SUMMARY
Mammalian neural stem cells (NSCs) have the capacity to both self-renew and to generate all the neuronal and glial cell-types of the adult nervous system. Global chromatin changes accompany the transition from proliferating NSCs to committed neuronal lineages, but the mechanisms involved have been unclear. Using a proteomics approach, we show that a switch in subunit composition of neural, ATP-dependent SWI/SNF-like chromatin remodeling complexes accompanies this developmental transition. Proliferating neural stem and progenitor cells express complexes in which BAF45a, a novel Krüppel/PHD domain protein and the actin-related protein BAF53a are quantitatively associated with the SWI2/SNF2-like ATPases, Brg and Brm. As neural progenitors exit the cell cycle, these subunits are replaced by the homologous BAF45b, BAF45c and BAF53b. BAF45a/53a subunits are necessary and sufficient for neural progenitor proliferation. Preventing the subunit switch impairs neuronal differentiation, indicating that this molecular event is essential for the transition from neural stem/progenitors to post-mitotic neurons. More broadly, these studies suggest that SWI/SNF-like complexes in vertebrates achieve biological specificity by combinatorial assembly of their subunits.
INTRODUCTION
The array of specialized neuronal and glial cell-types that characterize the adult central nervous system (CNS) originate from multipotent neuroepithelial (NE) precursor cells that line the ventricles of the developing spinal cord and brain (Gage, 2000; Roegiers and Jan, 2004; Temple, 2001). The decision of NE precursor cells to either self-renew or differentiate is regulated by both extrinsic and intrinsic mechanisms including environmental signals, cell-cell interactions and transcriptional events mediated, at least in part, by bHLH, STAT/Smad proteins and nuclear receptors (reviewed in Bertrand et al., 2002; Gotz and Huttner, 2005). Rapid nuclear reorganization and global changes in gene expression in newly-born neurons suggest that epigenetic changes at the level of chromatin structure might also be involved (Hsieh and Gage, 2005; Kondo, 2006). Epigenetic regulatory mechanisms, such as histone modification, polyADP-ribosylation, DNA methylation, regulatory noncoding RNAs and even retrotransposition have been linked with changes in neural plasticity and long-term memory (reviewed in Hsieh and Gage, 2005; Kandel, 2001). However, the role of chromatin-based epigenetic mechanisms in early neural development remains poorly understood.
The mammalian genome encodes 29 different SWI2/SNF2-like ATPases, several of which are assembled into mammalian SWI/SNF-like complexes suggesting functional diversification within this large family of complexes. Two of these, Brg and Brahma(Brm) are interchangeable subunits in a subfamily of 2 MDa chromatin remodeling complexes termed BAF (Brg/Brm associated factor) or mSWI/SNF (Khavari et al., 1993; Kwon et al., 1994; Lemon et al., 2001; Wang et al., 1996b). BAF complexes contain 10 core subunits, some of which are similar to those in yeast SWI/SNF, while others, such as BAF57 and BAF53, are similar to subunits of the yeast SWR1 and INO80 complexes (Mizuguchi et al., 2004; Shen et al., 2003). The subunits of BAF complexes are tightly associated with the Brg/Brm ATPases and have no detectable rate of exchange in vitro when challenged with purified subunits (Zhao et al., 1998). In vitro studies indicate that BAF complexes facilitate nucleosome exchange between chromosomal templates, increase the accessibility of DNA to sequence-specific transcription factors and nucleate the polymerization of actin filaments from β-actin, which is a core component of these complexes (reviewed in Cairns, 2005; Martens and Winston, 2003).
Evidence that mSWI/SNF (BAF) subunits might have non-redundant and dosage-sensitive roles in neural development has come from several observations. Mice heterozygous for either Brg or BAF155/Srg3 are predisposed to exencephaly, possibly by generation of too few neurons (Bultman et al., 2000; Kim et al., 2001). Similarly, the C. elegans psa-1/Brg and psa-4/BAF155 genes are required for asymmetric neurogenic divisions. Temperature-sensitive (ts) alleles of these genes in worms generate fewer neurons and favor non-neuronal cell types (Sawa et al., 2000). In vertebrates, BAF complexes in post-mitotic neurons are different from those in all other cell-types in that they contain the BAF53b Arp-4 like protein instead of BAF53a (Olave et al., 2002). Studies in Xenopus indicated that Brg is required for neuronal differentiation by mediating the transcriptional activities of the proneural bHLH Neurogenin or NeuroD proteins (Seo et al., 2005b). While Xenopus Brg was reported not to be required for neural stem cell (NSC) function, recent studies suggested that mouse Brg is essential for the repression of neuronal commitment in NSCs, as a means of permitting glial cell differentiation in response to gliogenic signals (Matsumoto et al., 2006). The discrepancy between these studies might be related to functional evolutionary diversification of these complexes or perhaps different roles of Brg/Brm at different times during neurogenesis. Indeed, the finding that post-mitotic neurons express complexes with BAF53b, rather than BAF53a, which is found in all other cell-types (Olave et al., 2002), suggests that the function of these complexes might change during neural development.
A distinctive characteristic of the vertebrate BAF complexes, when compared to the yeast SWI/SNF and Drosophila BAP complexes, is their combinatorial assembly from the products of gene families encoding their 10 core subunits (Lemon et al., 2001;Wang et al., 1996b). The position of the subunits within the complex can be occupied by one and only one family member (Wang et al., 1996a; Zhao et al., 1998). While earlier studies have suggested that combinatorial assembly of vertebrate BAF complexes might contribute to specificity (Bultman et al., 2000; Olave et al., 2002; Reyes et al., 1998), others have suggested that gene family members might be partially or fully redundant (Lickert et al., 2004; Strobeck et al., 2002; Yamamichi-Nishina et al., 2003). One interesting possibility is that structurally distinct BAF complexes create distinct patterns of chromatin accessibility; thereby imparting specialized regulatory functions to ubiquitously expressed transcriptional regulators.
We have investigated the biological significance of combinatorial assembly of mammalian ATP-dependent chromatin remodeling complexes using a proteomics approach in the developing nervous system. Purification of Brg and Brm-associated proteins revealed a new family of four stoichiometric subunits of mSWI/SNF or BAF complexes that we termed BAF45. Remarkably, we show that the transition from proliferating neural stem/progenitor cells to post-mitotic neurons requires a switch in subunit composition of these complexes. At this developmental transition, functional specificity of BAF complexes appears to arise from the combinatorial use of the family members of the 10 subunits like letters in a 10-letter word. Self-renewal and proliferative activities of neural stem/progenitor cells require npBAF complexes containing BAF45a and BAF53a and assembled on the Brg/Brm ATPases. The dynamic exchange of these subunits for the alternative BAF45b, BAF45c and BAF53b subunits in post-mitotic neurons orchestrates cell cycle withdrawal and the acquisition of neuronal properties.
RESULTS
Purification of ATP-Dependent Chromatin Remodeling Complexes from the Developing Murine CNS Reveals a New Family of Subunits
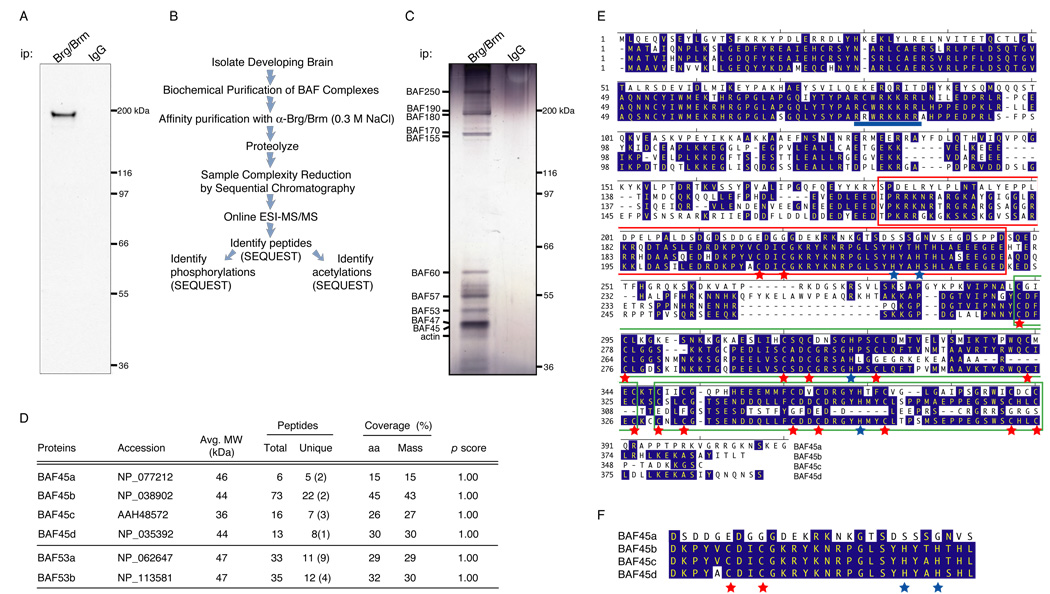
To understand the role of ATP-dependent chromatin remodeling complexes in neural development, we purified the endogenous complexes based on the Brg and Brm ATPases from newborn (P0) mouse brains using a combination of biochemical and affinity steps. We first generated affinity purified antibodies that recognized only Brg and Brm (Figure 1A). Affinity purification of complexes with these antibodies under stringent conditions (Figure 1B and 1C and supplemental experimental procedures) revealed a highly specific pattern of co-purifying proteins (Figure 1C). Brg/Brm associated proteins were subjected to proteolytic digestion, analyzed by multi-dimensional liquid chromatography Electrospray Ionization-Tandem Mass Spectrometry (ESI-MS/MS; Figure 1A) (Link et al., 1999) and identified using SEQUEST (Eng and McCormack, 1994). The purification based on the anti-Brg/Brm antisera yielded 47 unambiguously identified proteins with a minimal ProteinProphet probability of 0.9 (Nesvizhskii et al., 2003), including most (14 out of 15) family members of the 10 core subunits of BAF complexes (Table 1, Figure 1D–1F and unpublished results). No peptides from the BAF60b subunit were obtained, while ten peptides were identified from BAF60a and seven from BAF60c, suggesting that neural BAF complexes lack BAF60b (Table 1). In contrast, only 3 protein identifications were obtained from sequencing of the IgG eluates (minimal probability score of 0.8; hnRNPA3 (n=1 peptide), kininogen precursor (n=1 peptide) and α-1-antitrypsin precursor (n=6 peptides) (data not shown)).
