Abstract
Freshwater planarians exhibit a striking power of regeneration, based on a population of undifferentiated totipotent stem cells, called neoblasts. These somatic stem cells have several characteristics resembling those of germ line stem cells in other animals, such as the presence of perinuclear RNA granules (chromatoid bodies). We have isolated a Tudor domain-containing gene in the planarian species Schmidtea polychroa, Spoltud-1, and show that it is expressed in neoblast cells, germ line cells and central nervous system, and during embryonic development. Within the neoblasts, Spoltud-1 protein is enriched in chromatoid bodies. Spoltud-1 RNAi eliminates protein expression after 3 weeks, and abolishes the power of regeneration of planarians after 7 weeks. Neoblast cells are eliminated by the RNAi treatment, disappearing at the end rather than gradually during the process. Neoblasts with no detectable Spoltud-1 protein are able to proliferate and differentiate. These results suggest that Spoltud-1 is required for long term stem cell self renewal.
Keywords: Planarian, Schmidtea polychroa, Stem cells, Neoblasts, Regeneration, Tudor, Nuage, Chromatoid bodies, Germ line
Introduction
Planarian flatworms have an impressive power of regeneration. An undifferentiated population of totipotent stem cells known as neoblasts underlies this process (Baguñà et al., 1989; Handberg-Thorsager et al., 2008; Rossi et al., 2008; Sanchez Alvarado and Kang, 2005). These cells are able to give rise to all differentiated cell types in the adult planarian and are maintained throughout the animal's life. Neoblasts are defined on the basis of their proliferative capabilities and their morphological features, such as lack of differentiated structures, relatively uncondensed chromatin and high nucleocytoplasmic ratio. Neoblasts are scattered throughout the mesenchyme of the animal with the exception of the pharynx, located in the central region of the body, and the region anterior to the photoreceptors. Analysis of proliferation markers such as PCNA immunohistochemistry (Orii et al., 2005), mcm2 in situ hybridization (Salvetti et al., 2000), anti-phospho-histone 3, and bromodeoxyuridine labeling (Newmark and Sanchez Alvarado, 2000) show a similar pattern to that of neoblasts, since they are the only proliferative cell type in asexual planarians. Furthermore, neoblasts and all these markers are specifically eliminated after high doses of irradiation, a behavior typical of proliferative cells. Thus, proliferation markers are a powerful tool in order to identify neoblast cells.
A particular hallmark of neoblast cells is the presence of structures called chromatoid bodies, electron-dense perinuclear granules composed of RNA and protein (Coward, 1974; Hori, 1982; Morita and Best, 1984; Morita et al., 1969). Chromatoid bodies are also present in planarian germ cells and in blastomeres during embryonic development (Cardona et al., 2005b, 2006) and resemble germ granules, often called nuage, present in germ line cells from most animals (Eddy, 1975; Raz, 2000; Saffman and Lasko, 1999). The presence of these structures not only in the germ line but in totipotent somatic stem cells suggest a role for chromatoid bodies in establishing or maintaining totipotency or an undifferentiated state (Auladell et al., 1993). However, the precise role of chromatoid bodies in planarian stem cell biology is not clear yet.
Interestingly, apparent orthologues of several genes that encode components of nuage, or that required for germ line establishment in more complex metazoans, are required for planarian neoblast function. Smed-bruno-like (bruli) is a RNA-binding protein that has been recently shown to be required for stem cell maintenance in the planarian Schmidtea mediterranea (Guo et al., 2006). Previously, two genes belonging to the PIWI/Argonaute family, smedwi-1 and smedwi-2 were identified in the same species, and smedwi-2 was directly implicated in neoblast differentiation control (Reddien et al., 2005). An additional piwi gene with a similar function, smedwi-3, has been recently found (Palakodeti et al., 2008). A pumilio homologue, DjPum, was found in the planarian Dugesia japonica, and was also shown to be involved in planarian stem cell biology (Salvetti et al., 2005). All of these genes are expressed in mesenchymal cells with a pattern similar to that of neoblasts and in the germ line, and abolish regeneration capacity when inhibited through RNAi.
In Drosophila melanogaster, the germ line is specified through preformation (Extavour and Akam, 2003), meaning that it depends upon the formation of a specialized cytoplasm that is derived from the products of genes expressed maternally during oogenesis. In Drosophila, oskar RNA accumulates at the posterior pole of the developing oocyte, and nucleates the formation of pole plasm, which shares several components with nuage. Vasa and Tudor proteins are localized in this region in an Oskar-dependent manner and, like Oskar, both are essential for germ line specification (Boswell and Mahowald, 1985; Lasko and Ashburner, 1988). No homologue of oskar has been found in a non-dipteran species so far. However, a PL10 gene (closely related to vasa) was identified in the planarian species D. japonica, and its expression is described to be enriched in mesenchymal neoblasts (Shibata et al., 1999).
Tudor contains a number of repeated elements called Tudor domains (Thomson and Lasko, 2005). Different Tudor domain-containing proteins have been described in mice and humans and, among them, TDRD1, TDRD6 and TDRD7 have been shown to have a role in the germ line (Chuma et al., 2006; Hosokawa et al., 2007) although their orthology relationships are not clear. The Tudor domain binds symmetrical dimethylated arginines (Cote and Richard, 2005), and in D. melanogaster Tudor interacts with Valois, a component of the methylosome (Anne et al., 2007). Some protein arginine methyltransferases such as capsuleen in D. melanogaster and its homologue in mouse, PRMT5, have been demonstrated to have a role in germ line formation (Ancelin et al., 2006; Anne et al., 2007), suggesting that Tudor domains and symmetrical dimethylated arginines could have a crucial role in germ line biology.
Due to the close relationship between the metazoan germ line and planarian stem cells we performed Tudor domain searches in planarian databases in order to check their possible roles in neoblast cells. Here we present the isolation of spoltud-1, a Tudor domain-containing gene in planarians, and show that it is expressed in neoblast cells. Animals periodically injected with spoltud-1 dsRNA are able to regenerate during the first weeks but fail to produce blastemata after 2 months of treatment. Consistently, they dramatically lose their neoblasts at the end of this treatment, suggesting a role for spoltud-1 in the long-term maintenance of neoblasts.
Materials and methods
Animals
We selected S. polychroa as species of work. S. polychroa is convenient in order to study embryonic development and provides the possibility of comparing regeneration data to available embryonic development data (Martin-Duran et al., 2008; Solana and Romero, 2009). Furthermore, this species is closely related to S. mediterranea, for which there is an ongoing genome project. Sexual S. polychroa adults were collected in Sot de Ferrer (Castelló, Spain). Animals were kept in glass containers with 1:1 tap water:distilled water, at 17 °C in the dark, fed every week and starved at least 1 week prior to experiments. Sexually immature juvenile animals were selected for some experiments.
