excitation wavelengths were assumed to be known in this work and were obtained from literature values for canine limb [33]. The ring source configuration for the reflectance mode was modeled by a finite set of discrete sources, and an additive combination of the field generated by each of the sources was used to obtain the fluence distribution from the excitation ring. Similarly, for the implementation of the line source used in the transmission mode, a finite set of discrete sources was used in an additive manner.
Data calibration of the experimental Raman measurements was performed using the measurements from the tissue phantom with 0.5% Intralipid®, denoted as the reference data set. Measurements were simulated assuming known optical properties for the 0.5% Intralipid® phantom, and the average difference between the experimental and simulated data was used as a scaling factor in all reconstructions to compensate for model-data misfits. This average scaling factor was independent of the spatial heterogeneity present in the domain and presented a feasible way for data calibration.
3. Results
3.1 Tissue phantom
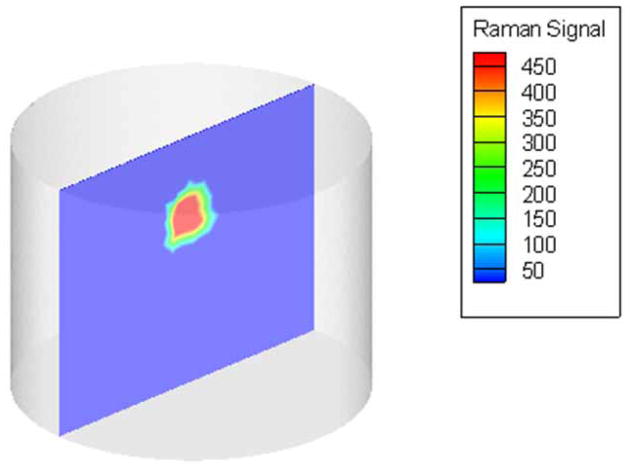
The experimental measurements from the tissue phantom with 1% Intralipid® background were calibrated and used for reconstructing IG-RS estimates of the Raman yield from the background and the Teflon® inclusion. A volumetric description of the phantom, generated using NETGEN [34], contained 5993 nodes and 28009 tetrahedral elements and was used to find approximate solutions to the coupled diffusion equations. A cross-section of the estimated Raman yield from the reconstructed volume is shown in Figure 3, where the yield values were scaled to produce a minimum of 1.0. The estimates of Raman yield showed a contrast of 460:1 between the Teflon® inclusion and the background agar/Intralipid® gel. The contrast for these estimates, which were reconstructed with prior knowledge of the spatial location of the inclusion, was much higher than for estimates reconstructed without spatial priors, where the contrast was only 7.4:1 [35]. The use of spatial priors in the image reconstruction allowed us to reconstruct Raman signal due to the Teflon® sphere in the expected location. The reconstruction algorithm converged in 6 iterations, requiring a computational time of 26 minutes on a standard Dell computer with a 2-GHz processor. These IG-RS estimates from the tissue phantom illustrate the ability to provide localized Raman spectroscopic values for the background gel and the Teflon® inclusion, as well as to indicate the differences in their chemical composition non-invasively.
Fig. 3.
Cross-section of the reconstructed volume showing IG-RS estimates of the tissue phantom imaged using the ring/disk probe. The Teflon® inclusion was recovered with a contrast of 460:1 with respect to the background agar/1% Intralipid® gel.
3.2 Canine tibia
A geometric description of the canine tibia was obtained using image segmentation of the CT images. To define the boundaries between bone and surrounding tissues, the CT images were segmented based on thresholding and region-growing algorithms using a commercial imaging software package (Mimics™, Materialise, Inc.). These segmented tissues were used to create a surface rendering of the canine limb (Figure 4). Using the surface mesh as a starting point, we created a volumetric grid for the limb with Spmesh, an in-house 3D mesh generator [36]. The mesh, which contained 8218 nodes and 40969 tetrahedral elements, was delineated into different materials corresponding to bone and skin using the image segmentations, and each material was labeled with the suitable optical properties [33]. The prepared mesh was used to find IG-RS estimates of Raman yield from bone and skin.
