Abstract
Recently, cases of severe respiratory illness in military and civilian populations have been associated with a new genomic variant of adenovirus (Ad) serotype 14, designated Ad14a. Compared to the Ad14 reference strain (de Wit), this new virus had a deletion of two amino acid residues in the fiber protein knob. Here we tested whether this mutation changed receptor usage of Ad14a compared to Ad14-de Wit. Competition studies with radio-labeled viruses revealed that both Ad14-de Wit and Ad14a used the same receptor which is hitherto unknown. We also found that recombinant fiber knobs only partially blocked attachment of Ad14a, indicating that virus capsid proteins other than the fiber are involved in infection.
Keywords: adenovirus, species B, tropism
Introduction
Human adenoviruses (Ads) have been classified into six species (A to F) currently containing 51 serotypes. Most Ad types utilize the coxsackie-adenovirus-receptor, CAR, as a primary attachment receptor. However, this is not the case for species B Ads. Species B Ads form two genetic clusters, B1 (Ad3, Ad7, Ad16, Ad21, and Ad50) and B2 (Ad11, Ad14, Ad34, and Ad35) (Wadell et al., 1980). Recently, we have suggested a new grouping of species B Ads based on their receptor usage (Tuve et al., 2008; Tuve et al., 2006). Group 1 B-Ads (Ad16, 21, 35, 50) use CD46 as a high-affinity attachment receptor; Group 2 B-Ads (Ad3, 7, 14-deWit) do not productively interact with CD46 (Persson et al., 2008) and instead utilize a yet non-identified receptor (receptor X) for infection; Group 3 B-Ad (Ad11p) preferentially interacts with CD46, but also utilizes receptor X if CD46 is blocked or absent. Species B, group 2 type 14, has been rarely identified or previously associated with severe clinical disease. After its original identification in acute respiratory tract infections among Dutch military recruits in 1955 (Van Der Veen and Kok, 1957), Ad14 was associated with pharyngoconjunctival fever in Great Britain in 1955, Uzbekistan in 1962, and Czechoslovakia in 1963 (Brandt et al., 1969; Chen et al., 2004; Cooper et al., 2000; Horwitz, 1996). Never previously documented in the United States, Ad14 was first reported in March and April 2006 during routine surveillance at several U.S. military-recruit training centers (Metzgar et al., 2007). Individual civilian cases of severe respiratory disease and deaths were identified in California and New York during this same period (Louie et al., 2008). During March – June of the following year, a total of 140 additional cases of confirmed Ad14 respiratory illness were identified in clusters of patients in Oregon, Washington and Texas. Thirty eight percent of these patients were hospitalized, including 17% who were admitted to intensive care units; 5% of patients died (http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5645a1.htm). Independent isolates of this new virus (designated Ad14a) from multiple locations were identical by genome restriction analysis and sequencing of the complete hexon and fiber genes, and were similar to but distinct from the Ad14 reference strain from 1955 (de Wit) (Louie et al., 2008).
Results and Discussion
Within the Ad capsid, the trimeric C-terminal fiber domain, called the fiber knob, is considered the moiety that binds to cellular receptors and mediates attachment of the viral particle to cells (Nicklin et al., 2005). Additionally, interaction between viral penton and cellular integrins, as secondary receptors, is required to trigger virus internalization (Wickham et al., 1993). The prototype Ad14, Ad14-de Wit, and Ad14a are identical in the fiber knob sequences except for a deletion of two amino-acids (Lys and Glu) at aa positions 251/252 found in Ad14a. This mutation is located within the F-G loop directly adjacent to the G-beta sheet (Fig. 1, black arrow). A comparison of Ad14a with other species B Ads revealed that 252Glu is conserved in species B serotypes. Furthermore, a 3D-model of Ad14a that we generated based on the published crystal structures of Ad3, Ad11, Ad16, and Ad35 (Durmort et al., 2001; Pache et al., 2008; Persson et al., 2007; Wang et al., 2007) indicated that the Lys/Glu deletion in Ad14a might change the structure of the FG loop and its proximity to the neighboring knob monomer. Taken these findings together, we speculated that the Lys/Glu mutation within Ad14a might change the fiber structure and thus the receptor usage of Ad14a, which in turn might account for the apparent higher virulence of Ad14a as compared the Ad14-de Wit. To test this, we amplified Ad14-de Wit and Ad14a (strain Portland 2971/2007) in A549 cells and prepared highly purified virus stocks by ultracentrifugation of cell lysates and ammonium-sulfate precipitated culture supernatant in CsCl gradients. Virus was titered as described previously (Lieber et al., 1996). Notably for Ad14a, we found more viral particles in culture supernatants of infected cells than in cell lysates, indicating that progeny Ad14a is more efficiently released from infected cells than Ad14-de Wit progeny. Virus titers were 2.9(+/−0.6) × 1012 viral particles (vp)/ml and 6.1 (+/−0.8) × 1012 vp/ml for Ad14-de Wit and Ad14a, respectively (3 preparations for each virus). The particle-to-pfu (plaque forming units) ratio was 16:1 for both viruses. Viral DNA from purified virus preparations was isolated and fiber knob and penton regions were PCR-amplified and sequenced. We confirmed the 251/252Lys/Glu deletion in Ad14a DNA isolated from purified particles. Additionally, we found an Asp→Asn substitution at Ad14a penton position 366 that has not been reported so far.