Figure 1. BAF45 family proteins are subunits of neural SWI/SNF-like BAF complexes.
A. Immunoprecipitation and Western blot analysis of Brg/Brm proteins in P0 mouse nuclear extracts using anti-Brg/Brm affinity purified antibodies. kDa, kilodaltons.
B. Strategy for purification and sequencing of neural SWI/SNF-like BAF complexes. Highly stringent conditions were used to isolate only the core subunits of the complexes plus tightly associated proteins (see supplemental experimental procedures). ESI-MS/MS, Electrospray Ionization- Tandem Mass Spectrometry.
C. Silver-stain analysis of immunopurified (anti-Brg/Brm) endogenous neural BAF complexes in P0 mouse brain nuclear extracts.
D. Peptides identified from the BAF45 and BAF53 families of subunits by mass spectrometry. The numbers in parentheses are the numbers of peptides corresponding to unique protein entries. MW, calculated molecular weight; aa, amino acids.
E. Conservation and divergence among mouse BAF45 family members. Conserved amino acids are highlighted in blue. The C2H2-type Krüppel-like domains are boxed in red and the PHD domains of the d4 domain are boxed in green. Red and blue stars indicate the conserved cysteine and histidine residues, respectively. The putative nuclear localization sequence is underlined in blue and is displaced 28 aa C-terminal in BAF45a. Note that only the first C3H motif of the PHD1 domain is conserved in BAF45c.
F. The C2H2-type Krüppel-like zinc finger motif is not conserved in BAF45a. The conserved cysteine and histidine residues are labeled with red and blue stars, respectively. Note their absence in BAF45a protein.
Table 1.
Core Subunits of the Neural BAF SWI/SNF-like ATP-dependent Chromatin Remodeling Complexes as Identified by a Proteomics Approach
| BAF subunits | # peptides differenta (uniqueb) |
Coverage (% mass) | Accessions | p scores |
|---|---|---|---|---|
| BAF250a/b;ARID1a/b/Osa1 | 75 (17) | - | c | 1.00 |
| ARID2 | 13 (8) | 15.4 | d | 1.00 |
| BAF190/Brm/Smarca2 | 10 (4) | 17.5 | NP_035546 | 1.00 |
| BAF190/Brg/Smarca4 | 50 (0) | 23.1 | NP_035547 | 1.00 |
| BAF180/polybromo-1 | 18 (4) | 20.9 | NP_240329e | 1.00 |
| BAF170/Smarcc2 | 34 (2) | 31.3 | NP_937803 | 1.00 |
| BAF155/Srg3/Smarcc1 | 15 (0) | 14.0 | NP_033237 | 1.00 |
| BAF60a/Smarcd1 | 10 (1) | 17.9 | NP_114030 | 1.00 |
| BAF60c/Smarcd3 | 7 (1) | 18.1 | NP_080167 | 1.00 |
| BAF57/Smarce1 | 15 (0) | 49.6 | NP_065643 | 1.00 |
| BAF53a/Actl6a | 11 (9) | 29.3 | NP_062647 | 1.00 |
| BAF53b/Actl6b | 12 (4) | 30.5 | NP_113581 | 1.00 |
| BAF45a/PHF10 | 5 (2) | 15.0 | NP_077212 | 1.00 |
| BAF45b/DPF1 | 22 (2) | 43.0 | NP_038902 | 1.00 |
| BAF45c/DPF3 | 7 (3) | 27.1 | AAH48572 | 1.00 |
| BAF45d/DPF2 | 8 (1) | 30.2 | NP_035392 | 1.00 |
| BAF47/Ini1/mSnf5/Smarcb1 | 8 (0) | 28.6 | NP_035548 | 1.00 |
| BAF42/actinf | 13 (4) | - | - | - |
Number of different peptides for each protein entry.
Number of peptides corresponding to a unique protein entry in the IPI human/mouse databases (Nesvizhskii et al., 2003).
The protein sequence of mouse ARID2 (AT-rich interactive domain 2) will be submitted to NCBI. ARID2 is an evolutionarily conserved protein of 1161 amino acids with a typical ARID DNA-binding domain characteristic of a protein family that includes Drosophila OSA/eyelid, yeast Swi1p, yeast Rsc9 and mammalian BAF250a/ARID1a and BAF250b/ARID1b. Drosophila BAP170, a recently identified subunit of the SWI/SNF-like BAP complex is 43% similar and 26% identical to human ARID2 and most likely represents its orthologue in flies. From an independent purification, human ARID2 was identified and termed BAF200 (Yan et al., 2005).
Rattus norvegicus Pb-1 protein sequence.
α-, β- and γ-actin family of proteins (J.L. and G.R.C, unpublished results).
Several peptides corresponding to 4 homologous proteins, not previously known to be part of SWI/SNF-like complexes were obtained. We referred to them as Brg/Brm associated factor of 45 kDa or BAF45a, b, c and d (Figure 1D–1F and Figure S1). Five different tryptic peptides were found from the previously uncharacterized open reading frame (ORF) of mouse BAF45a (15% mass coverage), twenty-two peptides from BAF45b (43% mass coverage), seven peptides from BAF45c (27% mass coverage) and eight peptides from BAF45d (30% coverage, Figure 1D and Table 1). BLAST searches identified substantial similarity to potential ORFs in several eukaryotic organisms including the worm, fly, pufferfish, zebrafish, frog, chicken, rodent and human (Figure S1 for BAF45a and data not shown). The 4 family members have a distinctive organization with a novel N-terminal domain of unknown function, a central C2H2-type Krüppel zinc finger motif with potential nucleic acid binding activity and a C-terminal d4 domain consisting of two tandem PHD fingers, with potential histone H3 trimethyl-lysine binding activity (Wysocka et al., 2006) (Figure 1E and Figure S1).
BAF45a, BAF45b and BAF45c are Largely Confined to 2 MDa BAF Complexes in Neural Cells
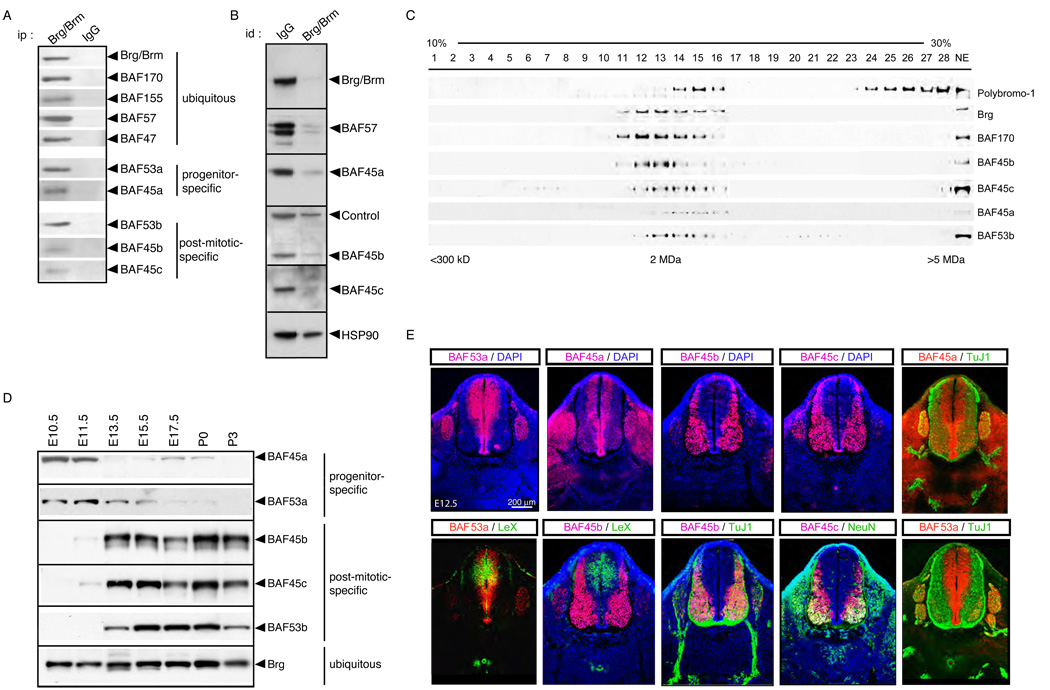
To determine whether the BAF45 proteins were simply associated with Brg/Brm-containing complexes in neural cells or were novel core subunits, we generated polyclonal rabbit antisera specific for each family member. Co-immunoprecipitation experiments confirmed their interaction with Brg and other core components of the complex under stringent conditions in neural cells (Figure 2A, Figure S2 and data not shown). BAF45 proteins are largely dedicated to neural BAF complexes since the affinity purified anti-Brg/Brm antibody depleted them from mouse brain (P0) nuclear extracts (Figure 2B). Glycerol gradient co-sedimentations of brain (E15.5) nuclear extracts demonstrated that virtually all of BAF45a, b and c co-fractionate with known subunits of BAF complexes such as Brg, BAF170 and the actin-related protein (Arp) BAF53b (Figure 2C) (Wang et al., 1996b). In contrast, the Polybromo-1/BAF180 protein, which is part of both P-BAF (Lemon et al., 2001) and kinetocore complexes (Nie et al., 2000), fractionated in two distinct molecular weight complexes in neural cells (E15.5; Figure 2C). The difference in the peak of mobility of the complexes with BAF45a, 45b and Brg probably reflects the combinatorial assembly of this polymorphic group of complexes in the E15.5 mouse brain, which consists of a mixture of stem/progenitor cells and differentiated neurons (Figure 2C). Low affinity biochemical interactions due to the relatively mild cell lysis and differential centrifugation conditions might also contribute to the size differences in the mobility of the complexes (see supplemental experimental procedures). As the BAF45 and BAF53 family proteins are near quantitatively associated with the neural Brg/Brm-containing complexes, we conclude that they are stoichiometric subunits of BAF complexes in neural cells and not simply associated proteins.