Isolation of spoltud-1
To identify S. polychroa genes containing Tudor domains we performed tBLASTn searches using several Tudor domains as query against the closely related species S. mediterranea genome. This ongoing genome project corresponds to a species sharing an extremely high similarity with S. polychroa. We found several candidate genes containing Tudor domains, based on their predicted tertiary structure, and on the basis of their reciprocal blast hits to Drosophila tudor and mammalian TDRD1 and TDRD6 we finally selected spoltud-1 and spoltud-2 for further experiments (Accession numbers FJ655915 and FJ655916, respectively). We designed specific primers against S. mediterranea and used them to amplify the genes in a S. polychroa cDNA, made from total RNA using MMLV reverse transcriptase (Promega). In order to obtain the full cDNA sequence of spoltud-1 RACE reactions were carried out adding a poly-A tail to the cDNA with TdT enzyme (New England Biolabs) and amplifying it by PCR by means of an anchor primer. The list and position of primers used (5′-3′) are indicated in Fig. S1. Probes for spoltud-1 in situ hybridization were generated from primers 3 and 4, dsRNA for RNAi was generated from primers 1 and 2.
In situ hybridization
Whole mount in situ hybridizations and in situ hybridization on paraffin sections were carried out as described previously (Cardona et al., 2005a; Umesono et al., 1997; Zayas et al., 2005). Fluorescent in situ hybridizations were incubated overnight with either antidigoxigenin-POD or antibiotin-POD (Roche) at a 1:100 dilution and developed with TSA system (Perkin Elmer). For double fluorescent in situ hybridizations slides were treated with 1% H2O2 in PBS for 1 h and washed 6 times for 15 min in PBS after first development prior to incubating with the second POD-labeled antibody. When in situ hybridization was followed by immunohistochemistry permeabilization was carried out by heating the slides at 95 °C for 20 min in citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0).
Immunohistochemistry
Slides were permeabilized with 0.4% pepsin in 0.1 N HCl at 37 °C or as described above, blocked in 5% non-fat dry milk in PBS 0.05% Tween 20, incubated overnight with primary antibodies in blocking solution, washed in PBS 0.05 Tween 20, and incubated for 4 h at room temperature with Alexa Fluor secondary antibodies (Molecular Probes) at a 1:400 dilution in blocking solution. The following primary antibodies were used: anti-PCNA (rabbit polyclonal kindly provided by Hidefumi Orii, 1:100) and anti-spoltud-1 (rabbit polyclonal against a BSA-conjugated peptide, BSA-[C]FKGSDSFSRKSESNF, peptide underlined in Fig. S1, produced by Sygma Genosys, 1:200). Nuclei were stained with TO-PRO 3 (Molecular Probes).
Immunohistochemistry for electron microscopy
Animals were fixed in 4% paraformaldehyde -1% glutaraldehyde in phosphate buffer, cryoprotected in saccharose, cryosubstituted in 0.5% uranyl acetate in methanol and cryoembedded in Lowicryl HM20 for 48 h at -50 °C and 24 h at RT. Ultrathin sections were obtained and collected on grids. Grids were blocked in 50 mM glycine in 0.1 M PBS for 4 min, washed in 0.1 PBS, blocked in 3% BSA in 0.1 PBS for 15 min and washed in 1% BSA for 3 min. Primary antibody was incubated for 2 h at a 1:10 dilution in 1% BSA 0.1 M PBS. Following incubation, grids were washed 5 times for 5 min in 0.1 M PBS 0.25% Tween 20, 3 min in 1% BSA 0.1 M PBS, incubated with gold-conjugated secondary antibody in 1% BSA 0.1 M PBS, washed 5 times for 5 min in 0.1 M PBS 0.25% Tween 20, washed extensively in deionized water, dried, and stained with uranyl acetate. Grids were observed with a JEOL 1010 electron microscope and images were captured with a Bioscan (Gatan) digital camera.
Imaging
Whole mount animals were observed with a Leica MZ16F stereomicroscope. Images were captured with a Leica DFC300FX camera. Fluorescent slides were mounted in Vectashield and imaged with a Leica TCS SPE confocal microscope. Images were analyzed with Image J and Adobe Photoshop software. Deconvolved images were produced with Huygens Essential software by Scientific Volume Imaging and using a theoretical point spread function.
Real time PCR
RNA was extracted using the Trizol reagent (Invitrogen). cDNA was obtained from 500 ng of total RNA by using MMLV Reverse Transcriptase (Promega). Elongation Factor 2 (EF2) was used as an internal control. Four animals were used per treatment and time point, and each sample was replicated three times in each real-time PCR experiment. PCR reactions were performed using the SYBR® Green RT-PCR kit (Applied Biosystems) and reactions were analyzed using an ABI Prism 7900HT (Applied Biosystems).
RNAi analysis
DsRNA was produced by in vitro transcription with T7 and SP6 enzymes (Roche) and injected into planarians as described before (Sanchez Alvarado and Newmark, 1999). Sexually immature juvenile animals were injected during 3 consecutive days and left for 11 days before starting a new round of injection, for a total of 2 months (4 rounds of injection). Control animals were injected with dsRNA of green fluorescent protein (GFP). Animals were kept at 20 °C and observed daily with a Zeiss Stemi SV6 scope and images of living animals were captured with a Nikon Coolpix E995.
Results
Isolation of Spoltud-1
tBLASTn searches in the S. mediterranea genome identified several contigs containing open reading frames that would encode Tudor domains, which generally share a low conservation at the amino acid level. On the basis of their similarity to well established Tudor domain-containing proteins involved in germ line control we selected two of them and designed specific primers to amplify the orthologous genes in S. polychroa. RACE reactions allowed us to isolate a full-length cDNA of 2482 bp which we named Spoltud-1.
The sequence of Spoltud-1 encodes a protein of 771 amino acids that contains an N-terminal RNA recognition motif (RRM1) and three Tudor domains (Fig. S1), and shares orthology with a previously described EST (05895_HH) from the planarian species D. japonica (Yoshida-Kashikawa et al., 2007). Spoltud-1 and EST 05895_HH have 65.0% identity and 88.0% similarity.
The partial clone corresponding to Spoltud-2 comprises a Tudor domain. The related sequence in the S. mediterranea genome encodes a putative protein of 793 amino acids and contains three Tudor domains, with a very low amino acid identity with Spoltud-1 (less than 10% throughout the whole protein) Although other Tudor domain-containing genes exist in the genome of the closely related species S. mediterranea we have not found clear paralogue genes with high similarity to Spoltud-1. Furthermore, only the sequences corresponding to Spoltud-1 and Spoltud-2 gave reciprocal BLAST hits to D. tudor and related mammalian genes involved in germ line biology.