Fig. 1. Comparison of amino acid sequence of Ad14-de Wit, Ad14a, and species B Ads 3, 7, 11, and 35.
The β-sheets are marked by a box. The mutation in Ad14a is marked by an arrow. The Genbank accession numbers were AAW33140 (Ad14-de Wit); P35774 (ad11p); AAF14128 (Ad7a); AAA66361 (Ad35p); CAA26029 Ad3.)
First, we performed competition studies for virus attachment to investigate whether Ad14-de Wit and Ad14a use the same receptor(s) (Fig. 2A). As a test cell line, we used 293 cells that are known to allow for efficient Ad14-de Wit attachment (Tuve et al., 2006). 293 cells were incubated on ice with 3H-labeled viruses and, following washing, cell-associated radioactivity was determined. We found that attachment of 3H-Ad14-de Wit (Fig. 2A, left panel) or 3H-Ad14a (Fig. 2A, right panel) was inhibited to the same degree by Ad14-de Wit and Ad14a. This suggests that Ad14-de Wit and Ad14a utilize the same receptor(s). This receptor is not CD46 because anti-CD46 antibodies that block interaction of Ad35 with CD46 (Tuve et al., 2006) did not compete for attachment of 3H-Ad14-de Wit and 3H-Ad14a. Furthermore, the group 1, CD46-interacting serotype Ad35 did not affect the binding of 3H-Ad14-de Wit. On the other hand, pre-incubation of cells with Ad3, a member of species B group 2 Ads, significantly reduced 3H-Ad14-de Wit binding (p<0.05). To further evaluate this, we performed the reverse competition experiment using 3H-labeled Ad3 (and as a control 3H-Ad35) (Fig. 2B). Attachment of 3H-Ad3 was efficiently blocked by pre-incubation with Ad14-de Wit but not by pre-incubation with Ad35 virus or anti-CD46 antibodies as shown before (Tuve et al., 2006). Along this line, Ad14-de Wit and Ad3 did not block binding of the CD46-interacting 3H-Ad35. In summary, these data show that Ad14-de Wit and Ad14a recognize the same receptor (which is not CD46) and that this receptor is also used by Ad3 (Tuve et al., 2008; Tuve et al., 2006). We therefore concluded that Ad14a uses the hitherto unidentified receptor X for attachment. To study quantitative differences in Ad14-de Wit and Ad14a binding to test cells, we measured the avidity of both viruses as described previously using Scatchard blots (Fig. 2C) (Tuve et al., 2006). The Ka for Ad14-de Wit and Ad14a were 5.438×109M−1 and 9.062×109M−1, respectively. Therefore, Ad14a binds to 293 cells at a 1.67-fold higher affinity than Ad14-de Wit.
Fig. 2. Competition of viruses for attachment on 293 cells.
A and B) 293 cells were pre-incubated with Ad14-de Wit, Ad14a, Ad3, Ad35 or 10μg/ml CD46 antibody (MEM-258, Serotec) for one hour on ice. After washing, 3H-Ad14-de Wit, (Fig 2A, left panel), 3H-Ad14a, (Fig. 2A, right panel), 3H-Ad3, (Fig. 2B, left panel) or 3H-Ad35 (Fig. 2B, right panel) was added for another hour on ice. After washing, cell-associated radioactivity was determined. N=3. Bars indicate the means. Standard deviation was less than 10% in all cases.