Figure 2. The subunit composition of neural BAF complexes changes as cells develop from neural progenitors to post-mitotic neurons.
A. Western blot analyses of immunopurified (anti-Brg/Brm) neural BAF complexes in P0 mouse brain nuclear extracts.
B. BAF45a, BAF45b and BAF45c are dedicated to neural BAF complexes. Serial immunodepletions were performed on anti-Brg/Brm columns in P0 brain nuclear extracts. Supernatants were blotted with anti-BAF antibodies. Hsp90, heat shock protein 90; id, immunodepletion.
C. BAF45a, BAF45b and BAF45c co-sediment with other components of neural BAF complexes. E15.5 brain nuclear extracts were separated by centrifugation on glycerol gradients (10 to 30 %) and isolated fractions analyzed by Western blotting.
D. Sequential expression of BAF45a, BAF53a and BAF45b, BAF45c and BAF53b during neurogenesis. Brain nuclear extracts were isolated at different developmental stages and blotted with BAF45, BAF53 and Brg specific antibodies.
E. BAF45a and BAF53a are expressed specifically in neural progenitors and are replaced by the BAF45b/c and BAF53b homologous subunits in post-mitotic neurons. Immunostaining on cross sections of E12.5 spinal cords together with progenitor-(LeX) and neuronal-specific (TuJ1 and NeuN) markers. TuJ1, antibody against β-tubulin type III; DAPI, 4′,6-Diamidino-2-phenylindole.
Mutually Exclusive Expression of the BAF45a/ BAF53a and BAF45b-45c/BAF53b Subunits in Neural Stem/Progenitor Cells and Post-Mitotic Neurons
Neural differentiation occurs by an anatomically ordered process in which progenitors for both neurons and glia originate in a proliferative zone lining the ventricles and migrate laterally as they exit mitosis and undergo differentiation (Gilbert, 2003; Noctor et al., 2004). Western blot analyses of mouse brain nuclear extracts isolated at different developmental stages (E10.5 to P3) demonstrated a switch in expression of BAF45 and BAF53 family proteins during neurogenesis. E10.5–E11.5 neural cells predominantly express the BAF45a and BAF53a subunits while BAF45b, BAF45c and BAF53b expression becomes detectable from E13.5 onwards (Figure 2D). In the developing spinal cord (E10.5 to E16.5), BAF45a and BAF53a are specifically expressed in proliferating neural progenitors of the ventricular zone, which are positive for LeX/CD15 and Ki-67 (Figure 2E and data not shown). In differentiated neurons of the post-mitotic zone, which express the neuron-specific type III β-tubulin (TuJ1) and NeuN markers, expression of the BAF45a and BAF53a subunits is replaced by the alternative homologous BAF45b, BAF45c and BAF53b subunits (Figure 2E and Figure S3) (Olave et al., 2002). In the developing forebrain and cerebellar primordium, BAF45a and BAF53a expression is also restricted to proliferating neuroepithelial progenitors and cerebellar granule precursors, while BAF45b, BAF45c and BAF53b are strictly expressed in post-mitotic neurons (Figures S4–S6 and data not shown). Reciprocal and mutually exclusive expression of BAF45a and BAF53a in the progenitor zone and BAF45b, BAF45c and BAF53b in the post-mitotic zone of the developing spinal cord was also observed by in situ hybridization (E10.5–E13.5; Figure S7 and data not shown). Although BAF45a and BAF53a are expressed selectively in neural progenitors of the developing neural tube, (Figure 2E and Figure S2), they are expressed in most adult tissues (data not shown) (Olave et al., 2002). In contrast, BAF250a, 180, 170, 155, 60a, 57, 47, 45d and ARID2 were expressed ubiquitously throughout the developing spinal cord, brain and other embryonic tissues at E10.5 to E16.5 (Table 1 and data not shown). Since BAF45a/53a and BAF45b/BAF53b are quantitatively associated with Brg (Figure 2B and 2C) in neural cells, these observations demonstrate that SWI/SNF-like BAF complexes switch subunits during neural development. For clarity, we refer to the SWI/SNF-like BAF complexes expressed in neural stem/progenitors cells and post-mitotic neurons as npBAF and nBAF, respectively.
BAF45a and BAF53a are Required for Proliferation of Neural Stem and Progenitor Cells
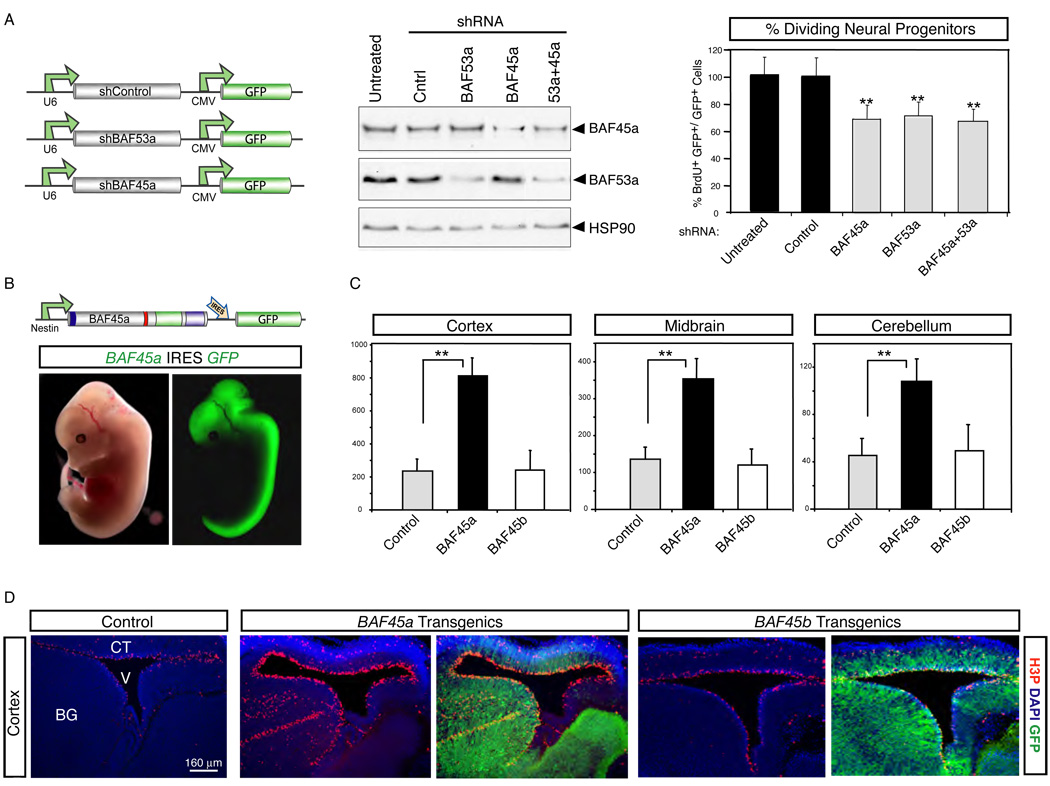
The studies described above indicated that proliferating neural stem/progenitor cells express 2 MDa npBAF complexes containing BAF45a/53a, but lacking BAF60b, which is expressed in all other cell types examined thus far (Wang et al., 1996b). To understand the roles of the npBAF complexes in neural progenitor function, we engineered shRNA constructs against the progenitor-specific BAF45a and BAF53a subunits (Figure 3A). Knockdown of both subunits (BAF45akd, BAF53akd) either individually or simultaneously in E13.5 cortical cultures specifically reduced their levels of expression by about 50 to 80% (Figure 3A). This reduction in protein levels significantly impaired neural stem/progenitor proliferation, as evidenced by a commensurate reduction in their rate of BrdU (5-bromodeoxyuridine) incorporation relative to controls (Figure 3A). These effects cannot be due to off-site targets of RNAi since either construct reduced the rate of neural stem/progenitor cell proliferation (Figure 3A). The decrease in neural stem/progenitor proliferation observed upon reducing BAF45a and BAF53a levels is not due to precocious and/or enhanced terminal differentiation as the percentage of transfected (GFP+) TuJ1+ cells was similar between the knockdown and control cultures (data not shown). Inactivation of both BAF45a and BAF53a in these cells did not affect their survival, as demonstrated by Annexin V and propidium iodide (PI) staining (data not shown). Together, these studies indicated that BAF45a/BAF53a and thereby the npBAF complexes with which they quantitatively associate are required for neural progenitor proliferation. The finding that a reduction in BAF45a/53a concentration produces a significant effect upon neural progenitor proliferation is consistent with the genetically dominant role of Brg in neural tube closure (Bultman et al., 2000).
Figure 3. BAF45a and BAF53a are necessary and sufficient for the proliferation of neural progenitors.
A. shRNA-mediated knockdown of BAF45a and/or BAF53a impairs the proliferative activity of neural progenitors in E14.5 adherent cortical cultures. Western blot analyses of protein extracts from these cultures (3 div) are shown. Hsp90 is a loading control. E14.5 cortical cells electroporated (Amaxa Inc, MD) with shRNA or control vectors were cultured in serum-containing media (4 div), stained with anti-LeX or anti-nestin and BrdU antibodies and analyzed by flow cytometry. BrdU was added 2 hrs before analysis. The percentage of dividing GFP+ BrdU+ neural progenitors in these cultures (n=3 independent experiments) is shown. Results are expressed as mean ±SD. **, p<0.01. See supplemental information for shRNA sequences. CMV, cytomegalovirus immediate early enhancer/promoter.
B. Nestin-driven BAF45a IRES-GFP expression in mouse E13.5 transient transgenics. Nestin, nestin-gene regulatory elements; IRES, internal ribosomal entry site. Note that the BAF45a and BAF45b expression patterns and levels were similar in all the BAF45a and BAF45b transient transgenic mice analyzed.
C. Quantification of the number of proliferating (H3P+) neural progenitors in the cortex, midbrain and cerebellum of BAF45a and BAF45b transient transgenic mice (n=5 and n=3 embryos, respectively) relative to control littermates. H3P+ cells (red) were manually counted using digitalized pictures of comparative brain sagittal sections stained with anti-H3P antibody (n>6 pictures per embryo). Results are expressed as mean ± SD. **, p<0.01. H3P, anti-phosphorylated Histone 3 (Ser10).