Spoltud-1 mRNA and protein expression
Spoltud-1 and Spoltud-2 mRNA expression was monitored by in situ hybridization. Spoltud-2 was found to be highly expressed in ovaries and weakly expressed in testes but not in neoblast cells (Fig. S2), and was therefore discarded for further experiments. In adult sexualized animals Spoltud-1 was expressed in testes and ovaries, central nervous system (CNS) and in the parenchyma with a pattern similar to that of neoblast cells (Fig. 1A). Some animals also showed expression in the eyes (not shown), as has been described for some other genes expressed in neoblasts such as nanos (Handberg-Thorsager and Salo, 2007). In situ hybridization on paraffin sections revealed that the expression in the testis was restricted to the peripheral area (Fig. 1B) which contains the spermatocytes and the spermatogonia, the germ line stem cells of the testis. Expression in the parenchyma was restricted to discrete cells (Fig. 1C) scattered throughout the body with the exception of the pharynx and the region anterior to the eyes. Juvenile animals lacking sexual structures only showed expression in the CNS and parenchyma (data not shown).
Fig. 1.

Spoltud-1 in situ hybridization on adult and juvenile planarians shows expression in the germ line, CNS and neoblasts. (A) spoltud-1 whole mount in situ hybridization on adult planarians. Anterior is to the left. Top: dorsal view. Bottom: ventral view. Expression in testes is seen as two dorsal rows indicated by asterisks. Expression in ovaries is indicated by arrows. Arrowheads indicate CNS expression in the brain and ventral nerve cords. (B, C) spoltud-1 in situ hybridization on paraffin sections of an adult planarian showing testes, indicated by arrows (B), and parenchymal neoblasts (C). (D-G) Confocal maximum projection of a double in situ hybridization and immunohistochemistry of neoblast markers. (D) spoltud-1 mRNA positive cells in green. (E) histone H2B mRNA positive cells in red. (F) Nuclear expression of PCNA protein in neoblast cells in blue. (G) Overlay image showing colocalization of Spoltud1 mRNA and both histone H2B mRNA and PCNA protein. te, testis. Scale bars: 1 mm in A; 50 μm in B,C; 10 μm in D-G.
Several neoblast markers have been described in recent years. One of them is Proliferative Cell Nuclear Antigen (PCNA), a protein expressed in proliferative cells. Since neoblasts are the only proliferative cell type in juvenile or asexual planarians, PCNA protein is an excellent marker to trace neoblasts, and its expression is well characterized in planarians (Orii et al., 2005). Another stem cellspecific gene is histone H2B (Guo et al., 2006), which has been shown to be irradiation sensitive and to localize to parenchymal cells in S. mediterranea asexual organisms. Our experiments have shown that both PCNA protein and histone H2B colocalize in S. polychroa neoblasts.
To further characterize parenchymal Spoltud-1 expressing cells we developed a protocol to perform double in situ hybridization followed by immunohistochemistry. In S. polychroa juveniles Spoltud-1 mRNA positive cells (Fig. 1D) expressed both histone H2B mRNA (Fig. 1E) and PCNA protein (Fig. 1F). Therefore, we conclude that spoltud-1 expressing cells in the parenchyma are neoblast cells. In adult sexualized animals histone H2B and PCNA also colocalized with spoltud-1 mRNA positive cells in the parenchyma and in testicular spermatocytes and spermatogonia (data not shown).
To address the subcellular localization of spoltud-1 protein we developed a polyclonal antibody against a peptide of the protein. Immunohistochemistry on paraffin sections of juvenile animals revealed that histone H2B positive cells (Fig. 2A) displayed spoltud-1 protein in perinuclear granules (Figs. 2B-D) in a pattern similar to that of chromatoid bodies and chromatoid body components such as DjCBC-1 (Yoshida-Kashikawa et al., 2007). Deconvolution of these images showed that these perinuclear particles surround the whole nucleus and lie completely outside of it (Fig. 2E). We then examined the localization of spoltud-1 at the electron microscopic level. Neoblasts are easily identified in electron microscopy because of the presence of chromatoid bodies, identified as electron-dense perinuclear structures. Immunohistochemistry on ultrathin sections revealed labeling in these structures (Figs. 2F-H). Therefore, we concluded that spoltud-1 protein localizes to chromatoid bodies in neoblasts.
Fig. 2.

Spoltud-1 immunohistochemistry on adult and juvenile planarians shows localization to chromatoid bodies in neoblasts and testes and perinuclear granules in CNS. (A-D) Confocal maximum projection of an in situ hybridization and immunohistochemistry of histone H2B and spoltud-1. (A) histone H2B mRNA. (B) spoltud-1 protein. (C) nuclei. (D) Overlay image. (E) Deconvolution of a confocal series of a neoblast immunolabeled with anti-spoltud-1. Labeled particles are perinuclear and are all around the nucleus. (F-H) Immunohistochemistry on electron microscope sections. (F) Low magnification of a neoblast cell displaying a chromatoid body. (G) Higher magnification of (F) showing an immunolabeled chromatoid body. (H) Detail of a chromatoid body immunolabeled with anti-spoltud-1. (I-K, M-O) Confocal maximum projection of an anti-spoltud-1 immunohistochemistry on brain sections (I-K) and testis section (M-O). (I, M) spoltud-1 protein. (J, N) Nuclei. (K, O) Overlay images. (L, P) Deconvolution of a confocal section of an anti-spoltud-1 immunohistochemistry on brain sections (L) and testis (P). Arrowheads in L indicate neural processes. nu, nucleus; cb, chromatoid body; mc, mitochondria; sg, spermatogonia; st, spermatids; sp, sperm cells. Scale bars: 5 μm in A-E; 1 μm in F; 400 nm in G; 100 nm in H; 20 μm in I-K; 7.5 μm in L;25 μm in M-O; 7 μm in P.
Then we addressed the distribution of spoltud-1 protein in CNS. Interestingly, the pattern was shown to be similar to the one in stem cells. spoltud-1 protein localizes to perinuclear particles surrounding the nuclei of neurons in the brain (Figs. 2I-K), a pattern similar to that of DjCBC-1. These particles are in general smaller than chromatoid bodies and more closely attached to the nucleus (Fig. 2L). Furthermore, some particles were observed in neuronal processes (Fig. 2L).
To address the expression in testes, we performed immunohistochemistry on paraffin sections of sexualized planarians. The protein was localized to perinuclear particles surrounding the nuclei of spermatocytes and spermatogonia (Figs. 2M-O). No expression was found in spermatids or sperm cells (Fig. 2P).
Spoltud-1 expression in developing embryos
We next addressed the expression of spoltud-1 in planarian embryos. Some Tudor domain-containing proteins, such as TUD in D. melanogaster, are expressed maternally and are necessary for germ line determination (Boswell and Mahowald, 1985; Thomson and Lasko, 2004, 2005). We therefore investigated if spoltud-1 was expressed during embryonic development. Planarian development is still poorly studied and is highly derived compared to more basal platyhelminthes and other model organisms (Cardona et al., 2005b, 2006). Briefly, several fertilized zygotes are deposited in egg capsules surrounded by yolk cells. Around each zygote a yolk syncytium forms by the fusion of yolk cells, and zygotes are cleaved into different blastomeres which freely spread through this syncytium without staying in touch to each other (Fig. 3A). In stage 2 an embryonic pharynx develops, as does the epidermis, covering the whole syncytium (Figs. 3B, C). In stages 3 to 5 the embryonic pharynx starts to ingest external, but not fused yolk cells. Due to this ingestion, a cavity is formed and the syncytium is restricted to the periphery of the embryo, which is henceforth called the germ band (Figs. 3D, E). In stage 5, embryonic cells start to differentiate massively into all definitive cell types (Fig. 3F). Due to the differentiation of muscle cells, embryos, which were previously spherical, adopt an ovoid worm-like shape (Fig. 3G). During stages 6-8 the germ band collapses and embryonic cells grow to form septa which would finally shape the gut and muscle system (Fig. 3H). A definitive pharynx develops de novo and CNS and muscle system complete their development (Fig. 3I).