Fig. 4.
Surface rendering of bone and surrounding tissues. The outer limb surface was used to generate a volumetric mesh for computation.
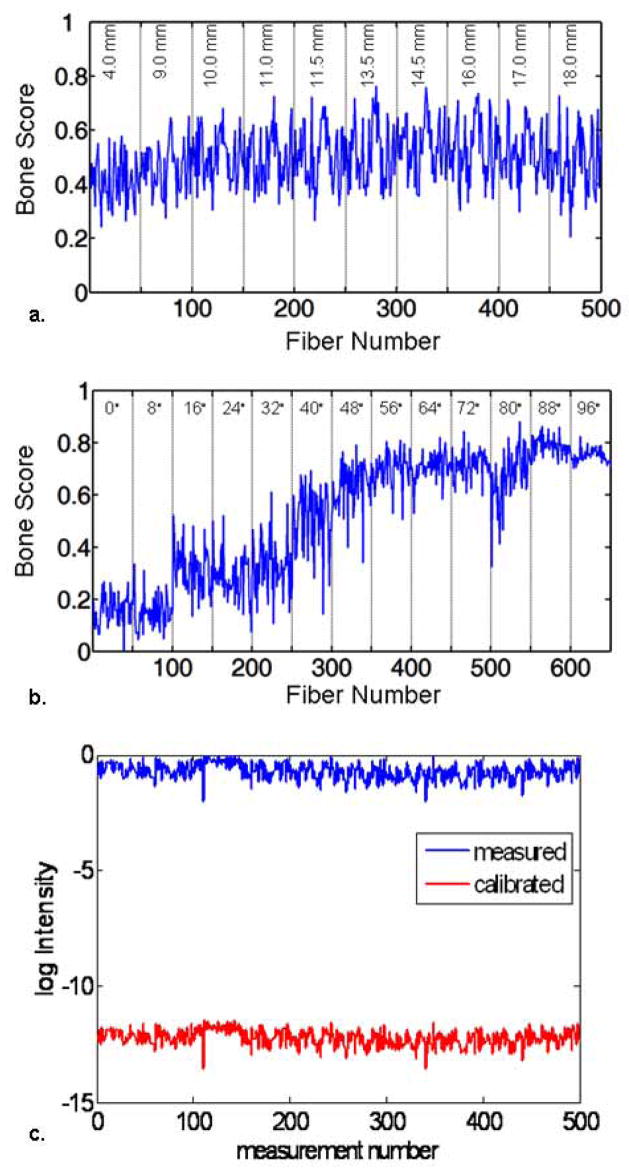
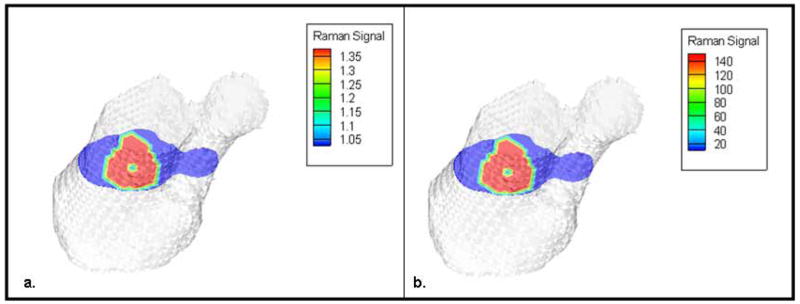
The reflectance measurements were processed using BTEM multivariate analysis to isolate the bone spectrum from the spectra of surrounding tissues. The recovered bone spectrum correlated well with the spectra obtained from exposed bone tissue[23]. The bone spectrum was used to compute the bone scores for both the reflectance and transmission measurements, and the corresponding score plots are shown in Figures 5a and 5b. The bone scores obtained in this manner were calibrated using the scaling factor computed from the reference tissue phantom. Figure 5c shows an example of this calibration for the reflectance data after the scaling factor was applied to the measurements. The calibrated Raman datasets from both geometries were used independently for the recovery of IG-RS estimates of Raman yield values from the bone and the skin. The reconstruction algorithm converged in 5 iterations using reflectance data (computation time = 38.4 min) and in 12 iterations using transmission data (computation time = 94.6 min). The IG-RS estimates were scaled to produce a minimum of 1.0, and these values were used to compare the two system geometries (Figure 6). The location of the bone was identical in both geometries and obtained from the CT images. However, the Raman yield values obtained from IG-RS differ in the two configurations. The IG-RS estimates of Raman yield from bone were much lower when using data from the reflectance geometry with a contrast of 1.4:1 than when using data from the transmission geometry with a contrast of 145:1. The reflectance values are likely lower, because Raman signal originating from deeper layers is not sufficiently collected in back-reflectance measurements that emphasize Raman signals arising from near the surface. The IG-RS estimates from bone and skin in the transmission measurements indicate that we can obtain a contrast between the bone and background of more than 100-fold with this technique.