D. Increased number of proliferating (H3P+) neural progenitors in the developing cortices and basal ganglia of E13.5 BAF45a transgenic embryos relative to BAF45b transgenic and control embryos. Comparable sagittal sections were stained with anti-H3P antibodies (red) and DAPI (blue). Representative images are shown. BG, basal ganglia; CT, cortex; V, ventricle.
BAF45a but not BAF45b is Sufficient to Enhance Neural Progenitor Proliferation
To further study the role of the newly identified progenitor-specific BAF45a subunit in regulating neural stem/progenitor function, we generated transgenic mice expressing BAF45a or the homologous BAF45b under the control of the neural-specific Nestin promoter (Chenn and Walsh, 2002). To facilitate detection of the cells expressing the transgenes, BAF45a and BAF45b cDNAs were linked to an internal ribosomal entry site (IRES) and GFP (Figure 3B). Using this strategy, we obtained high BAF45a or BAF45b expression (together with GFP) in neural stem/progenitor cells and differentiating neurons (although at relatively lower levels; Figures S5 and S6). As evidenced by anti-phosphorylated histone 3 (Ser10) reactivity (an M-phase marker), E13.5 BAF45a transgenic brains displayed a 2- to 4-fold increase in the number of mitotically active neural stem/progenitors compared to control littermates (Figure 3C and 3D). These proliferating (H3P+) progenitors were mostly confined to the apical/ventricular zone of the developing cortex and midbrain of the BAF45a transgenics, although many mitotic cells were observed outside the periventricular progenitor domains (i.e ganglionic eminences; Figure 3C and 3D). In the developing cerebellum, BAF45a transgenic mice showed a significant increase in the number of mitotically active granule cell progenitors in the external granular layer (EGL) relative to controls (E14.5; Figure 4B and 4C). M-phase H3P+ cells were also found in the internal differentiating zone of the developing cerebellum of BAF45a transgenics (Figure 4C). BrdU pulse-labeling experiments performed in vivo also revealed a marked increase in neural stem/progenitor cell proliferation in the midbrain of BAF45a transgenics (E14.5; Figure 4C). This increase in proliferation cannot be explained by a block to terminal differentiation or an alteration in cell death (Figures S8 and S9 and data not shown). Importantly, 3 transient transgenic lines expressing the homologous nBAF subunit BAF45b, which is normally expressed only in post-mitotic neurons, had normal numbers of M-phase (H3P+) neural stem/progenitor cells (Figure 3C and 3D) and their rate of BrdU incorporation was similar to that of controls (E14.5; data not shown). Possible explanations for this lack of effect is that BAF45b needs to be expressed in the context of the other post-mitotic specific BAF subunits (e.g. BAF53b) to assume its function and/or needs to be post-translationally modified by enzymatic moieties that are not present or active in the neural stem/progenitor compartment. Together, these results indicate that extended expression of one specific component of the neural progenitor (npBAF) complexes, BAF45a, is sufficient to extend the proliferative phase of neural stem/progenitor and cerebellar precursor cells in the developing CNS.
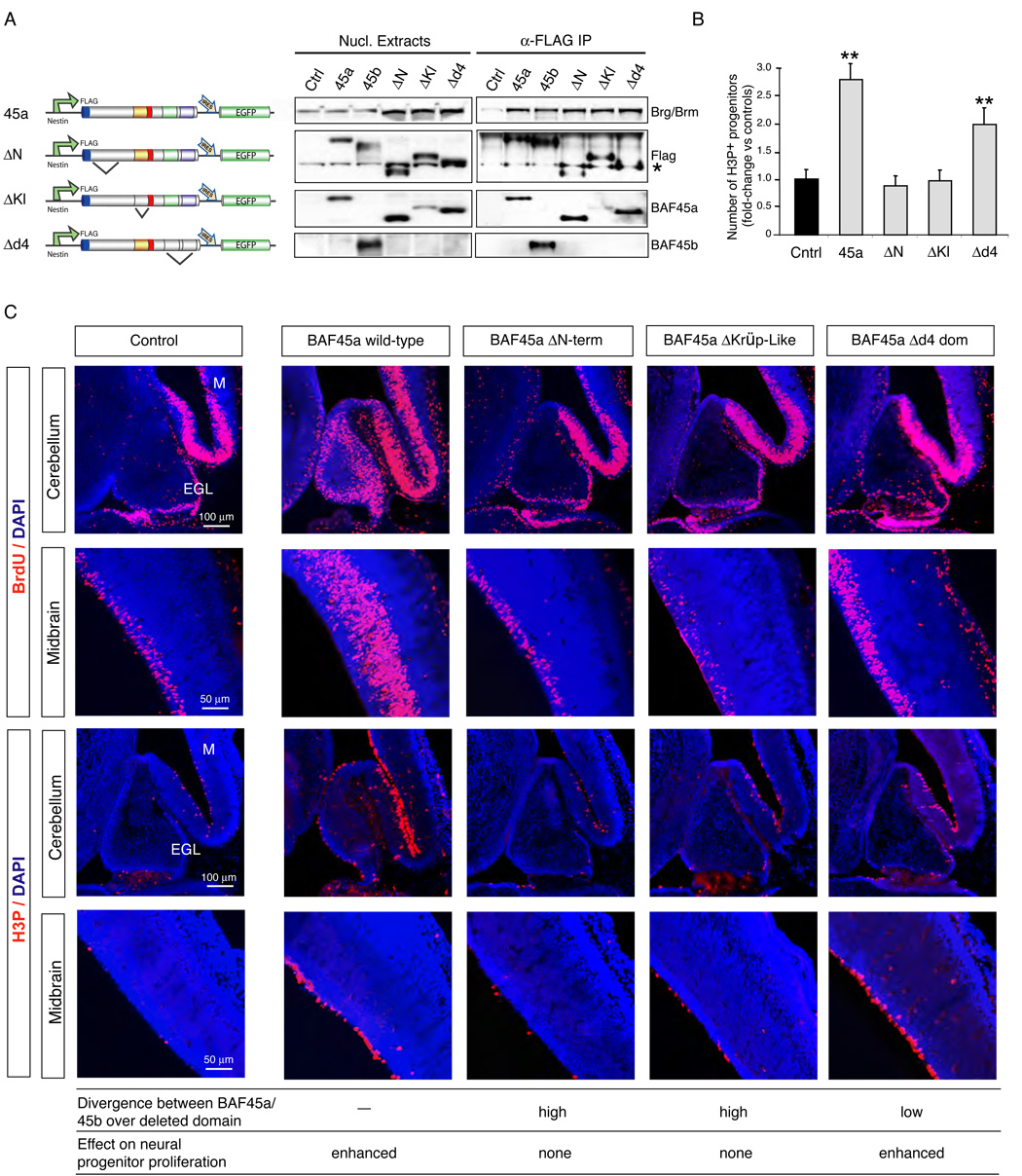
Figure 4. The N-terminus and Krüppel-like domains of BAF45a are essential to induce neural progenitor proliferation.
A. The N-terminal, Krüppel-like and d4 domains of BAF45a are not required for its assembly within npBAF complexes in P19 cells. Schematic representations of the transgenes are shown. Nuclear extracts of transfected P19 cells expressing similar levels of the FLAG-tagged proteins (β-actin promoter) were immunoprecipitated with M2 (anti-FLAG) agarose beads and analyzed by Western blotting. Note the lower detection level of the BAF45a ΔKrüppel-like protein (deletion aa 204–246) with the α-BAF45a antibody (raised against aa 180 to 282). *; non-specific band recognized by the FLAG antibody.
B. Number of M-phase (H3P+) cerebellar progenitors in E14.5 Nestin-driven BAF45a (n=3), BAF45a ΔN-terminal (n=2), BAF45a ΔKrüppel-like (n=3) and BAF45a Δd4 domain (n=3) transient transgenic mice relative to control littermates. H3P+ cells (red) were manually counted using digitalized pictures of comparative brain sagittal sections stained with anti-H3P antibody (n>6 pictures per embryo). Results are expressed as mean ± SD. **, p<0.01
C. Dividing (BrdU+ and H3P+) neural progenitors in the developing cerebellums and midbrains of Nestin-driven BAF45a and BAF45a deletion mutant transgenic mice (E14.5, 1 hr BrdU pulse). Comparable sagittal sections were stained with anti-BrdU (red) or anti-H3P (red) antibodies and DAPI (blue). Number of independent transgenic lines: BAF45a wild-type (n= 3), BAF45a ΔN-terminal (n= 2), BAF45a ΔKrüppel-like (n= 3) and BAF45a Δd4 domain (n=3). Representative images are shown. EGL, external granular layer; M, midbrain.
The N-terminal and Krüppel-like Domains Direct BAF45a to Neural Stem/Progenitor Proliferation in Transgenic Mice
The finding that BAF45a but not the homologous BAF45b subunit is sufficient to enhance neural stem/progenitor cell proliferation prompted us to define the regions of BAF45a that distinguish it from BAF45b in mediating this function. BAF45a and BAF45b are highly similar throughout the C-terminal double PHD (d4) domain (46% identity and 63% similarity at the amino acid level), however their N-terminal and C2H2-type Krüppel-like zinc finger domains are less conserved (Figure 1E and 1F). Based on these observations, we generated BAF45a deletion mutants (i.e. the N-terminal, Krüppel-like and d4 domains, Figure 4A) and tested their ability to drive neural stem/progenitor cell proliferation in transgenic mice. The constructs were FLAG-tagged and expressed under the control of the Nestin promoter with an IRES-GFP marker to allow comparison of transgene expression levels (Figure 4A and Figure S10). Each of the FLAG-tagged BAF45a deletion mutants incorporated as efficiently as the wild-type protein into npBAF complexes (Figure 4A). BAF45a transgenic mice expressing a deletion of the highly conserved d4 domain (Δd4; deletion aa 286–409) showed elevated rates of BrdU incorporation similar to the wild-type protein in neural progenitors of the midbrain and cerebellum (E14.5; Figure 4B and 4C). The number of dividing M-phase H3P+ progenitors was also increased in the ventricular zone of the midbrain of these mice relative to controls (Figure 4C). In the developing cerebellum, mitotic (H3P+) cells were observed within the internal differentiating zone (Figure 4C). In contrast, BAF45a mutants encoding either a deletion of the N-terminal (ΔN-term; deletion aa 1–97) or the Krüppel-like domain (ΔKrüppel-like; deletion aa 204–246) did not induce the proliferation of neural stem/progenitor cells (E14.5; Figure 4B and 4C). Together, these studies indicate that the N-terminal and Krüppel-like domains of the progenitor-specific BAF45a subunit are essential for its ability to induce neural stem/progenitor cell proliferation (Figure 4B and 4C). As these deletion mutant proteins associate with endogenous npBAF complexes (Figure 4A), the N-terminal and Krüppel-like domains of BAF45a likely direct the activity of the complex to a specific program of progenitor proliferation. Interestingly, these domains of BAF45a are the most divergent from the BAF45b and BAF45c neuronal-specific subunits (Figure 1E and Figure 3C), which lack the ability to induce neural stem/progenitor cell proliferation.
