Abstract
Background
Growth and division of Saccharomyces cerevisiae is dependent on the action of SNARE proteins that are required for membrane fusion. SNAREs are regulated, through a poorly understood mechanism, to ensure membrane fusion at the correct time and place within a cell. Although fusion of secretory vesicles with the plasma membrane is important for yeast cell growth, the relationship between exocytic SNAREs and cell physiology has not been established.
Methodology/Principal Findings
Using genetic analysis, we identified several influences on the function of exocytic SNAREs. Genetic disruption of the V-ATPase, but not vacuolar proteolysis, can suppress two different temperature-sensitive mutations in SEC9. Suppression is unlikely due to increased SNARE complex formation because increasing SNARE complex formation, through overexpression of SRO7, does not result in suppression. We also observed suppression of sec9 mutations by growth on alkaline media or on a non-fermentable carbon source, conditions associated with a reduced growth rate of wild-type cells and decreased SNARE complex formation.
Conclusions/Significance
Three main conclusions arise from our results. First, there is a genetic interaction between SEC9 and the V-ATPase, although it is unlikely that this interaction has functional significance with respect to membrane fusion or SNAREs. Second, Sro7p acts to promote SNARE complex formation. Finally, Sec9p function and SNARE complex formation are tightly coupled to the physiological state of the cell.
Introduction
Cell growth and division requires the addition of membrane and protein to the surface of the growing cell through the fusion of secretory vesicles with the plasma membrane [1], [2]. The molecules involved in membrane fusion are conserved from yeast to humans, and include the SNARE proteins, defined by a ∼70 amino-acid alpha-helical SNARE motif [3], [4]. The SNARE motif of SNARE proteins on vesicles and on the plasma membrane assemble into a very stable four-helix bundle called the SNARE complex. Although SNARE complex formation is thought to provide the driving force for membrane fusion, accessory proteins influence SNARE assembly and help couple SNARE assembly to fusion and ensure membrane traffic at the correct time and place within a cell.
The yeast exocytic SNAREs consist of the synaptobrevin homologues Snc1/2p on the secretory vesicle and the syntaxin homologues Sso1/2p and SNAP25 homologue Sec9p on the plasma membrane [5]. Analogous to the neuronal SNARE complex, Snc1/2p and Sso1/2p each contribute one helix to the SNARE complex, while Sec9p contributes two helices [6]. SEC9 is an essential gene originally identified through the isolation of recessive temperature-sensitive alleles, such as sec9-4, that cause defects in secretion at the non-permissive temperature [7]. The sec9-4 mutation encodes a Gly to Asp amino acid substitution in the N-terminal helical domain of Sec9p that reduces the ability of Sec9-4p to complex with Sso1/2p and Snc1/2p [8]. Another temperature-sensitive allele (sec9-7) results in a lower cut-off temperature, but does not affect SNARE complex formation in vitro [9] suggesting multiple functions for Sec9p. Snc1/2p and Sso1/2p are encoded by redundant yet essential genes: yeast lacking either Snc1p and Snc2p or Sso1p and Sso2p are defective in secretion and accumulate secretory vesicles [10], [11].
SNAREs are thought to constitute the core fusion machinery and considerable work has focused on the identification of additional components that may play a role in membrane fusion. One such component is the vacuolar H+ ATPase (V-ATPase), a multi-subunit complex whose primary function is acidification of intracellular organelles by coupling ATP hydrolysis with translocation of protons across membranes [12]. The V-ATPase is composed of two distinct and separable sectors: the V1 sector is cytosolic and contains the ATPase activity, while the trans-membrane V0 sector forms the proton translocation channel. Three lines of evidence support a role for the V-ATPase in membrane fusion. First, studies of homotypic vacuolar membrane fusion have suggested that the V0 sectors on opposing membranes can form a proteolipid fusion pore and that radial dissociation and expansion of V0 sectors results in membrane fusion [13], [14]. Second, genetic analysis in different model systems has suggested that the V-ATPase can contribute to membrane fusion, independent of vesicle acidification [15]–[17]. Finally, V-ATPase subunits and SNARE proteins have been shown to interact on synaptic vesicles, although the functional significance of this interaction has not been established. [15], [18]
Another possible regulator of SNARE function is Sro7p and its redundant homologue SRO77. Sro7p and Sro77p have been implicated in secretion based on genetic studies demonstrating decreased secretion of invertase and an accumulation of secretory vesicles in the absence of SRO7 and SRO77 [19]. SRO7 was initially isolated as a high-copy suppressor of rho3 mutants, suggesting a role for Sro7p in maintenance of actin polarity [20], [21]. However, further studies have established that the primary role for Sro7p is in membrane fusion. First, Sro7p binds directly to Sec9p, and the interaction between Sro7p and SNAREs is essential for Sro7p function [22], [23]. Second, Sro7p is an effector of the Rab GTPase Sec4p, which has multiple functions during secretion, one of which occurs after vesicle transport to sites of secretion [24]. Finally, tomosyn, which is closely related in sequence with Sro7p, has been implicated directly in vesicle fusion in different systems [25], [26]. While Sro7p is likely to be involved in membrane fusion through an interaction with Sec9p, a role for Sro7p in SNARE complex assembly has not been determined.