C) Affinity of Ad14-de Wit and Ad14a to 293 cells. To generate Scatchard blots, cells were incubated with increasing MOIs of 3H-Ad14-de Wit or 3H-Ad14a and the number of cells associated viral particles was measured after one hour of incubation on ice. The y-axis shows the ratio between bound particles to total input particles minus bound particles. The x-axis shows the number of bound particles. The binding affinities (Ka) of virus were calculated on the basis of the slope with standard Excel software as described previously (Tuve et al., 2006).
So far, in our competition studies we used complete virus particles. Recent studies by others and our laboratory indicate an interaction of Ad capsid proteins other than the fiber can mediate cell infection (Tuve et al., 2008; Waddington et al., 2008). We therefore studied the role of the fiber knob in Ad14-de Wit and Ad14a attachment. Recombinant fiber knobs of Ad35, A14-de Wit, and Ad14a were produced in E. coli. Knob protein was analyzed by polyacrylamide gel electrophoresis (Fig. 3A, left panel) and Western blot (Fig. 3A, right panel). All knobs formed trimeric proteins which could be disrupted into monomeric knob forms by heat denaturation. This study also excluded that the 251/252 Lys/Glu mutation in the Ad14a fiber knob affects fiber trimerization. After electrophoresis, knob proteins were blotted onto nitrocellulose membranes and incubated with soluble CD46 (sCD46). sCD46 binding was visualized using mouse anti-CD46 antibody followed by incubation with an anti-mouse HRP conjugate and ECL detection as described earlier (Wang et al., 2007). Only Ad35 fiber knob demonstrated detectable binding to sCD46 (Fig. 3A, right panel), corroborating our observation that Ad14-de Wit and Ad14a do not bind to CD46. We then used the recombinant Ad14-de Wit and Ad14a knobs as competitors in virus attachment studies to assess the role of the fiber knob in Ad14-de Wit and Ad14a attachment (Fig. 3B). Test cells were incubated with 0.4μg or 4μg knob protein for 1 hour on ice. Then, 3H-Ad14-de Wit or 3H-Ad14a were added at an MOI of 8,000 vp/cell for one hour. Cells were washed and cell-associated virus radioactivity was measured. Both Ad14-de Wit knob and Ad14a knob blocked attachment of Ad14-de Wit and Ad14a virus. In the presence of 0.4μg Ad14-de Wit and Ad14a knobs, Ad14-de Wit attachment was reduced by 65.1 and 68.3%, respectively. Ad14a attachment decreased 75.2 and 75.5% after Ad14-de Wit knob and Ad14a knob incubation, respectively. A 10-fold higher concentration of knobs (4μg) did not proportionally decrease Ad14-de Wit or Ad14a attachment. Notably, in previous studies with Ad5 and Ad35 viruses (Tuve et al., 2006), we found that the corresponding Ad5 and Ad35 knobs inhibited virus binding more than 95% at concentrations lower than 0.4 μg (Tuve et al., 2006). The inability of Ad14-de Wit and Ad14a knob to completely block Ad binding is similar to what we observed for Ad3 knob/Ad3 virus (Tuve et al., 2008). In a previous study, we found that the Ad3 fiber knob binds to cells only with low affinity, implying that the main contribution of high affinity Ad3 virus binding to cells is due to interaction of virus capsid proteins other than the fiber knob domain (such as the fiber shaft, penton base, or hexon) with the unidentified receptor X. The competition data in this study strongly indicate that Ad14-de Wit, as well as Ad14a, use a similar binding mechanism. We are currently attempting to identify this/these receptor/s X using whole Ad3, Ad14-de Wit and Ad14a viral particles for pull-down assays and subsequent mass spectrometry analysis as done earlier for the identification of CD46 as a species B Ad receptor (Gaggar, Shayakhmetov, and Lieber, 2003; Gaggar, Shayakhmetov, and Lieber, 2007).
Fig. 3. Fiber knob competition Ad14-de Wit and Ad14a attachment.
A) Analysis of recombinant Ad fiber knobs. Purified Ad35, Ad14-de Wit, and Ad14a knob protein (1μg/lane) was run as native protein (N) or after denaturation (D) on a polyacrylamide gel. Coomassie brilliant blue staining (left panel) revealed the trimeric knob form, which is converted into monomers after boiling. Western blots (right panel) were analyzed for CD46 binding to fiber knobs by subsequent incubation with sCD46, anti-CD46 Mab, and anti-mouse IgG-HRP.