Fig. 3.

Spoltud-1 expression in S. polychroa developing embryos. (A-I) Schematic representation of S. polychroa stages of development, see text for explanation. Grey: syncytium. Light grey: ingested yolk mass. Dark blue nuclei: syncytial nuclei. Green cells: Zygote-derived blastomeres and embryonic cells (A-E). Red: embryonic (A-G) and definitive (G-I) pharynx. Orange: gastrodermal cells and gastrodermis. Magenta: nerve cells and nervous system. Green: Muscle cells and muscular system. (J-R) spoltud-1 whole mount in situ hybridization in embryos. (J) Stage 2 embryo. (K) Stage 3 embryo. (L, M) Stage 4 embryos. (N) Stage 5 embryo. (O) Stage 6 embryo. (P) Stage 8 embryo, anterior is to the left, dorsal is to the top. (Q, R) Deconvolution of confocal sections of a spoltud-1 immunohistochemistry on embryo paraffin sections. (Q) Stage 4 embryo. (R) Stage 7 embryo. Scale bars: 300 μm in J-O; 150 μm in P; 7 μm in Q, R.
Whole mount in situ hybridization in S. polychroa embryos showed spoltud-1 expression in blastomeres and embryonic cells. spoltud-1 mRNA is detected in stage 2 embryos (Fig. 3J). In stage 3 positive embryonic cells are restricted to the germ band and are visible in the periphery of the embryo (Fig. 3K). During stage 4 embryos grow in size and the number of positive embryonic cells increases (Figs. 3L, M). In stage 5 embryonic cells highly diversify in size and shape coinciding with the onset of differentiation (Fig. 3N). During stages 6 and 7 the number of spoltud-1 positive cells is reduced (Figs. 3O, P) to finally conform a pattern similar to that of neoblast cells in the stage 8 prehatch juvenile worm (Fig. 3P). spoltud-1 immunohistochemistry on paraffin sections of embryos revealed that the distribution of the protein in stage 2-4 is in particles distributed throughout the cytoplasm of the embryonic cells but concentrated upon the surface of the multilobed nucleus (Fig. 3Q). In contrast, stages 6-8 differentiating positive cells display a more neoblast-like distribution, with perinuclear particles attached to the nuclear membrane of an unlobed nucleus (Fig. 3R).
Spoltud-1(RNAi) animals fail to regenerate after 7 weeks of treatment
To address the function of spoltud-1 in neoblast cells we silenced it using RNAi. Juvenile organisms were injected with spoltud-1 dsRNA and control animals were injected with GFP dsRNA, a gene not present in the planarian. Animals were injected during three consecutive days and cut on the fourth, and then allowed to regenerate for 10 days and injected again. Our results indicated that after the first three rounds of injection animals regenerated properly and displayed no obvious phenotypes, but after the fourth round of injection all animals (N =35) failed to regenerate (Fig. 4B), while this effect was not seen in control animals (N =35, Fig. 4A). This effect was considerably delayed compared to other RNAi phenotypes described in planarians for neoblast-related genes, in which usually animals fail to regenerate after one single round of injection (Guo et al., 2006; Reddien et al., 2005; Salvetti et al., 2005).
Fig. 4.

Spoltud-1 (RNAi) animals fail to regenerate after 7 weeks of treatment. (A, B) Representative animals of the RNAi experiments after four rounds of injection and 7 days post-amputation (A) gfp(RNAi) control animal. (B) Spoltud-1 (RNAi) animal. (C-E) Head regeneration of representative animals that could initiate a regenerative blastema but failed to regenerate completely. (C) gfp(RNAi) control animal. (D, E) Different Spoltud-1 (RNAi) representative animals. Blastemata 4 days post-amputation are indicated by arrowheads. Eye rudiment in one individual is indicated by arrow. Anterior is to the top in A, B, and to the right in C-E. Scale bars: 120 μm.
Interestingly, some spoltud-1 (RNAi) animals (N = 14) were able to initiate a regenerative process after the fourth round of injection, but failed to finish it properly. Two representative animals are shown in Figs. 4D, E, along with a control animal (Fig. 4C). Four days after the cut the animals have developed a blastema. After 5 days, control animals show visible eyes. The animal depicted in Fig. 4D never developed eyes, while the animal in Fig. 4E displayed a rudiment of a right eye 7 days post-amputation. In S. polychroa regenerants eye development is not completely synchronic; some animals develop one eye hours before the other. The animal in Fig. 4D never developed the right eye completely and never displayed any rudiments of the left eye. Interestingly, spoltud-1 (RNAi) animals pigmented the aberrant blastemata after 12-16 days, and these blastemata never regressed, suggesting that these cells were effectively differentiating. Smedwi-2 RNAi and Smed-bruli RNAi animals are able to produce small regeneration blastemata that regress after some time (Guo et al., 2006; Reddien et al., 2005), indicating an effect in differentiating and differentiated cells that come from inhibited neoblasts. Our results show that during spoltud-1 RNAi, differentiated cells coming from inhibited neoblasts are stable over time. These results strongly suggest that the phenotype observed is caused by an effect in neoblast cells rather than in differentiated cells.
Due to their larger size and the difficulty to deliver dsRNA for such a long time, sexually mature animals were not tested in order to test a possible effect in ovaries and testes. Similarly, the effect in embryos could not be tested due to the lack of feasible experimental procedures to perform RNAi analyses during planarian embryonic development, although significant progress has been recently produced (Martin-Duran et al., 2008).
Spoltud-1 RNAi leads to a decrease in Spoltud-1 mRNA and protein levels
Since the phenotype observed in Spoltud-1 (RNAi) animals was considerably delayed, we examined if the RNAi was effectively inhibiting the function of the gene. In order to address this question we performed immunohistochemistry with anti-Spoltud-1 in injected animals. Animals after one round of injection still showed Spoltud-1 expression in chromatoid bodies (data not shown) but after a second round of injection much of Spoltud-1 protein expression was lost both in neoblast cells (Figs. 5G-I, compare with control in Figs. 5A-C) and brain (Figs. 5J-L, compare with control in Figs. 5D-F). After a third round of injection no Spoltud-1 protein was detected (Fig. S3). These results suggest that the lack of phenotype during the first weeks can be explained by a persistence of the protein but the latency observed between the disappearance of the protein and the onset of the phenotype has to be explained by other means.
Fig. 5.