Fig. 5.
(a) Bone scores computed from reflectance measurements. (b) Bone scores computed from transmission measurements. The score plots indicate the correlation between the spectra obtained at each collection fiber and the bone spectra. (c) The bone scores were calibrated before the reconstruction using a scaling factor obtained from the phantom data, which compensated for the misfit between the FEM model and the measured data.
Fig. 6.
A cross-section of the reconstructed IG-RS estimates using (a) reflectance measurements from the ring/disk configuration and (b) transmission measurements using the rectangular array of collection fibers. Recovered contrast between the bone and background skin was more than 100-fold higher using transmission measurements than using reflectance data, as seen from the scale for reconstructed Raman yield.
4. Discussion
The availability of high-efficiency fiber-optic probes and systems have radically improved the use of Raman spectroscopy in clinical settings; an increasing number of research studies are evaluating molecular and biochemical contrast mechanisms with origins in Raman scattering and vibrational spectroscopy [21, 37]. Raman spectroscopy is a promising technique, because it has known biochemical specificity that can be attributed to the spectral constituents of materials/tissues [38] and because it provides the possibility for non-invasive, low-cost, spectrally-rich measurements in vivo, without the need for contrast agent injection. Our study demonstrates the feasibility of linking Raman measurements with CT imaging of bone and subsequently examining both the structure and biochemical content of intact bone in vivo.
This study on Raman tomography indicated that Raman scattering measurements can provide spatial maps of changes in tissue composition. Measurements obtained in transmission mode showed a superior depth penetration as compared to reflectance mode. These data were consistent with trends observed in fluorescence studies using reflectance mode which have shown that the penetration depth and accurate localization may be limited to a maximum of 10 mm [39]. In previous Raman studies, this depth was quantified at 6.5 mm for canine tissue [18]. Clearly, the reflectance mode is more useful for characterizing subsurface features within a limited depth of approximately 10 mm. The transmission geometry is more effective for deeper tissue penetration, as our previous study of canine tissue imaging demonstrated[23].
The key limitation with imaging in the diffuse optical regime, whether using reflectance or transmission geometries, is the spatial resolution, which is limited to the millimeter scale due to high scattering. Increasingly, researchers are investigating multi-modality image-guidance to provide high resolution characterization of tissue, as well as to improve the accuracy of computational models. In these multi-modality systems, the exact tissue structure is obtained from the conventional micron-resolution imaging techniques such as CT or MRI with optical techniques providing unique molecular and functional signatures of the tissues. Towards this end, successful hybrid systems have combined MRI [2], x rays [3] and ultrasound [4] with diffuse optical imaging in clinical applications. We have extended this concept of multimodality image-guidance for providing localized and non-invasive tissue characterization to include Raman spectroscopy. This work, to the best of our knowledge, presents the first IG-RS estimates from biological tissue in situ.
As the field of molecular contrast agents and biomarkers expands, the need to increase the contrasts observable from relevant tissue boundaries becomes increasingly important. Typically, measurable and quantifiable contrasts between diseased and normal tissues degrade exponentially when going from ex-vivo and in-vitro experiments to in-vivo data. Hence, an inherently high-contrast imaging technique is particularly attractive for use in molecular tissue characterization. Our results on canine tibia indicate that a hundred-fold contrast can be observed from changes in the biochemical composition of tissue. This technique will have potential applications in observing changes in the mineral and matrix compositions that occur in bone tissue, while providing high resolution via image-guidance from MRI or CT. Future clinically-relevant studies will evaluate the ability of IG-RS to monitor biochemical changes occurring from disease.