B) Fiber knob competition. Cells were pre-incubated with 0.4μg or 4μg knob protein on ice for 1 hour. Then 3H-Ad14-de Wit or 3H-Ad14a virus was added and cell- associated radioactivity was measured after 1 hour incubation. N=3. Bars indicate the means. Standard deviation was less than 10% in all cases.
There is one report suggesting that Ad14 can bind to and infect CD46-expressing cells (Marttila et al., 2005). Although this is possible on cells with high CD46 density, it appears that the Ad14 knob–CD46 interaction has a low affinity and is unstable, indicating that Ad14 is unlikely to use CD46 productively for infection. (Persson et al., 2008). Because a series of studies suggest a crucial role of an arginine in position 279 of Ad fiber knob in conferring stable binding to CD46 (Gustafsson et al., 2006; Persson et al., 2008), we tested whether we can convert Ad14-de Wit and Ad14a into CD46-binding viruses by substituting 279Gln (see Fig. 1, empty arrow) with Arg. In Western blots we were unable to detect sCD46 binding of both Ad14-de Wit and Ad14a knobs containing the Gln279Arg mutation (data not shown). Furthermore, these mutants did not compete with 3H-Ad35 for binding and the level of binding inhibition of 3H-Ad3, 3H-Ad14-de Wit, and 3H-Ad14a did not differ from that of corresponding non-mutated knobs. These data suggest that stable interaction of a fiber knob with CD46 involves structural features other than the Arg in position 279.
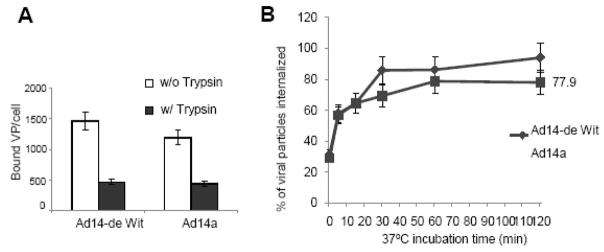
In this study, we also tested how the higher affinity of Ad14a affects subsequent infection steps. For Ad5, and apparently also for species B Ads, following initial attachment, RGD motifs within the penton base interact with cellular integrins triggering endocytosis of Ad particles (Murakami et al., 2007; Wickham et al., 1993). To study Ad14-de Wit and Ad14a internalization, test cells were incubated 3H-Ad14-de Wit or 3H-Ad14a virus on ice to allow for attachment. Then, cells were moved to a 37°C incubator for the indicated time periods (Fig. 4). Cells were then washed with PBS and incubated with trypsin at 37°C for 20 minutes to remove surface bound virus. The number of internalized viral particles was measured based on cell associated radioactivity. A preliminary study showed that trypsin treatment of cells that were incubated with virus on ice efficiently removed attached particles (Fig. 4A). Overall, our internalization study shown in Fig. 4B did not reveal significant differences between Ad14-de Wit and Ad14a. Taken together, our data indicate that the 251Lys/252Glu deletion in the Ad14a fiber and the 366Asp→Asn mutation in the Ad14a penton did not significantly influence the attachment and internalization of this virus. Most likely, differences in post-internalization steps or in the ability to elude the host immune response account for the higher virulence of Ad14a compared to Ad14-de Wit. Based on the findings of this study we group Ad14a into species B group 2, which so far consisted of Ad3, Ad7p and Ad14-de Wit.
Fig. 4. Internalization of Ad particles.
A) Cells were detached from culture dishes by incubation with versene, washed with PBS, and chilled on ice for 45 minutes. A total of 2 × 105 cells per tube were resuspended in 150μl of ice-cold adhesion buffer containing 8,000VP/cell of 3H-labeled wild-type Ad14-de Wit or Ad14a virus. After 60 minutes of incubation on ice, cells were washed with PBS to remove unbound virus, one set of samples were incubate with trypsin at 37°C for 20 minutes (w/Trysin) the other sample with PBS (w/o Trypsin). Cells were washed and cell associated radioactivity was measured. B) Cells were incubated with 3H-Ad14-de Wit or 3H-Ad14a virus for 10 minutes on ice and then at 37°C for the indicated time. Non-internalized virus particles were removed by 20 minutes of trypsin treatment of cells and cell-associated radioactivity was measured. The Y-axis shows the percentage of viral particles internalized compared to the number of viral particles bound per cell. (N=3)
Materials and Methods
Cell lines
293 (Microbix, Toronto, Ontario, Canada), A549, and HeLa cells (American Type Culture Collection, ATCC) were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and with 2mM L-glutamine, 100 U penicillin/ml, and 100μg streptomycin/ml (Pen-Strep).