Spoltud-1 protein is lost after 3 weeks of treatment. (A-L) Anti-Spoltud-1 immunohistochemistry in control and Spoltud-1 (RNAi) animals after second round of injection (3 weeks). (A-C) Parenchyma of a gfp(RNAi) control animal. (D-F) Brain of a gfp(RNAi) control animal. (G-I) Parenchyma of a Spoltud-1 (RNAi) animal. (J-L) Brain of a Spoltud-1 (RNAi) animal. (A, D, G, J) Nuclei staining (blue). (B, E, H, K) anti-Spoltud-1 (green). (C, F, I, L) Overlay images. Anterior is to the bottom left. Scale bars: 30 μm.
Next, we analyzed Spoltud-1 mRNA levels during the RNAi treatment by means of real-time PCR. Spoltud-1 mRNA levels rapidly drop from the first dsRNA injection (Fig. 6A) and remain low during the whole treatment. The total amount of Spoltud-1 mRNA in Spoltud-1 (RNAi) animals is approximately 5% of the amount in controls.
Fig. 6.
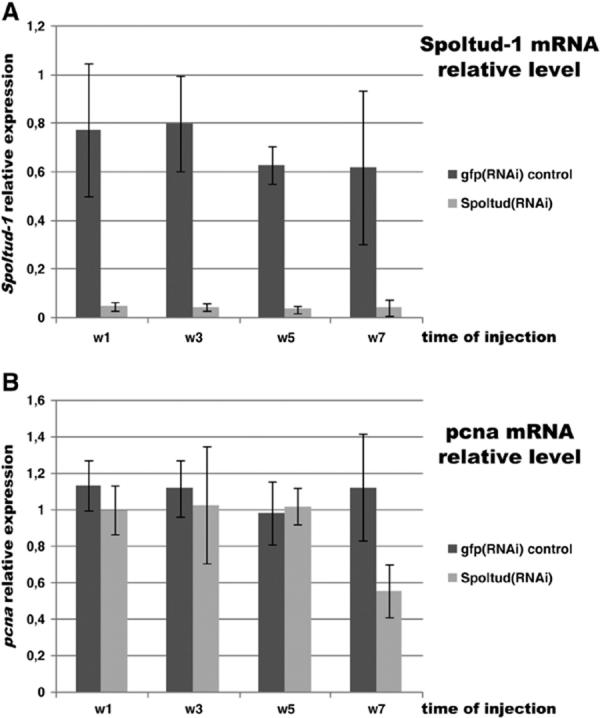
Spoltud-1 and pcna mRNA expression levels in Spoltud-1(RNAi) animals. (A) Spoltud-1 mRNA relative levels by means of real time PCR during RNAi treatment, showing the efficiency of the dsRNA injections. (B) pcna mRNA levels during RNAi treatment. Pcna levels in Spoltud-1(RNAi) animals are not significantly different from control animals during the first 5 weeks. In the seventh week there is a decrease in pcna mRNA levels. w1, week 1; w3, week 3; w5, week 5; w7, week 7.
Neoblasts are rapidly lost after 7 weeks of treatment
In order to analyze neoblasts in RNAi animals we performed His-tone H2B in situ hybridization and anti-PCNA immunohistochemistry. After three rounds of injections animals showed a normal pattern of neoblasts (Figs. 7E-H, compare with control in Figs. 7A-D). No differences were detected either in animals after the first and second rounds of injection (data not shown). Nevertheless, after the fourth round of injection the number of neoblasts was dramatically reduced (Figs. 7I-L), coinciding with the moment in which animals lost their regeneration capability. These results indicate that, although protein function might be absent from the third week on, neoblast cells are not affected at that time and continue to proliferate and differentiate until the fourth round of injection, when a rapid decay in neoblast number is seen. Taking together all these results, we suggest that during Spoltud-1 RNAi neoblast self-renewal is affected rather than neoblast proliferation or differentiation. Animals after 7 weeks of injection are not able to produce blastemata or produce abnormal blastemata that are not able to complete regeneration because they might contain insufficient neoblasts, but the abnormal blastemata produced are still able to generate differentiated cells, strongly suggesting that proliferation and differentiation are not affected.
Fig. 7.

Neoblasts are rapidly lost after 7 weeks of treatment. (A-L) Confocal maximum projection of a Histone H2B in situ hybridization and anti-PCNA immunohistochemistry in control and Spoltud-1(RNAi) representative animals. (A-D) gfp(RNAi) control after third round of injection (5 weeks). (E-H) Spoltud-1(RNAi) animal after third round of injection (5 weeks). (I-L) Spoltud-1(RNAi) animal after fourth round of injection (7 weeks). (M-R) Confocal maximum projection of an anti-PCNA immunohistochemistry in sagital sections of control and Spoltud-1(RNAi) representative animals. (M-O) gfp(RNAi) animal. (P-R) Spoltud-1(RNAi) animal. (A, E, I, M, P) Nuclei staining (blue). (B, F, J, N, Q) anti-PCNA (green). (C, G, K) Histone H2B fluorescent in situ hybridization (red). (D, H, L, O, R) Overlay images. Anterior is to the left. Scale bars: 50 μminA-H; 30 μmin I-L; 200 μmin M-R.
Interestingly, some animals lost anterior neoblasts before losing their posterior counterparts (Figs. 7P-R, compare with control in Figs. 7M-O), a pattern similar to the one shown after inhibition of smedinx-11, a neoblast gap junction gene the inhibition of which leads to neoblast loss and lack of regeneration (Oviedo and Levin, 2007). Nevertheless, animals after a fifth round of injection (N = 10) lost nearly all neoblasts (data not shown). These results suggest that anterior neoblasts are lost consistently before posterior ones during RNAi of different stem cell genes, perhaps reflecting a difference in between these neoblast subpopulations. During smedinx-11 RNAi treatment (Oviedo and Levin, 2007) neoblasts are lost in an anterior- posterior progression. A mitotic gradient exists in planarians, with anterior neoblasts having more mitotic activity than posterior ones (Baguñà, 1976; Oviedo and Levin, 2007). We also observed a specific loss of anterior neoblasts prior to posterior, suggesting that the more mitotically active cells are first lost due to a failure in stem-cell maintenance capability.
We analyzed pcna mRNA levels in RNAi animals to confirm the presence and function of neoblasts by means of real time-PCR (Fig. 6B). mRNA in Spoltud-1(RNAi) animals did not significantly differ from control animals during the first 5 weeks. Nevertheless, in the seventh week, and coinciding with the onset of the neoblast loss, a significant decrease in pcna mRNA levels is detected. Consistent with the phenotype observed the pcna mRNA levels of Spoltud-1 (RNAi) animals in the seventh week do not reflect a complete disappearance of neoblast but an ongoing process of loss. These results confirm a late disappearance of neoblast cells in Spoltud-1 (RNAi) animals.