Here, we describe genetic and physiological influences on SNARE complex formation. A forward genetic selection was performed to isolate mutations that suppress the temperature-sensitive phenotype of sec9-4 mutants. This screen revealed that disruption of the V-ATPase suppresses the temperature-sensitive growth phenotype caused by mutations in SEC9. Suppression was not accompanied by increased SNARE complex formation, suggesting that suppression involves a mechanism other than restoration of SNARE complex formation between Sso1/2p, Snc1/2p, and mutant alleles of Sec9p. Improved growth of sec9 mutants was also observed under conditions in which SNARE complex assembly and the growth rate of wild-type cells was reduced. Thus, suppression is likely the result of lowering the secretory demands of the cell to match the reduced level of Sec9p function. Furthermore, our results suggest that SNARE complex formation is highly responsive to the physiological state of the cell.
Results
Disruption of the V-ATPase suppresses sec9-4
The sec9-4 mutation disrupts the first SNARE-forming helix of Sec9p, preventing the formation of dimeric SNARE complexes between Sec9-4p and Sso1/2p and partially inhibiting the formation of trimeric SNAREs [8]. The phenotypic consequences of the sec9-4 mutation is a temperature-sensitive growth phenotype as sec9-4 cells display wild-type growth characteristics at 25°C, but are unable to form colonies at 35°C on rich media [7], [8]. We reasoned that mutations that restore growth to sec9-4 cells at the non-permissive temperature might do so by increasing SNARE complex formation. To isolate such mutations, we performed a forward genetic selection for extragenic suppressors of the sec9-4 temperature-sensitive phenotype [27]. Suppressors were obtained by plating sec9-4 cells on rich media and selection of colonies that formed at the non-permissive temperature. These colonies were then screened for secondary phenotypes to facilitate cloning of the suppressor gene. The isolated strains were backcrossed to determine if the suppressor was genetically linked to the secondary phenotype and to separate the suppressor from the sec9-4 mutation. Two independent suppressing mutations shared the secondary phenotypes of cold-sensitivity and poor growth on glycerol in SEC9 cells. In both cases the suppressor locus was tightly linked to the secondary growth defects and segregated 2∶2 during tetrad analysis, indicating that they were the result of a single mutation. Furthermore, both the suppression and the secondary phenotypes were found to be genetically recessive to wild type, suggesting that they are the result of a loss-of-function mutation.
Although the phenotypes caused by these two mutations were indistinguishable, they complemented each other indicating that they represent alleles of different loci (data not shown). The wild-type allele of each mutant gene was isolated and identified by complementation cloning: mutants were transformed with a genomic library and colonies able to grow on glycerol were selected. The plasmids conferring rescue were recovered and the genomic fragment contained within the rescuing plasmid was identified by restriction analysis and sequencing. From each strain, the rescuing plasmid contains a genomic fragment encoding a subunit of the V-ATPase (VMA1 and VMA16), suggesting that mutations in V-ATPase genes are able to suppress sec9-4.