Viruses
Ad3 (GB strain) and Ad35 (Holden strain) were obtained from the ATCC. Ad14a (strain Portland 2971/2007) and Ad14-de Wit were provided by the Center for Disease Control and Prevention (Atlanta, GA). Ads were propagated in A549 cells, methyl-3H thymidine-labeled, purified, dialyzed and stored in aliquots as described elsewhere (Lieber et al., 1996; Shayakhmetov et al., 2000). Wild-type Ad particle (viral particle, VP) concentrations were determined spectrophotometrically by measuring the optical density at 260 nm (OD260) and plaque titering (plaque forming units, pfu) was performed using 293 cells as described elsewhere (Shayakhmetov et al., 2000). The virion-specific radioactivity was measured by a liquid scintillation counter and was always in the range of 10−5 to 10−4 cpm per virion.
Sequencing of Ad14a fiber and penton genes
The genes for Ad14a fiber knob and penton were PCR-amplified using the following primers.
Ad14 fiber knob forward:
P1:
5′ CAGACTGGATCCAATTCAAACAACATTTGCATTGATGACAATATTAACACC
Ad14 fiber knob reverse:
P2:
5′ AGACTAAGCTTTCAGTCGTCTTCTCTGATGTAGTAAAAGGTAAATGGGGAGGTAACTAGG
Ad14 penton forward:
5′ AAAGGAAAACTCACCAAGGCCATGGCGACGAGCGTACG
Ad14 penton reverse:
5′ GTTTGCGTGCGCCTCCGTACATCTTGCTTGGAG
PCR products was cloned into pGEM-T Easy (Promega, Madison, WI) and sequenced.
Virus attachment assays
Cells were detached from culture dishes by incubation with Versene and washed with PBS. A total of 105 cells/tube were resuspended in 100μl of ice-cold adhesion buffer containing 100,000 vp/cell of “cold” competitor Ad, 10μg/ml of monoclonal antibody directed against CD46 (clone MEM-258; Serotec), or 0.4 or 4 μg of Ad fiber knob. Cells were incubated with competitors for one hour on ice. After washing with ice-cold PBS, 3H-thymidine labeled Ads in adhesion buffer were added at an MOI of 8,000 vp/cell and cells were incubated for one hour on ice. Following washing, cell-associated radioactivity was determined. Background scintillation was determined using cells that were not incubated with 3H-labeled Ad. Background scintillation was subtracted from scintillation of 3H-labeled Ad incubated samples.
Scatchard blots
Virus affinity was measured as described elsewhere (Tuve et al., 2006). In brief, 293 cells were incubated on ice for one hour with different amounts of 3H-Ad14-de Wit or 3H-Ad14a virus (10,000; 20,000; 50,000; 100,000; 200,000; 400,000; and 800,000 vp/cell). Scatchard plots were constructed. The binding affinities (Ka) of virus were calculated on the basis of the slope with standard Excel software.
Production of Ad14 fiber knobs
The coding sequence of the Ad14 fiber knob was obtained by PCR from Ad14-de Wit or Ad14a viral DNA using the primers P1 and P2. The PCR product was cloned into pQE30 (QIAGEN, Valencia, CA) BamH/HindIII site. To introduce the Q279R mutation into the Ad14a and Ad14-deWit fiber knob sequence, a two-step PCR amplification was performed. In the first step P1 and P3 (5′-TCTTATTGCTCTTCGGTTAAGCATGACAGATATGTCAATG-3′) or P2 and P4 (5′-CCATTGACATATCTGTCATGCTTAACCGAAGAGCAATAAGAGCTGATAC-3′) was used as primer and Ad DNA as template. The corresponding two PCR products were purified and combined as template for a second PCR, using P1 and P2 as primers. The knob domains were produced in E. coli with N-terminal tags of six consecutive histidine residues (6-HIS) and purified by Ni-NTA agarose chromatography as described elsewhere (Wang et al., 2007). The fiber knob proteins were dialyzed against 20mM Hepes, 200mM NaCl, 17% glycerol.