Brg is Essential for Neural Progenitor Development
Previous studies reported conflicting results concerning the role of the Brg ATPase subunit in the neural stem cell (NSC) population. In Xenopus, Brg was suggested not to be required for NSC function (Seo et al., 2005a, 2005b) while murine Brg was reported to be necessary for the expression of NSC markers but could not be detected in the developing neural tube before E14 (Matsumoto et al., 2006). In contrast, we detected Brg in periventricular stem/progenitor cells of the developing neural tube from E10.5 onwards using Brg-specific antibodies (H-88 and G-7; Figure 2 and Figure 5B and data not shown). Since Brg is quantitatively associated with BAF45a and BAF53a in neural stem/progenitor cells (Figure 2), we reasoned that we could more thoroughly evaluate the role of the npBAF complexes by Cre-mediated deletion of Brg specifically in neural stem/progenitor cells. To this aim, we bred mice containing a LoxP-flanked allele of Brg (Brgflox) to mice expressing Cre recombinase under the control of the Nestin promoter (Figure 5A) (Sumi-Ichinose et al., 1997; Tronche et al., 1999).
Figure 5. Brg is essential for the self-renewal and maintenance of neural stem/progenitor cells.
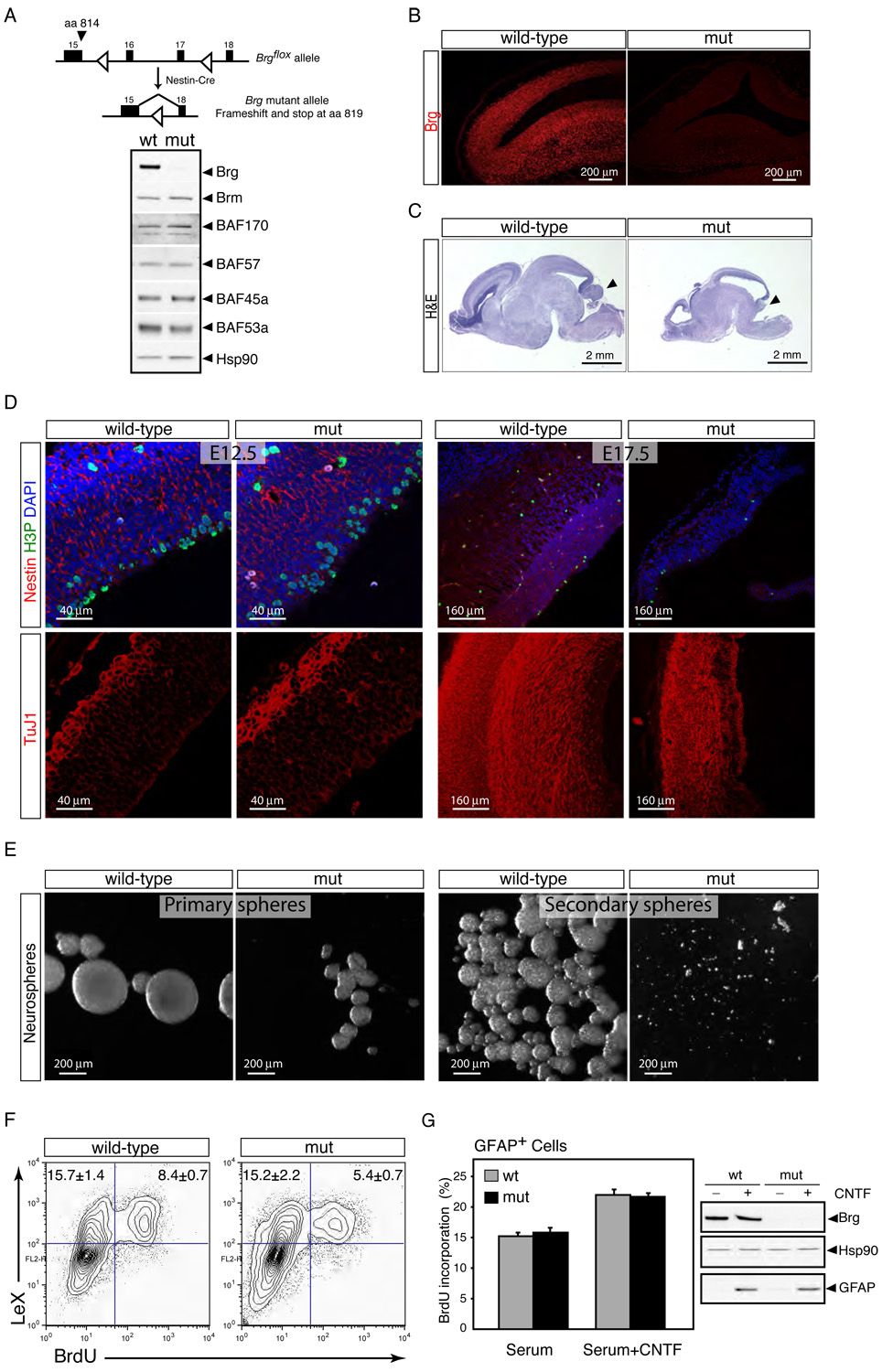
A. Brg deletion does not affect the expression of Brm and other npBAF subunits. Strategy for deletion of Brg in neural progenitors. Genomic structure of the floxed Brg allele from exon 15 through 18 encoding part of the conserved ATPase domain (Sumi-Ichinose et al., 1997). Two LoxP sites (empty arrowheads) were inserted in introns 15 and 17. The Cre recombinase expressed in neural progenitors by the Nestin-Cre transgene recombines the two LoxP sites and results in the deletion of exons 16 and 17. If exon 15 splices to exon 18, translation of the Brg protein stops at aa 819. Western blot analysis of BAF subunits in cellular extracts from E13.5 control and Brg mutant cortices. Hsp90 is a loading control.
B. Brg is deleted in neural stem/progenitor cells by E12.5. Sagittal sections of E12.5 control (wt) and Brg mutant frontal cortices were immunostained with specific anti-Brg antibodies (red). mut, mutant.
C. Brg mutant brains are smaller and lack a cerebellum (arrows). Hematoxylin and eosin (H&E) staining of sagittal sections of E18.5 wt and Brg mutant brains.
D. Progressive depletion of cortical neural progenitors in the Brgf/f;Nestin-Cre mice during embryogenesis. Upper panels: comparable sections of E12.5 and E17.5 cortices stained with anti-Nestin (red), anti-H3P (green) antibodies and DAPI (blue). Note the near absence of Nestin+ cells in E17.5 Brg mutant cortices. Lower panels: similar cortical regions stained with TuJ1 antibody (red).
E. Defective neurosphere formation by Brg-deficient cortical cells. Primary neurospheres were cultured from dissociated E13.5 control and mutant cortices for 7 div. Secondary spheres were cultured from dissociated primary spheres (2 wt neurospheres and 20 Brg mutant neurospheres) for another 4 div.
F. Brg-deficient progenitors proliferate slower than wild-type progenitors. Brg mutant and control E14.5 cortical cells were cultured in serum-containing media for 15 hrs (1 div), stained with anti-LeX and BrdU antibodies and analyzed by flow cytometry. BrdU was added 3 hrs before the analysis. Representative images of n=3 independent experiments performed in duplicate are shown.
G. Brg is not required for glia proliferation in vitro. Brg mutant and control E16.5 cortical cells were cultured in serum-containing media (3 div) with or without CNTF (100 ng/ml), stained with anti-GFAP and BrdU antibodies and analyzed by flow cytometry. Results are expressed as mean ± SD. Western blot analyses of Brg (H-88) and GFAP protein levels in these cultures are shown. CNTF, ciliary neurotropic factor; GFAP, glial fibrillary acidic protein.
In Brgf/f;Nestin-Cre mice, the Brg gene is deleted at E10.5 and protein levels are undetectable by immunostaining in neural stem/progenitors from E12.5 onwards (Figure 5B and data not shown). Most other npBAF subunits (as well as Brm) are expressed at normal levels in the absence of Brg (Figure 5A) and probably incorporated into Brm-based complexes. All Brgf/f;Nestin-Cre mice died at birth without breathing. We found reduced brain size, severe thinning of the neocortex and midbrain and nearly complete absence of the cerebellum in Brgf/f;Nestin-Cre embryos (Figure 5C). Exencephaly was also observed in 6 of 43 Brg heterozygotes at E12.5 (data not shown), indicating that Brg and the family of npBAF complexes that contain Brg have non-redundant and dosage-sensitive roles in neural development.