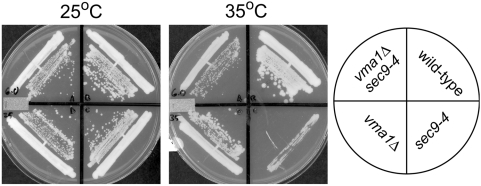
To directly test whether disruption of the V-ATPase is able to suppress mutations in SEC9, we generated a deletion allele of the V1 sector VMA1 and asked whether the deletion allele suppresses sec9-4 mutations [28]. Trans-heterozygous diploids were sporulated and subject to meiotic analysis and spores from tetra-type tetrads scored for growth at the sec9-4 permissive and non-permissive temperatures. At the permissive temperature, all four meiotic products displayed growth and were able to form individual colonies (Figure 1). At the non-permissive temperature, sec9-4 mutant cells grew very poorly and failed to form individual colonies, while sec9-4 vma1Δ double mutant cells grew and formed colonies. The suppression is not specific for disruption of the V1 ATPase sector of the V-ATPase, because disruption of VMA16, which encodes a subunit of the V0 transmembrane sector [29], is also able to suppress sec9-4 (data not shown). These results indicate that disruption of the V-ATPase suppresses the temperature-sensitive growth phenotype of sec9-4.
Figure 1. Disruption of the V-ATPase suppresses the sec9-4 temperature-sensitive phenotype.
Isogenic strains of the indicated genotype were plated on YPD media and incubated at 25°C or 35°C for 3 days. At 25°C, cells of all genotypes display growth and are able to form individual colonies. By contrast, at 35°C, sec9-4 cells grow poorly and do not form individual colonies while sec9-4 vma1Δ cells show increased growth and colony formation.
Suppression is not due to defects in vacuolar proteolysis
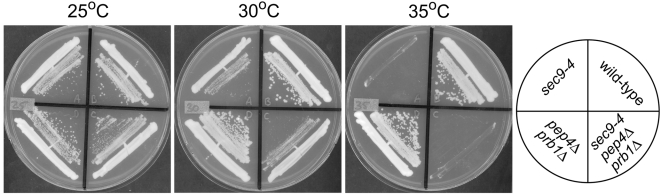
The loss of vacuole acidification in V-ATPase mutants prevents zymogen activation and causes defects in the proteolytic function of the vacuole [30], [31]. If the suppression of sec9-4 by disruption of the V-ATPase is due to defects in vacuolar proteolysis, then mutations that prevent zymogen activation should also suppress. To test this hypothesis, we obtained deletion mutations in PEP4 and PRB1, which are required for the activation of vacuolar proteases, and tested whether these deletions are able to suppress sec9-4 [32]–[34]. At the sec9-4 semi- and non-permissive temperatures the growth of sec9-4 cells was indistinguishable from sec9-4 pep4Δ or sec9-4 prb1Δ double mutants, indicating that disruption of vacuolar proteolysis is unable to suppress sec9-4. In addition, we compared sec9-4 cells to sec9-4 pep4Δ prb1Δ triple mutants (Figure 2) and did not observe any differences in growth or colony formation at any temperature. These results suggest that suppression of sec9-4 by disruption of the V-ATPase is not due to defects in the proteolytic function of the vacuole.
Figure 2. Suppression of sec9-4 is not due to defects in vacuolar proteolysis.
Strains of the indicated genotype were plated on YPD media and incubated at the indicated temperature for 3 days. At all temperatures, the growth characteristics of sec9-4 cells are identical to sec9-4 pep4Δ prb1Δ cells.
Growth under alkaline conditions suppresses sec9 temperature-sensitive alleles
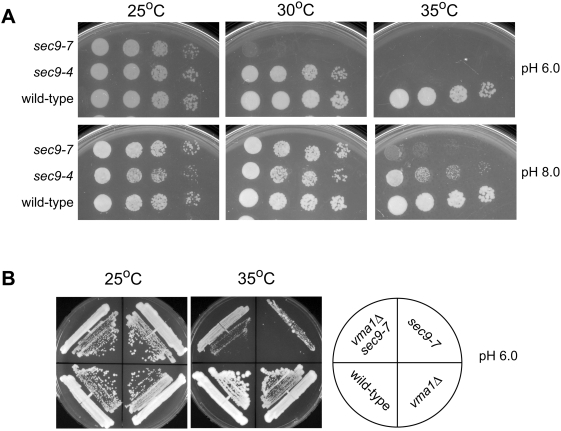
The V-ATPase has been implicated in cytosolic pH homeostasis in yeast and in vitro experiments have shown that the rate of SNARE complex formation is influenced by pH [35], [36]. While it is difficult to translate in vitro assembly results into in vivo effects, the improved growth of sec9-4 mutant cells upon loss of the V-ATPase could be due to changes in SNARE complex formation caused by alterations of intracellular pH. To test this, we assessed whether the pH of the medium affects the growth of sec9 mutants. Cells with temperature-sensitive mutations in sec9 were plated on media buffered at different pH and incubated at different temperatures (Figure 3a). Wild-type cells grew and formed individual colonies independent of pH or temperature. In contrast, sec9-4 mutants showed improved growth at the semi- and non-permissive temperature when grown on medium buffered at pH 8.0 compared to pH 6.0. These results suggest that suppression of sec9-4 in the absence of the V-ATPase and at alkaline pH could be caused by the same mechanism, possibly the result of altered pH homeostasis.