Western blot
Recombinant knobs (1μg respectively) were separated by polyacrylamide gel electrophoresis. Protein samples were loaded in loading buffer (50mM Tris-HCl, pH6.8, 100mM dithiothreitol, 2% sodium dodecyl sulfate, 10% glycerol, 0.2% bromophenol blue) with or without boiling. Gels were either stained with Coomassie blue or transferred onto nitrocellulose membranes. To detect whether recombinant Ad knobs bind to CD46, the blot was incubated with sCD46 in TBS (10mM Tris-HCl, pH 7.5, 150mM NaCl) and 3% blotting grade milk (BIO-RAD, Hercules, CA) for 1h at room temperature (RT) and then washed three times for 10 min in TBS-0.05% Tween20 (TBS-T) buffer. The blot was then incubated with anti-CD46 antibody (clone J4.48; Fitzgerald, Concord, MA) (1:50) in TBS and 3% milk for 1h at RT and then washed three times for 10 min in TBS-T buffer. To visualize binding, the blot was incubated with goat anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP) (BD Pharmingen) (1:1000) in TBS and 3% dry milk for 1h at RT. Filters were washed three times for 10 min in TBS-T buffer and subjected to enhanced chemiluminescence substrate (Pierce, Rockford, IL).
Virus internalization
To study Ad14 internalization, test cells were incubated with 10,000 vp/cell of 3H-Ad14-de Wit or 3H-Ad14a virus for 10 minutes on ice to allow for attachment. Then, cells were moved to a 37°C incubator for the indicated time periods. Cells were then washed with PBS and incubated with trypsin at 37°C for 20 minutes to remove surface bound virus. Cells were washed twice to remove trypsin and unbound virus and the number of internalized viral particles was measured based on cell associated radioactivity.
Acknowledgments
This work was supported by NIH grant HLA078836. We thank Daniel Stone for help with adenovirus 3D-models and Roma Yumul for editing the manuscript.
The opinions expressed by the authors contributing to this manuscript do not necessarily reflect the opinions of the Centers of Disease Control and Prevention.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brandt CD, Kim HW, Vargosko AJ, Jeffries BC, Arrobio JO, Rindge B, Parrott RH, Chanock RM. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. I. Adenovirus pathogenicity in relation to serologic type and illness syndrome. Am J Epidemiol. 1969;90 (6):484–500. doi: 10.1093/oxfordjournals.aje.a121094. [DOI] [PubMed] [Google Scholar]
- Chen HL, Chiou SS, Hsiao HP, Ke GM, Lin YC, Lin KH, Jong YJ. Respiratory adenoviral infections in children: a study of hospitalized cases in southern Taiwan in 2001--2002. J Trop Pediatr. 2004;50 (5):279–84. doi: 10.1093/tropej/50.5.279. [DOI] [PubMed] [Google Scholar]
- Cooper RJ, Hallett R, Tullo AB, Klapper PE. The epidemiology of adenovirus infections in Greater Manchester, UK 1982–96. Epidemiol Infect. 2000;125(2):333–45. doi: 10.1017/s0950268899004550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmort C, Stehlin C, Schoehn G, Mitraki A, Drouet E, Cusack S, Burmeister WP. Structure of the fiber head of Ad3, a non-CAR-binding serotype of adenovirus. Virology. 2001;285 (2):302–12. doi: 10.1006/viro.2001.0967. [DOI] [PubMed] [Google Scholar]
- Gaggar A, Shayakhmetov D, Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nature Medicine. 2003;9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- Gaggar A, Shayakhmetov D, Lieber A. Identifying functional adenovirus-host interactions using tandem mass spectrometry. Methods Mol Med. 2007;131:141–55. doi: 10.1007/978-1-59745-277-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson DJ, Segerman A, Lindman K, Mei YF, Wadell G. The Arg279Gln [corrected] substitution in the adenovirus type 11p (Ad11p) fiber knob abolishes EDTA-resistant binding to A549 and CHO-CD46 cells, converting the phenotype to that of Ad7p. J Virol. 2006;80 (4):1897–905. doi: 10.1128/JVI.80.4.1897-1905.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz MS. Adenoviruses. In: Fields BN, Knipe DM, Howley PM, editors. Virology. Vol. 2. Lippincott; Philadelphia: 1996. pp. 2149–2171. [Google Scholar]