Brg is Essential for the Self-Renewal/Maintenance of Neural Stem/Progenitor Cells
The cortical thinning observed in Brgf/f;Nestin-Cre mice suggested that the multipotent neural stem/progenitor cells were unable to supply sufficient neurons and/or glial cells to the developing CNS. At E12.5, 12–24 hours after the disappearance of Brg protein, we observed a mild increase in the number of H3P+ M-phase progenitors in the ventricular zone of the Brg-deficient cortices relative to controls (Figure 5D, left panel). This increase in mitotic figures was not due to an M-phase arrest since the percentage of LeX+ cells with a 4N DNA content was not increased in Brg-deficient cultures (data not shown). However, by E15.5, the number of M-phase progenitors in the periventricular zone of Brgf/f;Nestin-Cre cortices was clearly reduced and by E17.5, neural progenitors were virtually absent, as evidenced by absence of H3P and Nestin staining (Figure 5D, right panel). E17.5 Brg mutant cortices were thinner than controls and consisted mostly of post-mitotic differentiated (TuJ1+) neurons (Figure 5D, right panel). A modest increase in the number of apoptotic cells in the ventricular zones of Brg-deficient cortices was also noted at both E12.5 and E15.5 relative to controls (Figure S11). We also observed a significant reduction in the number of GFAP+ astrocytes in Brg mutant cortices at late embryonic stages (i.e. E17.5 and later; data not shown), suggesting a defect at the level of the multipotent neural stem cell population to both neurons and glia. Together, these studies suggest that Brg and the npBAF complexes are initially dispensable for the division of the multipotent neural stem cells (NSCs), but are essential for their self-renewal and/or maintenance.
To directly examine the role of Brg and by inference npBAF complexes in regulating the self-renewal and proliferative capacities of neural stem cells (NSCs), we studied neurosphere formation from single NSCs. Strikingly, when analyzed after 7 days in vitro (div), both the number and size of the neurospheres were severely reduced in Brg-deficient cultures compared to controls, indicating that the proliferative potential of the neurosphere forming-NSC (nf-NSC) is impaired in the absence of Brg (E13.5; Figure 5E, left panels). The number of secondary neurospheres generated by subcloning single primary neurospheres is considered an estimate of self-renewal (expansionary) divisions of the nf-NSC (Reynolds and Weiss, 1996). To determine whether the ability of the Brg-deficient nf-NSCs to self-renew was impaired, individual primary colonies were dissociated and observed for their ability to form secondary neurospheres. In contrast to controls, secondary colonies were rarely generated from the Brg-deficient subcultures (Figure 5E, right panels). These observations are consistent with the progressive depletion of neural progenitors observed in Brgf/f;Nestin-Cre mice and indicate that the Brg-containing npBAF complexes are essential for the self-renewal/proliferative capacity of the multipotent neural stem cells.
To determine if Brg is required for neural stem/progenitor cell proliferation, we measured the rate of BrdU incorporation (3 hrs pulse) in Brg-deficient E14.5 cortical cultures. The percentage of BrdU+ LeX+ neural stem/progenitor cells was significantly reduced in Brg-deficient cultures compared to controls (1 div; Figure 5F). BrdU pulse-labeling experiments performed in vivo provided similar results (data not shown). This indicated that the impaired self-renewal potential of Brg-deficient NSCs might be partially explained by a slower cell cycle. These findings are surprising since both Brg and Brm were previously shown to restrain cellular proliferation by activating p21Cip1/Waf1 and p15INK4b and repressing cyclin gene expression (Coisy et al., 2004; Dunaief et al., 1994; Kang et al., 2004; Strobeck et al., 2000).
To determine if Brg had a specific role in regulating neural stem/progenitor cell proliferation, we analyzed the proliferation rate of cultured Brg-deficient astrocytes (GFAP+) under similar culture conditions as those for neural stem/progenitor cells (E16.5 cortical cultures at 3 div; 4-hour pulse of BrdU). Surprisingly, Brg-deficient astrocytes proliferated normally, even though these cells are the direct descendants of the neural stem/progenitor cells that require Brg for their proliferation (Figure 5G). This suggests that npBAF complexes selectively regulate the self-renewal/proliferation of neural progenitors, but have little role in the proliferation of glia after they have committed to this lineage. In other studies, we find that glia express BAF complexes with a different subunit composition (Figure S4 and GRC and JL, unpublished observation). This specialized function of Brg in neural stem/progenitors is likely mediated through its interaction with the progenitor-specific BAF45a and BAF53a subunits of npBAF complexes, which are both necessary and sufficient for neural stem/progenitor cell proliferation.
Brg Directly Regulates the Expression of Components of the Notch and Sonic Hedgehog Pathways Involved in Neural Development
To understand the molecular mechanism by which Brg and the 2MDa npBAF complexes regulate the self-renewal/proliferation of the NSCs, we defined Brg-dependent genes 12–24 hours after its disappearance, when neural stem and progenitor cells are the predominant cell-types in the nervous system (Figure 6A). RNA from three sets of E12.5 Brgf/f;Nestin-Cre mutant and littermate control telencephalons were prepared and hybridized to transcript arrays. Student’s t-test comparison yielded 440 transcripts with more than 2-fold difference in expression levels and a p value <0.01 (253 genes increased and 187 genes decreased out of 39,000 spots; Figure 6A and data not shown).
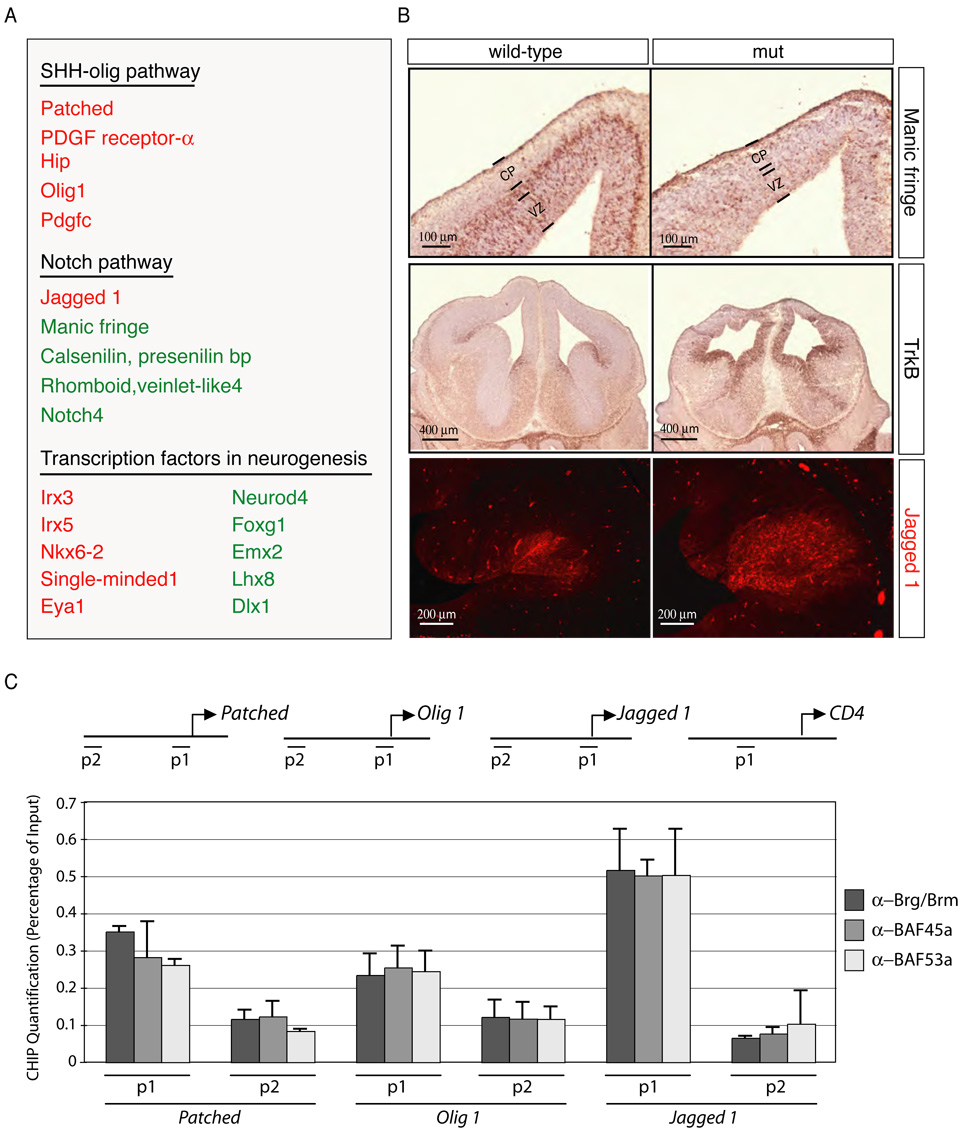
Figure 6. Brg-containing npBAF complexes regulate the transcription of Sonic Hedgehog (SHH) and Notch signaling components in neural development.
A. Microarrays (Mouse 430 2.0 arrays (Affymetrix) containing 39,000 transcripts) were probed with cRNA generated from RNAs isolated from 3 control and 3 Brg-deficient E12.5 telencephalons. Default settings were used to normalize the data and for comparison. Selected genes changed more than two-fold (p<0.01) are shown. Genes that are downregulated in the absence of Brg appear in green and genes that are up-regulated are in red.
B. In situ hybridization and immunostaining analyses of transcriptional targets of npBAF complexes in the E13.5 mouse brain. Positive cells are dark brown (coronal sections). Upper panel: Expression of Manic fringe in newly-born neurons requires Brg. VZ: ventricular zone; CP: cortical plate. Middle panel: Repression of TrkB expression in neural progenitors requires Brg. Lower panel: Increased Jagged1 protein levels in Brg-deficient basal forebrain.
C. BAF45a, BAF53a, Brg and npBAF complexes are present at the promoters of components of the SHH and Notch signaling pathways (P1). P2 regions are ~5 kb upstream. Anti-Brg/Brm, BAF45a and BAF53a antibodies were used for chromatin immunoprecipitation from E13.5 mouse forebrain lysates followed by quantitative PCR. A genomic element of the murine CD4 gene was used as a negative binding control (Chi et al., 2002). The relative binding abundance of each region is presented as percentage of the input DNA material. The data were normalized to the relative abundances of the CD4 P1 region and DNA precipitated by an anti-GFP antibody (negative control) (n=3 independent experiments). Primer sequences are available upon request.
This analysis revealed that Brg is necessary for the normal expression of several components of the Notch signaling pathway, which are essential for the maintenance of the stem/progenitor cell population in the nervous system (Henrique et al., 1997; Hitoshi et al., 2002) (Figure 6A and 6B). Brg is also necessary for the repression of several components as well as downstream targets of the Sonic Hedgehog (SHH) pathway involved in neural development (Figure 6A and 6B) (Chiang et al., 1996; Rowitch et al., 1999). Finally, the expression of several transcription factors involved in neurogenesis, such as NeuroD4, Foxg1, and Emx2, was perturbed in Brg-deficient progenitors (Figure 6A and 6B).