Acknowledgments
This work was supported through NIH grants R01-CA109558 and R01-CA120368 to B.W.P. and R01-AR055222 to M.D.M.
Footnotes
OCIS codes: (170.5660) Raman spectroscopy; (170.3880) Medical and biological imaging.
References and links
- 1.Poplack SP, Paulsen KD, Hartov A, Meaney PM, Pogue BW, Tosteson TD, Grove MR, Soho SK, Wells WA. Electromagnetic breast imaging: average tissue property values in women with negative clinical findings. Radiology. 2004;231:571–580. doi: 10.1148/radiol.2312030606. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter C, Pogue BW, Jiang S, Dehghani H, Wang X, Paulsen KD, Wells WA, Forero J, Kogel C, Weaver J, Poplack SP, Kaufman PA. Image-guided optical spectroscopy provides molecular-specific information in vivo: MRI-guided spectroscopy of breast cancer hemoglobin, water & scatterer Size. Opt Lett. 2007;32:933–935. doi: 10.1364/ol.32.000933. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Q, Brukilacchio TJ, Li A, Stott JJ, Chaves T, Hillman E, Wu T, Chorlton M, Rafferty E, Moore RH, Kopans DB, Boas DA. Coregistered tomographic x-ray and optical breast imaging: initial results. J Biomed Opt. 2005;10:024033–0240339. doi: 10.1117/1.1899183. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Q, Cronin EB, Currier AA, Vine HS, Huang M, Chen N, Xu C. Benign versus malignant breast masses: optical differentiation with US-guided optical imaging reconstruction. Radiology. 2005;237:57–66. doi: 10.1148/radiol.2371041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis SC, Dehghani H, Wang J, Jiang S, Pogue BW, Paulsen KD. Image-guided diffuse optical fluorescence tomography implemented with Laplacian-type regularization. Opt Express. 2007;15:4066–4082. doi: 10.1364/oe.15.004066. [DOI] [PubMed] [Google Scholar]
- 6.Brooksby B, Dehghani H, Pogue BW, Paulsen KD. Near infrared (NIR) tomography breast image reconstruction with apriori structural information from MRI: algorithm development for reconstructing heterogeneities. IEEe J Sel Top Quantum Electron on Lasers in Medicine and Biology. 2003;9:199–209. [Google Scholar]
- 7.Dehghani H, Pogue BW, Shudong J, Brooksby B, Paulsen KD. Three-dimensional optical-tomography: resolution in small-object imaging. Appl Opt. 2003;42:3117–3128. doi: 10.1364/ao.42.003117. [DOI] [PubMed] [Google Scholar]
- 8.Robichaux-Viehoever A, Kanter E, Shappell H, Billheimer D, Jones H, III, Mahadevan-Jansen A. Characterization of Raman spectra measured in vivo for the detection of cervical dysplasia. Appl Spectrosc. 2007;61:986–993. doi: 10.1366/000370207781746053. [DOI] [PubMed] [Google Scholar]
- 9.Matousek P, Stone N. Prospects for the diagnosis of breast cancer by noninvasive probing of calcifications using transmission Raman spectroscopy. J Biomed Opt. 2007;12:024008, 024001–024008. doi: 10.1117/1.2718934. [DOI] [PubMed] [Google Scholar]
- 10.Lambert JL, Pelletier CC, Borchert M. Glucose determination in human aqueous humor with Raman spectroscopy. J Biomed Opt. 2005;10(031110):031111–031118. doi: 10.1117/1.1914843. [DOI] [PubMed] [Google Scholar]
- 11.Enejder AMK, Scecina TG, Oh J, Hunter M, Shih W, Sasic S, Horowitz GL, Feld MS. Raman spectroscopy for noninvasive glucose measurements. J Biomed Opt. 2005;10(031114):031111–031119. doi: 10.1117/1.1920212. [DOI] [PubMed] [Google Scholar]