- Lieber A, He CY, Kirillova I, Kay MA. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J Virol. 1996;70 (12):8944–60. doi: 10.1128/jvi.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie JK, Kajon AE, Holodniy M, Guardia-LaBar L, Lee B, Petru AM, Hacker JK, Schnurr DP. Severe pneumonia due to adenovirus serotype 14: a new respiratory threat? Clin Infect Dis. 2008;46 (3):421–5. doi: 10.1086/525261. [DOI] [PubMed] [Google Scholar]
- Marttila M, Persson D, Gustafsson D, Liszewski MK, Atkinson JP, Wadell G, Arnberg N. CD46 is a cellular receptor for all species B adenoviruses except types 3 and 7. J Virol. 2005;79 (22):14429–36. doi: 10.1128/JVI.79.22.14429-14436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzgar D, Osuna M, Kajon AE, Hawksworth AW, Irvine M, Russell KL. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J Infect Dis. 2007;196 (10):1465–73. doi: 10.1086/522970. [DOI] [PubMed] [Google Scholar]
- Murakami S, Sakurai F, Kawabata K, Okada N, Fujita T, Yamamoto A, Hayakawa T, Mizuguchi H. Interaction of penton base Arg-Gly-Asp motifs with integrins is crucial for adenovirus serotype 35 vector transduction in human hematopoietic cells. Gene Ther. 2007;14 (21):1525–33. doi: 10.1038/sj.gt.3303019. [DOI] [PubMed] [Google Scholar]
- Nicklin SA, Wu E, Nemerow GR, Baker AH. The influence of adenovirus fiber structure and function on vector development for gene therapy. Mol Ther. 2005;12 (3):384–93. doi: 10.1016/j.ymthe.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Pache L, Venkataraman S, Reddy VS, Nemerow GR. Structural variations in species B adenovirus fibers impact CD46 association. J Virol. 2008;82(16):7923–31. doi: 10.1128/JVI.00754-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson BD, Muller S, Reiter DM, Schmitt BBT, Martilla M, Sumowski CV, Schweizer S, Scheu U, Ochsenfeld C, Stehle T. An arginine switch in the species B adenovirus knob determines high-affinity engagement of the cellular receptor CD46. Journal of Virology. 2008 doi: 10.1128/JVI.01967-08. publication ahead of print, Nov 5, 2008, doi: 10.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson BD, Reiter DM, Marttila M, Mei YF, Casasnovas JM, Arnberg N, Stehle T. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat Struct Mol Biol. 2007;14 (2):164–6. doi: 10.1038/nsmb1190. [DOI] [PubMed] [Google Scholar]
- Shayakhmetov DM, Papayannopoulou T, Stamatoyannopoulos G, Lieber A. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J Virol. 2000;74 (6):2567–83. doi: 10.1128/jvi.74.6.2567-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuve S, Wang H, Jacobs JD, Yumul RC, Smith DF, Lieber A. Role of cellular heparan sulfate proteoglycans in infection of human adenovirus serotype 3 and 35. PLoS Pathog. 2008;4 (10):e1000189. doi: 10.1371/journal.ppat.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuve S, Wang H, Ware C, Liu Y, Gaggar A, Bernt K, Shayakhmetov D, Li Z, Strauss R, Stone D, Lieber A. A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. J Virol. 2006;80(24):12109–20. doi: 10.1128/JVI.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Veen J, Kok G. Isolation and typing of adenoviruses recovered from military recruits with acute respiratory disease in The Netherlands. Am J Hyg. 1957;65(2):119–29. doi: 10.1093/oxfordjournals.aje.a119860. [DOI] [PubMed] [Google Scholar]
- Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H, Pink R, Buckley SM, Greig JA, Denby L, Custers J, Morita T, Francischetti IM, Monteiro RQ, Barouch DH, van Rooijen N, Napoli C, Havenga MJ, Nicklin SA, Baker AH. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132(3):397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Wadell G, Hammarskjold ML, Winberg G, Varsanyi TM, Sundell G. Genetic variability of adenoviruses. Ann N Y Acad Sci. 1980;354:16–42. doi: 10.1111/j.1749-6632.1980.tb27955.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Liaw YC, Stone D, Kalyuzhniy O, Amiraslanov I, Tuve S, Verlinde CL, Shayakhmetov D, Stehle T, Roffler S, Lieber A. Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob. J Virol. 2007;81 (23):12785–92. doi: 10.1128/JVI.01732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73 (2):309–19. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]