Chromatin immunoprecipitation followed by quantitative polymerase chain reaction (PCR) experiments indicated that Brg, BAF45a, BAF53a and by inference npBAF complexes specifically occupy the promoters of several components of the SHH and Notch signaling pathways including Patched, Olig1 and Jagged1 but not that of the CD4 gene, a direct and functional transcriptional target of BAF complexes in T cells (Chi et al., 2002) (E13.5; P1 regions; Figure 6C and data not shown). Binding was specific since regions only 5 kb further upstream provided a far weaker signal (P2 regions; Figure 6C). These results suggest that Brg-containing npBAF complexes may promote neural stem cell self-renewal/proliferation by enhancing Notch-dependent proliferative signals, while concurrently making the NSC insensitive to SHH-dependent differentiating cues.
Switching Subunits of ATP-Dependent Chromatin Remodeling Complexes is Essential for Neural Development
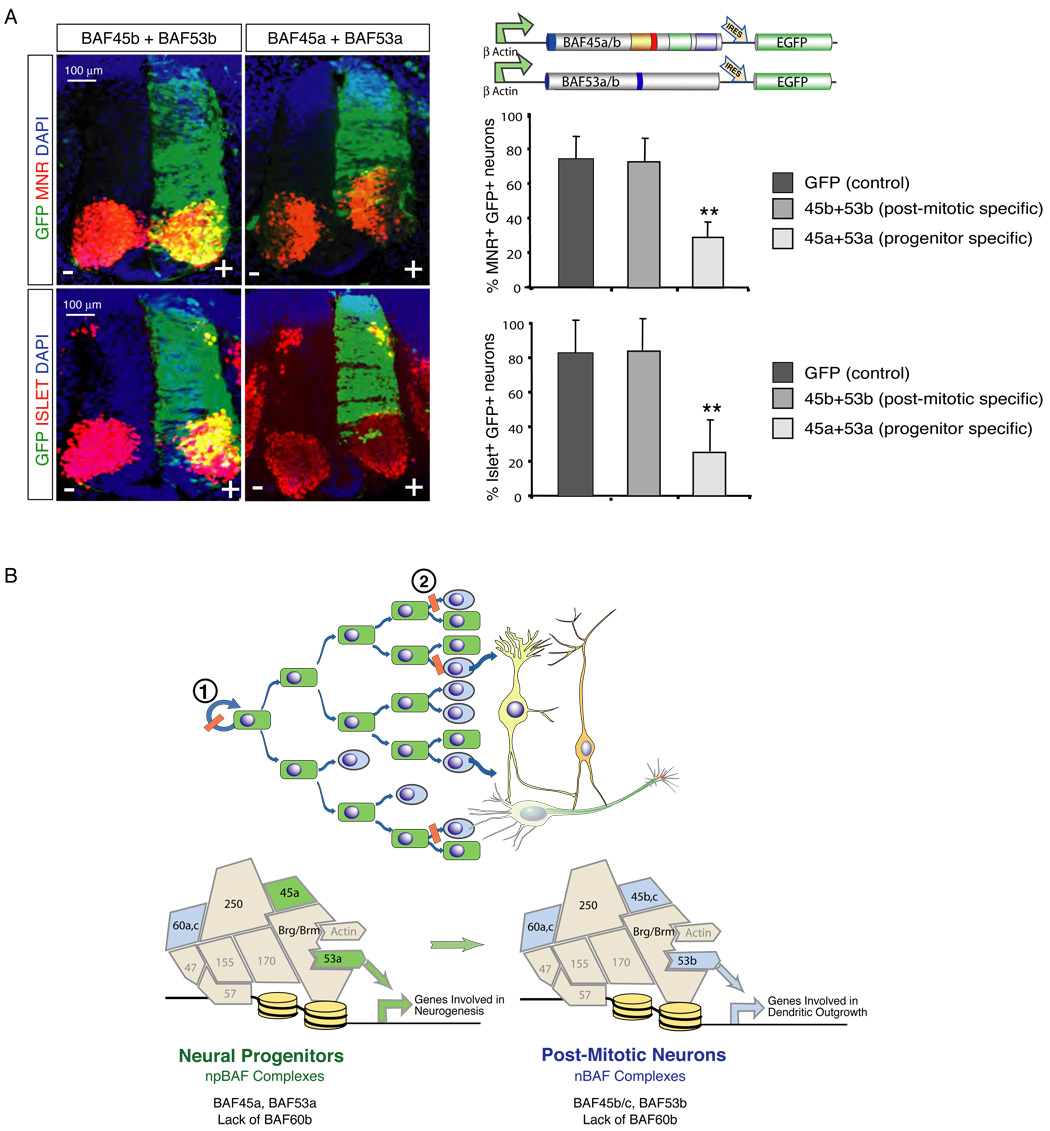
To determine if the exchange of npBAF and nBAF components is essential for neural development, we extended the expression of the progenitor-specific subunits past the time at which they are normally repressed. This was done by directing the expression of BAF45a and BAF53a (with IRES-GFP) under the control of the β-actin promoter in the developing chicken neural tube by in ovo electroporation. The differentiation outcome of multipotent electroporated (GFP+) neural progenitors was analyzed in Hamburger-Hamilton (HH) stage 22–24 embryos (48–50 hrs post-electroporation). Immunoprecipitation experiments revealed that the transfected FLAG-tagged subunits were readily incorporated into endogenous BAF complexes (data not shown). Extending the expression of the BAF45a and BAF53a progenitor-specific subunits after cell-cycle exit was incompatible with the differentiation of specific classes of neurons since GFP-positive cells rarely expressed the MNR and Islet1 determinants of motor neuron differentiation (n= 9 of 10 embryos; Figure 7A). This block of neuronal differentiation was specific to the progenitor subunits since embryos expressing the neuronal subunits, BAF45b and BAF53b had similar numbers of GFP+ MNR and Islet 1 post-mitotic neurons (n=10 of 10 embryos; Figure 7A). The failure of appearance of specific classes of neurons was paralleled by a reduction in the size of the spinal cord on the transfected side of the BAF45a/BAF53a embryos (n= 9/10 embryos), but not of the BAF45b/BAF53b embryos (n=10/10 embryos; Figure 7A and Figure S12). The number of GFP+ cells coexpressing the TuJ1 and NeuN pan-neuronal markers was not significantly affected by ectopic expression of BAF45a and BAF53a in post-mitotic neurons (data not shown). TUNEL assays indicated that these changes cannot be explained by increased cell death (Figure S12). Therefore, suppression of the progenitor-specific BAF45a and BAF53a subunits after cell cycle exit is essential for differentiation of specific classes of neurons in the chick neural tube (Figure 7A).
Figure 7. Repression of BAF45a and BAF53a is essential for neuronal differentiation.
A. Left panels: expression of the post-mitotic-specific BAF53b and BAF45b subunits in neural progenitors of the embryonic chicken spinal cord does not interfere with neuronal differentiation. Representative images are shown. Note the high number of (yellow) double positive GFP+ and MNR+ or Islet+ neurons. Right panels: forced expression of the npBAF progenitor-specific BAF45a and BAF53a subunits in post-mitotic neurons is incompatible with neuronal differentiation. Note the low number of (yellow) double-positive GFP+ and MNR+ or Islet+ neurons and the reduced size of the spinal cord on the transfected side. MNR, Motor neuron reactive cells; Isl, Islet-1/2 reactive cells. A quantification of GFP+ /MNR+ or Islet+ double positive neurons is shown. Results are expressed as mean ± SD. **, p<0.01.
B. A switch in subunit composition of SWI/SNF-like complexes underlies vertebrate neural development. We propose that Brg and npBAF complexes are required for neural stem cell self-renewal and proliferation (1). The exchange of npBAF and nBAF components is essential for the transition from proliferating neural stem/progenitor cells to post-mitotic neurons (2). BAF complexes are drawn in jigsaw puzzle configuration to denote the apparent fit of the subunits within the complexes. For example, BAF53 and actin directly interact with the ATPase domain of Brg (Zhao et al., 1998). However, the position of the other BAF subunits within the complexes has not been experimentally defined.
DISCUSSION
An Epigenetic Switch Controlling Neural Development: the Lexicon of Chromatin Remodeling
Our studies define a previously unknown, but essential epigenetic mechanism underlying the transition from proliferating neural stem/progenitor cells to post-mitotic neurons. We show that the progenitors to both neurons and glia are maintained in a proliferative state by a specialized SWI/SNF-like npBAF complex that contains both BAF45a and BAF53a (Figure 7B, 1). The transition from proliferative to neurogenic divisions requires the exchange of these subunits for the homologous BAF45b or BAF45c and BAF53b proteins (Figure 7B, 2). Remarkably BAF45a, but not the related BAF45b, is sufficient to direct progenitor proliferation within the context of the developing murine nervous system. Extending expression of BAF45a and BAF53a in the neuronal lineage prevents the differentiation of specific classes of neurons, while expression of the homologous BAF45b and 53b subunits in progenitors is compatible with neuronal differentiation. Our studies also show that the developing neural tube lacks BAF60b, which is present in all other tissues examined to date. These findings are consistent with the concept that a specific combination of subunits (i.e. the “word”) rather than the simple presence of a specific subunit (i.e. the “letter”) provides functional specificity to neural progenitor (npBAF) and neuronal (nBAF) complexes based on the Brg ATPase in the developing nervous system.