Discussion
Spoltud-1 is a component of chromatoid bodies in germ cells and neoblasts
Several proteins with key functions in the germ line in different animals have been recently shown to be related to neoblast biology in planarians (Guo et al., 2006; Reddien et al., 2005; Salvetti et al., 2005; Shibata et al., 1999). The similarity between neoblasts and the germ line also extends to the presence of chromatoid bodies in planarian stem cells. These structures resemble the structures present in the germ line of most animals (P granules in C. elegans, polar granules in D. melanogaster and chromatoid body in mammals, often called nuage) and its presence in planarian somatic stem cells implies a relationship with maintenance of the undifferentiated state and genome integrity. In asexual species of planaria, reproduction is carried out exclusively by regeneration. Thus the neoblasts must possess properties similar to those of germ line stem cells in other animals. Different Tudor domain-containing proteins have been related to the germ line in many different organisms (Chuma et al., 2003, 2006; Hosokawa et al., 2007; Ikema et al., 2002). Our results demonstrate that Spoltud-1 protein is a component of chromatoid bodies in planarian stem cells.
Spoltud-1 expression in CNS
Contrary to what was proposed for the ortholog EST 05895_HH from D. japonica in a recent study (Yoshida-Kashikawa et al., 2007), we have found that Spoltud-1 is consistently expressed in CNS. This difference can be explained either by species variability or different sensitivities of techniques. Interestingly, several genes expressed in neoblasts have been also shown to be in CNS (Guo et al., 2006; Salvetti et al., 2005). Furthermore, 39 out of 60 putative RNA-binding proteins were described as expressed in neoblasts and CNS in the above mentioned study (Yoshida-Kashikawa et al., 2007). Only 6 out of 60 were described as expressed specifically in neoblasts and not in CNS, including the ortholog EST of Spoltud-1. These data strongly suggest a relationship between neoblast and neuron gene expression patterns. Some proteins classically related to germ line development have been recently found to be involved in neuronal function and development. Translational regulators such us Nanos and Pumilio, for example, are involved in neuronal excitability, dendrite morphogenesis, and long-term memory in D. melanogaster (Mee et al., 2004; Menon et al., 2004; Muraro et al., 2008; Schweers et al., 2002; Ye et al., 2004). Although our experiments with Spoltud-1 did not reveal any obvious nervous defects we cannot discard the existence of a neuronal function for the protein. Similarly, none of studies of genes described to be expressed in neoblast cells and CNS resulted in a nervous system RNAi phenotype (Guo et al., 2006; Salvetti et al., 2005).
DjCBC-1, described in the study by Yoshida-Kashikawa and coworkers, is a chromatoid body component with an expression pattern extremely similar to that of Spoltud-1. It corresponds to a Me31B homologue which belongs to the DEAD-box RNA helicases (Ladomery et al., 1997; Nakamura et al., 2001; Weston and Sommerville, 2006). Interestingly, it has been recently shown that Me31B in D. melanogaster forms a complex together with TUD (Thomson et al., 2008). DjCBC-1 is also expressed in the brain, in perinuclear particles surrounding the nuclei of neurons. At the ultrastructural level, DjCBC-1 can be seen to be localized to electron dense chromatoid body-like structures attached to the neuron nucleus (Oosaki and Ishii, 1965; Yoshida-Kashikawa et al., 2007). These structures might reflect the close relationship between neoblasts and neurons. Neuronal granules have been described in many animals and are thought to be responsible for the transport of silenced mRNAs from the cell bodies to the synaptic surface where they can be specifically translated. They share, together with germ granules and other types of RNA granules (Anderson and Kedersha, 2006), a common machinery including several RNA-binding protein such as Staufen (Ramasamy et al., 2006; Thomas et al., 2005), translation factors (Amiri et al., 2001; Kedersha et al., 2002, 2005), and many more (Thomson et al., 2008).
The embryonic origin of neoblasts
Our results show that during the embryonic development of S. polychroa there are two different types of Spoltud-1 expression. The first type exists during the first stages of development (stages 1-4) and is present in blastomeres and embryonic cells derived from them. They are relatively big in size and possess a multilobed nucleus. Spoltud-1 protein is present within them in granules distributed throughout the cytoplasm but concentrated on the periphery of the multilobed nucleus. It has been recently proposed that these early stages could represent a cryptic larval stage that feeds on maternal resources (Cardona et al., 2006). This putative larval stage extends over the period in which the first type of Spoltud-1 expression is detected.
During stage 5 a wave of differentiation occurs within the developing embryo and Spoltud-1 expression is seen as a second type of expression. Most of the cells generated during this stage are smaller and differently shaped, perhaps corresponding to the onset of their differentiation. During stages 6 and 7 Spoltud-1 expressing cells display a more neoblast-like pattern of distribution of Spoltud-1. Nevertheless, most of these cells might correspond to differentiating cells rather than neoblasts, although an undifferentiated population must exist during these stages. It is important to emphasize that during stage 8 only a restricted pool of cells displays Spoltud-1 protein and mRNA. This pool might correspond to the neoblast cells present in a hatchling.
Spoltud-1 is required for neoblast long-term self maintenance
Consistent with the expression of Spoltud-1 in neoblast cells we found that Spoltud-1(RNAi) animals failed to regenerate after a long treatment, but could effectively regenerate during the first weeks. Furthermore, these cells are lost at the end of the treatment (Fig. 8). These results suggest that Spoltud-1 function in neoblasts is important for their long-term maintenance. The fact that in the absence of detectable Spoltud-1 protein neoblasts were able to proliferate and differentiate strongly suggests a function in stem cell self maintenance. Furthermore, some Spoltud-1(RNAi) animals after the fourth round of injection were able to produce blastemata that did not conclude a proper regeneration, but the cells present in these blastemata were able to differentiate organs such as the eye and ultimately acquire pigmentation, a sign of terminal differentiation. Taken together, these results suggest that in the absence of Spoltud-1 protein cells are able to proliferate and differentiate but not able to self-renew, ultimately reaching a state in which nearly all neoblasts have differentiated and there are no stem cells left for a proper conclusion of regeneration. Nevertheless, we cannot rule out a possible redundancy in function by other Tudor domain-containing proteins present in planarian species, though this possibility is unlikely due to the low similarity exhibited by them. Spoltud-2, another Tudor-domain containing gene similar to D. tudor and related mammalian gene, was found to be not expressed in neoblast cells, and, furthermore, its similarity to Spoltud-1 is very low (less than 10%).
Fig. 8.

Schematic drawing of Spoltud-1 effect on juvenile planarians. Relative levels of Spoltud-1 mRNA (blue) and protein (green) compared to neoblast number (red). Time points indicated on bottom, regeneration, proliferation and differentiation on top. Two different periods of protein persistence and latency are defined.
The late phenotype seen in Spoltud-1(RNAi) opens the door for a new understanding of neoblast biology. RNAi-mediated knockdown of other genes expressed in planarian stem cells result in no obvious phenotypes on the first weeks of treatment, but these analyses did not extend for as long a time as ours (Reddien et al., 2005; Yoshida-Kashikawa et al., 2007). We speculate that some of these other knockdowns might in fact result in a phenotype upon a lengthier exposure to RNAi, in a manner similar to Spoltud-1.