- 12.Wilson BC. Detection and treatment of dysplasia in Barrett’s esophagus: a pivotal challenge in translating biophotonics from bench to bedside. J Biomed Opt. 2007;12(051401):051401–051422. doi: 10.1117/1.2795688. [DOI] [PubMed] [Google Scholar]
- 13.Draper ERC, Morris MD, Camacho NP, Matousek P, Towrie M, Parker AW, Goodship AE. Novel assessment of bone using time-resolved transcutaneous Raman spectroscopy. J Bone Miner Res. 2005;20:1968–1972. doi: 10.1359/JBMR.050710. [DOI] [PubMed] [Google Scholar]
- 14.Bailey AJ, Wotton SF, Sims TJ, Thompson PW. Post-translational modifications in the collagen of human osteoporotic femoral head. Biochem Biophys Res Commun. 1992;185:801–805. doi: 10.1016/0006-291x(92)91697-o. [DOI] [PubMed] [Google Scholar]
- 15.Bailey B, Farkas DL, Taylor DL, Lanni F. Enhancement of axial resolution in fluorescence microscopy by standing wave excitation. 1993;366:44–48. doi: 10.1038/366044a0. [DOI] [PubMed] [Google Scholar]
- 16.Knott L, Bailey AJ. Collagen cross-links in mineralizing tissues: A review of their chemistry, function, and clinical relevance. Bone. 1998;22:181–187. doi: 10.1016/s8756-3282(97)00279-2. [DOI] [PubMed] [Google Scholar]
- 17.Paschalis EP, Shane E, Lyritis G, Skarantavos G, Mendelsohn R, Boskey AL. Bone fragility and collagen cross-links. J Bone Miner Res. 2004;19:2000–2004. doi: 10.1359/JBMR.040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulmerich MV, Dooley KA, Vanasse TM, Goldstein SA, Morris MD. Subsurface and transcutaneous Raman spectroscopy and mapping using concentric illumination rings and collection with a circular fiber-optic array. Appl Spectrosc. 2007;61:671–678. doi: 10.1366/000370207781393307. [DOI] [PubMed] [Google Scholar]
- 19.McCreadie BR, Morris MD, Chen T-c, Sudhaker Rao D, Finney WF, Widjaja E, Goldstein SA. Bone tissue compositional differences in women with and without osteoporotic fracture. Bone. 2006;39:1190–1195. doi: 10.1016/j.bone.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Carter JC, Angel SM, Lawrence-Snyder M, Scaffidi J, Whipple RE, Reynolds JG. Standoff detection of high explosive magterials at 50 meters in ambientl light conditions using a small Raman instrument. Appl Spectrosc. 2005;59:769–775. doi: 10.1366/0003702054280612. [DOI] [PubMed] [Google Scholar]
- 21.Johnson WR, Wilson DW, Fink W, Humayun M, Bearman G. Snapshot hyperspectral imaging in ophthalmology. J Biomed Opt. 2007;12:014036–014037. doi: 10.1117/1.2434950. [DOI] [PubMed] [Google Scholar]
- 22.Poplack SP, Tosteson TD, Wells WA, Pogue BW, Meaney PM, Hartov A, Kogel CA, Soho SK, Gibson JJ, Paulsen KD. Electromagnetic breast imaging: results of a pilot study in women with abnormal mammograms. Radiology. 2007;243:350–359. doi: 10.1148/radiol.2432060286. [DOI] [PubMed] [Google Scholar]
- 23.Schulmerich MV, Srinivasan S, Cole JH, Kreider J, Dooley KA, Goldstein SA, Pogue BW, Morris MD. Non-invasive Raman tomographic imaging of canine cortical bone tissue. JBO Lett. 2007;13(020506):020501–020503. doi: 10.1117/1.2904940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulmerich MV, Dooley KA, Vanasse TM, Goldstein SA, Morris MD. Subsurface and Transcutaneous Raman Spectroscopy and Mapping Using Concentric Illumination Rings and Collection with a Circular Fiber-Optic Array. Appl Spectrosc. 2007;61:671–678. doi: 10.1366/000370207781393307. [DOI] [PubMed] [Google Scholar]