The npBAF Complex is Essential for Neural Stem Cell Self-Renewal
Neural stem cells (NSCs) are characterized by their ability to generate, in a controlled fashion, all neuronal and glial cell lineages and maintain their original pool through self-renewal. The process of stem cell self-renewal is probably distinguished from conventional proliferation by the need to replicate the genome in a multipotent epigenetic state. Our studies indicate that a chromatin remodeling complex containing Brg, BAF45a and BAF53a (npBAF complex) is essential for the inheritance and maintenance of this unique multipotent epigenetic state. Deletion of the Brg ATPase which is quantitatively associated with BAF45a and BAF53a in neural stem/progenitor cells results in a progressive depletion of the NSC population and subsequent reduction in the number of both neurons and glia. The role of Brg and the npBAF complexes in regulating the self-renewal and proliferative activity of the multipotent NSCs is also illustrated by the reduced number and size of primary neurospheres in Brg-deficient cortical cultures and their inability to form secondary neuropheres when replated. Interestingly, Brg is not a general regulator of cellular proliferation (e.g. glia derived from NSCs proliferate normally in its absence as do mouse embryonic fibroblasts (MEFs) (Bultman et al., 2000)) and has a tumor suppressive role in many cell types (Bultman et al., 2000; Dunaief et al., 1994; Hendricks et al., 2004). The specific role of Brg in regulating the proliferation of neural stem/progenitor cells is likely due to the presence of BAF45a and BAF53a in the complexes, which are both necessary and sufficient for their proliferation (Figure 3).
How might npBAF complexes specifically promote neural stem cell self-renewal and proliferation? We find that the ability of the progenitor-specific BAF45a subunit to induce neural progenitor proliferation requires the Krüppel-like domain and the N-terminus, but not the PHD (d4) domains. These essential domains are the most divergent with the neuronal-specific BAF45b subunit which, although highly homologous, lacks this ability. The Krüppel-like domain of BAF45a probably does not bind DNA, since residues previously shown to be critical for DNA binding are not conserved. BAF53a, the other progenitor subunit, is an actin-related protein (Arp) and is also unlikely to contact DNA (Wang et al., 1996b). Thus, we favor a mechanism in which BAF45a and BAF53a function to allow association of the npBAF complexes with key regulators of neural stem cell self-renewal/proliferation. Bmi-1, a member of the Polycomb-group gene (PcG) chromatin-remodeling family was recently shown to be essential for the long-term maintenance/self-renewal of both neural (Molofsky et al., 2003) and hemopoietic stem cells (Lessard and Sauvageau, 2003; Park et al., 2003). An interesting possibility is that Bmi1-based complexes and npBAF complexes cooperate to establish a chromatin landscape that favors self-renewal and ensures identical multipotency and proliferative capacities to progeny cells.
The function of npBAF complexes in neural stem cell self-renewal/proliferation is likely related to a direct role of these complexes in the coordinated control of cell cycle regulators as well as components of the Notch and Sonic hedgehog (SHH) signaling pathways. Work by Kroll and colleagues indicated that Xenopus Geminin maintains the undifferentiated neural progenitor state by antagonizing Brg’s function in mediating the transcriptional activity of the proneural bHLH Neurogenin (Ngn) and NeuroD proteins (Seo et al., 2005a, 2005b). However, our high stringency purification of the neural BAF complexes did not reveal Geminin, NeuroD or Ngn peptides (data not shown), suggesting either a weak/transient interaction or evolutionary differences between the frog and mouse neural BAF complexes. Alternatively, our studies suggest that Notch signaling, which is essential for the self-renewal/proliferation of stem cells in the nervous system (Henrique et al., 1997; Hitoshi et al., 2002), is altered in Brg−/− multipotent neural progenitors (Figure 6). Moreover, our observation that several components and targets of Sonic Hedgehog signaling are directly repressed by Brg in E12.5 telencephalic cells suggests that npBAF complexes might prevent neural stem cells from sensing SHH signals that would otherwise induce neuronal differentiation and patterning (Chiang et al., 1996; Rowitch et al., 1999).
One of the goals of modern developmental biology is to recapitulate pathways of development for the regeneration of tissues and cell-types. We have demonstrated that a switch from a stem/progenitor to a post-mitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. This switch is driven by chromatin remodeling complexes with distinctive and specialized roles in neural progenitors (this study) and post-mitotic neurons (Wu et al., submitted). These findings raise the possibility that the switch could be reversed by activating progenitor subunits (BAF45a or 53a) or inactivating neuronal subunits (BAF45b or 53b) in adult neurons, thereby allowing them to develop stem/progenitor cell characteristics.
EXPERIMENTAL PROCEDURES
Antibody Production
Polyclonal antibodies against Brg/Brm (J1) and BAF45 (a, b and c) proteins were raised in rabbits using MBP fusion proteins encoding amino acids (aa) 1257 to 1338 of hBrg, aa 96 to 207 or aa 180 to 282 (M2–34) of mBAF45a (NP_077212), aa 95 to 187 of mBAF45b (NP_038902) and aa 95 to 188 of mBAF45c (NP_478119). BAF45a mouse monoclonal antibodies (clones 46 and 75) were raised against aa 180 to 282 of mBAF45a.
Cortical Cell Cultures
E13.5 to E16.5 mouse cortices were trypsinized and dissociated to single cells. Serum-free media (DMEM supplemented with B-27, N2, NAC) was used in neurosphere cultures (with 20 ng/ml FGF2). Media was supplemented with 5% serum for the neural progenitor and astrocyte proliferation assays. Astrocyte differentiation was induced with 100 ng/ml CNTF for 3 days.
Immunohistochemistry and TUNEL Assays
Timed mouse pregnancies were determined by plugging date as day 0.5. Paraffin or frozen sections were stained with anti-Brg (G7 or H-88, Santa Cruz), anti-Nestin (BD; clone rat 401), TuJ1 (Covance), anti-phosphorylated Histone 3 (Ser 10; Upstate), Ki-67 (BD) and anti-NeuN (Chemicon) antibodies and visualized using a Leica SP2 AOBS confocal microscope. TUNEL assays were performed using an In situ Cell Death Detection kit (Roche).
Chicken in ovo Electroporation
Supercoiled plasmid DNA was injected into the lumen of the neural tube of Hamburger-Hamilton (HH) stage 10–12 chicken embryos at 1.0 µg/µl with 50 ng/µl Fast-Green. Electrodes were placed flanking the neural tube and pulsed 5 times at 21V for 50.6 msec. using an ECM 830 Electro Square electroporator (BTX Genetronics). Embryos were harvested 48–50 hrs later (HH stage 22–24), fixed in 4% PFA, incubated in PBS for 15 hrs, cryoprotected in 30% sucrose and embedded in O.C.T compound. 20 µm thick sections were stained using: anti-MNR2 and anti-Islet (Developmental Studies Hybridoma Bank) and Alexa Fluor-594 (Molecular Probes) secondary antibodies. For BrdU incorporation assays, 100 µM BrdU was applied to chick embryos in ovo and incubated 4 hrs at 38.3°C.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to P. Chambon for providing the Brg floxed mouse, A. Imamoto for the Nestin-Cre transgenics and A. Chenn for the Nestin promoter construct. We thank L. Chen and A. Kuo for their technical assistance, S. Donohoe of the ISB proteomics facility for processing the samples, E. Gallo for FACS analysis and R. Vries for critically reading the manuscript. G.R.C is an investigator of the Howard Hughes Medical Institute (HHMI). J.L. is supported by a long-term post-doctoral fellowship from the Human Frontier Science Program (HFSP). These studies have been funded by grants from the National Institutes of Health (NIH) to R.A. (National Heart, Lung and Blood Institute; N01-HV-28179) and to G.R.C. (NS046789).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat.Rev.Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol.Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr.Opin.Genet.Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Coisy M, Roure V, Ribot M, Philips A, Muchardt C, Blanchard JM, Dantonel JC. Cyclin A repression in quiescent cells is associated with chromatin remodeling of its promoter and requires Brahma/SNF2alpha. Mol.Cell. 2004;15:43–56. doi: 10.1016/j.molcel.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, et al. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in protein databases. J.Am.Soc.Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. In Developmental Biology. Seventh Edition. Sunderland, Massachusetts: Sinauer Associates, INC; 2003. The Emergence of The Ectoderm: The Central Nervous System and the Epidermis. [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat.Rev.Mol.Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumor suppression. Mol.Cell Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, Hirsinger E, Adam J, Le R, I, Pourquie O, Ish-Horowicz D, Lewis J. Maintenance of neuroepithelial progenitor cells by Delta-Notch signalling in the embryonic chick retina. Curr.Biol. 1997;7:661–670. doi: 10.1016/s0960-9822(06)00293-4. [DOI] [PubMed] [Google Scholar]
- Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der KD. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr.Opin.Cell Biol. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol.Cell Biol. 2004;24:1188–1199. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal.Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, Nam JS, Kim H, Chung H, Lee HW, Park SD, Seong RH. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol.Cell Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T. Epigenetic alchemy for cell fate conversion. Curr.Opin.Genet.Dev. 2006;16:502–507. doi: 10.1016/j.gde.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von B, I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., III Direct analysis of protein complexes using mass spectrometry. Nat.Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr.Opin.Genet.Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev.Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal.Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol.Cell Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat.Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev.Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- Roegiers F, Jan YN. Asymmetric cell division. Curr.Opin.Cell Biol. 2004;16:195–205. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Jacques B, Lee SM, Flax JD, Snyder EY, McMahon AP. Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J.Neurosci. 1999;19:8954–8965. doi: 10.1523/JNEUROSCI.19-20-08954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H, Kouike H, Okano H. Components of the SWI/SNF complex are required for asymmetric cell division in C. elegans. Mol.Cell. 2000;6:617–624. doi: 10.1016/s1097-2765(00)00060-5. [DOI] [PubMed] [Google Scholar]
- Seo S, Herr A, Lim JW, Richardson GA, Richardson H, Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005a;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005b;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- Strobeck MW, Knudsen KE, Fribourg AF, DeCristofaro MF, Weissman BE, Imbalzano AN, Knudsen ES. BRG-1 is required for RB-mediated cell cycle arrest. Proc.Natl.Acad.Sci.U.S.A. 2000;97:7748–7753. doi: 10.1073/pnas.97.14.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck MW, Reisman DN, Gunawardena RW, Betz BL, Angus SP, Knudsen KE, Kowalik TF, Weissman BE, Knudsen ES. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J.Biol.Chem. 2002;277:4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol.Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat.Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996a;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996b;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yamamichi-Nishina M, Ito T, Mizutani T, Yamamichi N, Watanabe H, Iba H. SW13 cells can transition between two distinct subtypes by switching expression of BRG1 and Brm genes at the post-transcriptional level. J.Biol.Chem. 2003;278:7422–7430. doi: 10.1074/jbc.M208458200. [DOI] [PubMed] [Google Scholar]
- Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.