Function of Spoltud-1 in planarian chromatoid bodies
We have demonstrated that Spoltud-1 activity is essential for neoblast maintenance. Nevertheless, neoblasts retain their function in the absence of Spoltud-1 protein for an extended period of time. The timing of this effect occurs in a very long fashion. Our experiments indicate that during the first 3 weeks of treatment Spoltud-1 protein is still present but after the third round of injection the protein is no longer detectable (Fig. 7). Thus, the persistence of the protein in the first weeks can account for the lack of phenotype during this period. Nevertheless, the appearance of the phenotype after a fourth round of injection has to be explained by other means.
Chromatoid bodies are believed to be very stable structures which contain RNA and are sites of translational regulation. Our results indicate that Spoltud-1 protein, a component of chromatoid bodies, is highly stable over time and this fact might be related to chromatoid body stability. mRNAs contained in these structures are likely to be highly stable as well, as classical studies have described that juvenile planarians are able to regenerate in presence of Actinomycin D, a potent RNA synthesis inhibitor (Coward, 1968; Gabriel and Lemoigne, 1971; Le Moigne and Gabriel, 1971a,b). This could be explained if chromatoid bodies stably contain sufficient pre-synthesized RNAs for one round of regeneration.
In D. melanogaster TUD is required for localization of mitochondrial RNAs to polar granules (Amikura et al., 2001a). It has been speculated that translation in polar granules, nuage and similar structures might be dependent on mitochondrial translational machinery. Indeed, mitochondrial ribosomes have been observed in D. melanogaster polar granules (Amikura et al., 2001b) and polar granule formation is impaired by use of inhibitors of prokaryotic-type translation such as chloramphenicol (Amikura et al., 2005). This importance of mitochondrial translation in germ line cells has been described in other animals, such as Xenopus and humans (Kashikawa et al., 2001; Kobayashi et al., 1998; Villegas et al., 2002). In planarians, chromatoid bodies have been described to be closely associated with nuclear pores and mitochondria (Coward, 1974; Hay and Coward, 1975; Hori, 1982). Though we cannot confirm an association of Spoltud-1 expression and mitochondria, some Spoltud-1 particles were not closely attached to the nucleus (data not shown).
Taking together all this data a possible role of Spoltud-1 in transport of RNA to or from the chromatoid bodies might be considered, either affecting mitochondrial RNA or a different class of RNA transport. In this scenario, a possible explanation for the latency period observed between the extinction of the protein and the loss of neoblast arises. After the disappearance of Spoltud-1 protein, RNA income or stability in the chromatoid bodies is impaired, although the RNA therein might be highly stable. This RNA may consist of determinants for neoblast differentiation and the presence of this RNA in chromatoid bodies may constitute a determinant of undifferentiated state. While Spoltud-1 function is affected, these RNAs may be slowly mobilized and translated cytoplasmically. After RNA in chromatoid bodies is exhausted, neoblast cells are no longer able to self-maintain an undifferentiated state and after some time differentiate without renewing the neoblast pool. Thus, animals are depleted of stem cells.
Supplementary Material
Acknowledgments
We are grateful to: Francesc Cebrià and Aziz Aboobaker for critical review of the manuscript, Jon Permanyer for valuable help with initial BLAST searches, Mette Handberg-Thorsager for help in developing the double in situ hybridization technique, Hidefumi Orii for kindly providing anti-DjPCNA antibody, and members of the Romero and Lasko lab for helpful discussion and for kindly sharing reagents and samples. Electron and Confocal microscopy and Real-Time PCR were performed at the Serveis Cientificotècnics of the University of Barcelona and under the supervision of their professional experts, in particular, Manel Bosch, Carmen López-Iglesias, Gema Martínez, Elisenda Coll, Ramón Seminago and Amaya Amador. J.S is supported by a BRD grant from the University of Barcelona and a travel fellowship from Development journal. This work was supported by a BFU07-63209 grant to R.R.
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2009.01.043.
References
- Amikura R, Hanyu K, Kashikawa M, Kobayashi S. Tudor protein is essential for the localization of mitochondrial RNAs in polar granules of Drosophila embryos. Mech. Dev. 2001a;107:97–104. doi: 10.1016/s0925-4773(01)00455-5. [DOI] [PubMed] [Google Scholar]
- Amikura R, Kashikawa M, Nakamura A, Kobayashi S. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc. Natl. Acad. Sci. U. S. A. 2001b;98:9133–9138. doi: 10.1073/pnas.171286998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikura R, Sato K, Kobayashi S. Role of mitochondrial ribosome-dependent translation in germline formation in Drosophila embryos. Mech. Dev. 2005;122:1087–1093. doi: 10.1016/j.mod.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE, Strome S. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. 2001;128:3899–3912. doi: 10.1242/dev.128.20.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- Auladell C, García-Valero J, Baguñà J. Ultrastructural-localization of RNA in the chromatoid bodies of undifferentiated cells (Neoblasts) in planarians by the RNase gold complex technique. J. Morphol. 1993;216:319–326. doi: 10.1002/jmor.1052160307. [DOI] [PubMed] [Google Scholar]
- Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n. sp. I. Mitotic studies during growth, feeding and starvation. J. Exp. Zool. 1976;195:65–80. [Google Scholar]
- Baguñà J, Saló E, Auladell C. Regeneration and pattern formation in planarians III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development. 1989;107:77–86. [Google Scholar]
- Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- Cardona A, Fernandez J, Solana J, Romero R. An in situ hybridization protocol for planarian embryos: monitoring myosin heavy chain gene expression. Dev. Genes Evol. 2005a;215:482–488. doi: 10.1007/s00427-005-0003-1. [DOI] [PubMed] [Google Scholar]
- Cardona A, Hartenstein V, Romero R. The embryonic development of the triclad Schmidtea polychroa. Dev. Genes Evol. 2005b;215:109–131. doi: 10.1007/s00427-004-0455-8. [DOI] [PubMed] [Google Scholar]
- Cardona A, Hartenstein V, Romero R. Early embryogenesis of planaria: a cryptic larva feeding on maternal resources. Dev. Genes Evol. 2006;216:667–681. doi: 10.1007/s00427-006-0094-3. [DOI] [PubMed] [Google Scholar]
- Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- Coward SJ. Effects of Actinomycin D on regeneration give evidence of sequential gene activation. Nature. 1968;219:1257–1258. doi: 10.1038/2191257a0. [DOI] [PubMed] [Google Scholar]
- Coward SJ. Chromatoid bodies in somatic cells of the planarian: observations on their behavior during mitosis. Anat. Rec. 1974;180:533–545. doi: 10.1002/ar.1091800312. [DOI] [PubMed] [Google Scholar]
- Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamune K, Nakatsuji N. Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mech. Dev. 2003;120:979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Gabriel A, Lemoigne A. Action of Actinomycin-D on cellular differentiation during regeneration in planarians. 1. Morphological, histological and ultrastructural studies on regeneration capacity in presence of antibiotic. Z. Zellforsch. Mikrosk. Anat. 1971;115:426–441. [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA. A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Handberg-Thorsager M, Salo E. The planarian nanos-like gene Smednos is expressed in germline and eye precursor cells during development and regeneration. Dev. Genes Evol. 2007;217:403–411. doi: 10.1007/s00427-007-0146-3. [DOI] [PubMed] [Google Scholar]
- Handberg-Thorsager M, Fernandez E, Salo E. Stem cells and regeneration in planarians. Front. Biosci. 2008;13:6374–6394. doi: 10.2741/3160. [DOI] [PubMed] [Google Scholar]
- Hay ED, Coward SJ. Fine structure studies on the planarian, Dugesia. I. Nature of the “neoblast” and other cell types in noninjured worms. J. Ultrastruct. Res. 1975;50:1–21. doi: 10.1016/s0022-5320(75)90003-9. [DOI] [PubMed] [Google Scholar]
- Hori I. An ultrastructural study of the chromatoid body in planarian regenerative cells. J. Electron. Microsc. (Tokyo) 1982;31:63–72. [Google Scholar]
- Hosokawa M, Shoji M, Kitamura K, Tanaka T, Noce T, Chuma S, Nakatsuji N. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev. Biol. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- Ikema Y, Hiyoshi M, Daiyasu H, Toh H, Mori M, Takamune K. Two novel genes expressed in Xenopus germ line: characteristic features of putative protein structures, their gene expression profiles and their possible roles in gametogenesis and embryogenesis. Mol. Reprod. Dev. 2002;62:421–430. doi: 10.1002/mrd.90003. [DOI] [PubMed] [Google Scholar]
- Kashikawa M, Amikura R, Kobayashi S. Mitochondrial small ribosomal RNA is a component of germinal granules in Xenopus embryos. Mech. Dev. 2001;101:71–77. doi: 10.1016/s0925-4773(00)00553-0. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Amikura R, Mukai M. Localization of mitochondrial large ribosomal RNA in germ plasm of Xenopus embryos. Curr. Biol. 1998;8:1117–1120. doi: 10.1016/s0960-9822(98)70466-x. [DOI] [PubMed] [Google Scholar]
- Ladomery M, Wade E, Sommerville J. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res. 1997;25:965–973. doi: 10.1093/nar/25.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Le Moigne A, Gabriel A. Action of Actinomycin D on cellular differentiation during regeneration in planarians Part 2 auto radiographic histological and ultrastructural studies on the action of the antibiotic on the RNA synthesis. Z. Zellforsch. Mikrosk. Anat. 1971a;115:442–460. [PubMed] [Google Scholar]
- Le Moigne A, Gabriel A. Action of Actinomycin D on cellular differentiation during regeneration in planarians Part 3 ultrastructural study of tritiated actinomycin penetration into regenerating cells. Z. Zellforsch. Mikrosk. Anat. 1971b;122:26–35. [PubMed] [Google Scholar]
- Martin-Duran JM, Duocastella M, Serra P, Romero R. New method to deliver exogenous material into developing planarian embryos. J. Exp. Zool. B Mol. Dev. Evol. 2008;310:668–681. doi: 10.1002/jez.b.21243. [DOI] [PubMed] [Google Scholar]
- Mee CJ, Pym EC, Moffat KG, Baines RA. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J. Neurosci. 2004;24:8695–8703. doi: 10.1523/JNEUROSCI.2282-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Morita M, Best JB. Electron microscopic studies of planarian regeneration. IV. Cell division of neoblasts in Dugesia dorotochepala. J. Exp. Zool. 1984;229:425–436. [Google Scholar]
- Morita M, Best JB, Noel J. Electron microscopic studies of planarian regeneration. I. Fine structure of neoblasts in Dugesia dorotocephala. J. Ultrastruct. Res. 1969;27:7–23. [PubMed] [Google Scholar]
- Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, Baines RA. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J. Neurosci. 2008;28:2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sanchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Oosaki T, Ishii S. Observations on the ultrastructure of nerve cells in the brain of the planarian, Dugesia gonocephala. Z. Zellforsch. Mikrosk. Anat. 1965;66:782–793. doi: 10.1007/BF00342956. [DOI] [PubMed] [Google Scholar]
- Orii H, Sakurai T, Watanabe K. Distribution of the stem cells (neoblasts) in the planarian Dugesia japonica. Dev. Genes Evol. 2005;215:143–157. doi: 10.1007/s00427-004-0460-y. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Levin M. smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development. 2007;134:3121–3131. doi: 10.1242/dev.006635. [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S, Wang H, Quach HN, Sampath K. Zebrafish Staufen1 and Staufen2 are required for the survival and migration of primordial germ cells. Dev. Biol. 2006;292:393–406. doi: 10.1016/j.ydbio.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Raz E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-3-reviews1017. REVIEWS1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Rossi L, Salvetti A, Batistoni R, Deri P, Gremigni V. Planarians, a tale of stem cells. Cell. Mol. Life Sci. 2008;65:16–23. doi: 10.1007/s00018-007-7426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman EE, Lasko P. Germline development in vertebrates and invertebrates. Cell. Mol. Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Deri P, Batistoni R. An MCM2-related gene is expressed in proliferating cells of intact and regenerating planarians. Dev. Dyn. 2000;218:603–614. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1016>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Lena A, Batistoni R, Deri P, Rainaldi G, Locci MT, Evangelista M, Gremigni V. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132:1863–1874. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Alvarado A, Kang H. Multicellularity, stem cells, and the neoblasts of the planarian Schmidtea mediterranea. Exp. Cell Res. 2005;306:299–308. doi: 10.1016/j.yexcr.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Schweers BA, Walters KJ, Stern M. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics. 2002;161:1177–1185. doi: 10.1093/genetics/161.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Solana J, Romero R. SpolvlgA is a DDX3/PL10-related DEAD-box RNA helicase expressed in blastomeres and embryonic cells in planarian embryonic development. Int. J. Biol. Sci. 2009;5:64–73. doi: 10.7150/ijbs.5.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell. 2005;16:405–420. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Tudor and its domains: germ cell formation from a Tudor perspective. Cell Res. 2005;15:281–291. doi: 10.1038/sj.cr.7290297. [DOI] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 2008;125:865–873. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K. A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev. Growth Differ. 1997;39:723–727. doi: 10.1046/j.1440-169x.1997.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- Villegas J, Araya P, Bustos-Obregon E, Burzio LO. Localization of the 16S mitochondrial rRNA in the nucleus of mammalian spermatogenic cells. Mol. Hum. Reprod. 2002;8:977–983. doi: 10.1093/molehr/8.11.977. [DOI] [PubMed] [Google Scholar]
- Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Yoshida-Kashikawa M, Shibata N, Takechi K, Agata K. DjCBC-1, a conserved DEAD box RNA helicase of the RCK/p54/Me31B family, is a component of RNA-protein complexes in planarian stem cells and neurons. Dev. Dyn. 2007;236:3436–3450. doi: 10.1002/dvdy.21375. [DOI] [PubMed] [Google Scholar]
- Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