- 25.Schulmerich MV, Morris MD, Vanasse TM, Goldstein SA. Transcutaneous Raman spectroscopy of bone global sampling and ring/disk fiber optic probes. Advanced Biomedical and Clinical Diagnostic Systems V SPIE. 2007:643009–643008. [Google Scholar]
- 26.Schulmerich MV, Finney WF, Fredericks RA, Morris MD. Subsurface Raman Spectroscopy and Mapping Using a Globally Illuminated Non-Confocal Fiber-Optic Array Probe in the Presence of Raman Photon Migration. Appl Spectrosc. 2006;60:109–114. doi: 10.1366/000370206776023340. [DOI] [PubMed] [Google Scholar]
- 27.Schulmerich MV, Srinivasan S, Kreider J, Cole JH, Dooley KA, Goldstein SA, Pogue BW, Morris MD. Raman tomography of tissue phantoms and bone tissue. Biomedical optical spectroscopy, (SPIE. 2008 [Google Scholar]
- 28.Harris FJ. On the Use of Windows for Harmonic Analysis with the Discrete Fourier Transform. Proc IEEE. 1978;66:51–83. [Google Scholar]
- 29.Lieber CA, Mahadevan-Jansen A. Automated method for subtraction of fluorescence from biological Raman spectra. Appl Spectrosc. 2003;57:1363–1367. doi: 10.1366/000370203322554518. [DOI] [PubMed] [Google Scholar]
- 30.Carden A, Morris MD. Application of vibrational spectroscopy to the study of mineralized tissues (review) J Biomed Opt. 2000;5:259–268. doi: 10.1117/1.429994. [DOI] [PubMed] [Google Scholar]
- 31.Kohn DH, Sahar ND, Hong SI, Golcuk K, Morris MD. Local Mineral and Matrix Changes Associated with Bone Adaptation and Microdamage. Mater Res Soc Symp Proc. 2006;898E:1–11. [Google Scholar]
- 32.Paithankar DY, Chen AU, Pogue BW, Patterson MS, SevickMuraca EM. Imaging of fluorescent yield and lifetime from multiply scattered light reemitted from random media. Appl Opt. 1997;36:2260–2272. doi: 10.1364/ao.36.002260. [DOI] [PubMed] [Google Scholar]
- 33.Alexandrakis G, Rannou FR, Chatziioannou AF. Tomographic bioluminescence imaging by use of a combined optical-PET (OPET) system: a computer simulation feasibility study. Phys Med Biol. 2005;50:4225–4241. doi: 10.1088/0031-9155/50/17/021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schöberl J. NETGEN - An advancing front 2D/3D-mesh generator based on abstract rules [Google Scholar]
- 35.Srinivasan S, Schulmerich MV, Pogue BW, Morris MD. Biomedical Optics. Optical Society of America; 2008. 3-D image-guided raman characterization in a phantom study. BWD5. [Google Scholar]
- 36.Zhang JQ, Sullivan JM, Jr, Ghadyani H, Meyer DM. MRI guided 3D mesh generation and registration for biological modeling. Journal of computing and information science in engineering. 2005;5:283–290. [Google Scholar]
- 37.Morris MD, Berger A, Mahadevan-Jansen A. Infrared and Raman spectroscopy. J Biomed Opt. 2005;10:031101. doi: 10.1117/1.1906246. [DOI] [PubMed] [Google Scholar]
- 38.Movasaghi Z, Rehman S, Rehman IU. Raman spectroscopy of biological tissues. Appl Spectrosc. 2007;42:493–541. [Google Scholar]
- 39.Kepshire DS, Davis SC, Dehghani H, Paulsen KD, Pogue BW. Subsurface diffuse optical tomography can localize absorber and fluorescent objects but recovered image sensitivity is nonlinear with depth. Appl Opt. 2007;40:1669–1678. doi: 10.1364/ao.46.001669. [DOI] [PubMed] [Google Scholar]