Figure 3. Growth under alkaline conditions suppresses temperature-sensitive alleles of sec9.
(A) Serial dilutions of sec9-7, sec9-4, or wild-type cell suspensions were spotted onto YPD media buffered at pH 6.0 or pH 8.0 and incubated at the indicated temperature for 3 days. On pH 6.0 media at 35°C, neither sec9-7 nor sec9-4 cells are able to grow and at 30°C sec9-7 cells grow very poorly, relative to wild type. On media buffered to pH 8.0, both sec9-4 and sec9-7 cells show increased growth characteristics at non-permissive temperatures. At 30°C, sec9-7 cells display wild-type growth, while at 35°C, sec9-4 cells show increased growth and are able to form individual colonies. (B) Strains of the indicated genotype were struck onto YPD plates buffered at pH 6.0 and incubated at permissive (25°C) or non-permissive (35°C) temperatures. At 35°C, sec9-7 mutants showed poor growth and failed to form individual colonies, while sec9-7 vma1Δ double mutants displayed increased growth and colony formation. The large colonies in sec9-7 sectors are likely extragenic reverants that are noticeably absent in sec9-7 vma1Δ sectors.
We tested whether suppression is specific to the sec9-4 allele by assaying the growth properties of sec9-7 cells under alkaline conditions and in the absence of the V-ATPase. At 30°C, sec9-7 cells grew very poorly on media that was buffered to pH 6.0 yet displayed near wild-type growth on pH 8.0 media (Figure 3a). The growth properties of sec9-7 mutants were also increased in the absence VMA1. At 35°C, sec9-7 mutants showed poor growth and failed to form individual colonies, while sec9-7 vma1Δ double mutants grew and formed individual colonies (Figure 3b). These results indicate that suppression is not specific to the sec9-4 allele. Although suppression is not sec9 allele specific, it is specific for mutations within sec9. Other sects alleles that affect late stages in post-Golgi secretion were analyzed and none of the tested mutations were suppressed by either vma1Δ or growth at elevated pH (Table 1). Together these results demonstrate that suppression is specific to mutations in sec9, which suggests that suppression reflects a bypass or restoration of a specific function that is impaired when SEC9 is mutated, rather than general defects in secretion.
Table 1. Summary of growth characteristics of sects mutants at elevated pH and in combination with vma1Δ.
| Allele | Growth at pH 8.0 | Growth with vmaΔ |
| sec1-1 | no effect | no effect |
| sec2-41 | no effect | no effect |
| sec3-2 | no effect | not determined |
| sec4-8 | no effect | no effect |
| sec5-24 | no effect | no effect |
| sec6-4 | no effect | slightly worse |
| sec8-9 | no effect | not determined |
| sec10-2 | no effect | not determined |
| sec15-1 | worse | not determined |
| sec9-4 | better | better |
| sec9-7 | better | better |
Increased SNARE complex formation does not result in suppression
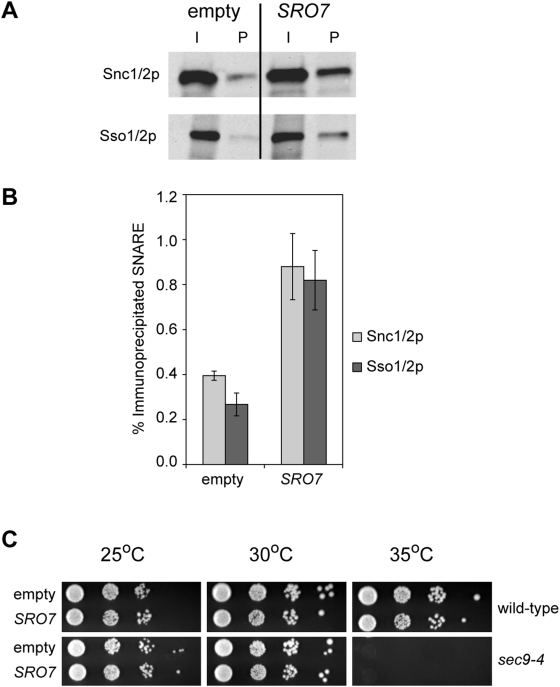
One possible mechanism to explain the increased growth of sec9 mutations in the absence of the V-ATPase or on alkaline media is through an elevation of SNARE complex formation under suppressing conditions. If this were the case, then increasing SNARE complex formation by other means would be expected to also result in suppression of sec9-4. To identify conditions that increase SNARE complex formation, we tested whether overproduction of a protein that interacts with Sec9p influences the levels of in vivo SNARE complexes. Sro7p is an effector of Sec4p that binds directly to Sec9p, and can assemble into a ternary Sro7p-Sec4p-Sec9p complex [24], [23]. In addition, Sro7p plays a positive role in secretion based on phenotypic analysis of loss-of-function mutants as well as genetic studies demonstrating suppression of a broad range of secretory mutations by SRO7 overexpression [19]. We assayed SNARE complex formation in strains transformed with high-copy number plasmids containing SRO7 or empty vector. Increased expression of SRO7 from a high-copy plasmid did not have any measurable effect on the steady-state levels of Sso1/2p or Snc1/2p (Figure 4 and data not shown). SNARE complex formation was assayed by immunoprecipitation of HA-Sec9p and probing the pellets for Sso1/2p and Snc1/2p by western blot (Figure 4a and b). When compared to empty-vector controls, strains bearing SRO7 plasmids showed an increase of approximately two-fold in the amount of Snc1/2p and Sso1/2p that co-precipitated with HA-Sec9p. We next asked whether increased SNARE complex formation is associated with suppression by testing SRO7 overexpression plasmids for suppression of the sec9-4 growth defect. The growth characteristics of sec9-4 strains harboring SRO7 on a high-copy number plasmid were indistinguishable from empty vector controls at permissive and non-permissive temperatures (Figure 4c). These results demonstrate that increased SNARE complex formation does not result in suppression of sec9-4 and therefore suggest that the suppression of sec9 mutations, by loss of VMA function or increased pH of the media, is unlikely the result of increased SNARE assembly.
Figure 4. Increased SNARE complex formation does not result in suppression of sec9-4.
(A) Overexpression of SRO7 causes an increase in SNARE complex formation. Immunoprecipitates of HA-SEC9 strains harboring high-copy number SRO7 or empty vector were probed by western blot with antibodies against Sso1/2p or Snc1/2p. The lanes labeled P are the immunoprecipitation pellets, while the input lanes represent 5% of the lysate from which the immunoprecipitate was derived. (B) Quantification of the immunoprecipitation in A. Integrated intensity of input and pellet bands for each genotype was determined by infrared imaging and used to determine the percent of immunoprecipitated SNARE. There is an increase in the amount of Sso1/2p and Snc1/2p that is immunoprecipitated in strains with SRO7 overexpression plasmids compared to empty-vector controls (P<0.05, two-tailed T-test, n = 3). Columns represent the mean+/−SEM. Similar levels of HA-Sec9p were immunoprecipitated from both genotypes. (data not shown). (C) Overexpression of SRO7 does not result in suppression of sec9-4. Serial dilutions of wild-type and sec9-4 cells harboring the indicated overexpression plasmid were spotted on selective media (SC-Ura) and incubated at the indicated temperature for 3 days. The growth characteristics of sec9-4 cells harboring SRO7 on an overexpression plasmid were indistinguishable from empty vector controls at all temperatures tested.
Suppressing conditions are not associated with increased SNARE complex formation
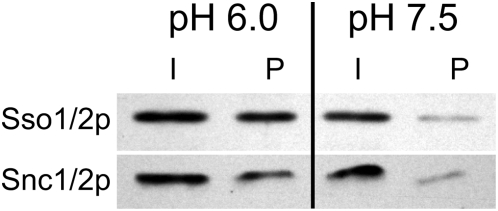
To directly test whether pH-dependent suppression is due to increased SNARE complex formation, we analyzed in vivo SNARE complex formation in cells grown in media buffered at different pH (Figure 5). Strains expressing HA-Sec9p were grown in media that was buffered at low (6.0) or high (7.5) pH. The amount of Sso1/2p and Snc1/2p that was pulled down with Sec9p was dramatically decreased in cells grown at pH 7.5 when compared to cells grown at pH 6.0. This result demonstrates that suppression of sec9-4 is not due to an increase in SNARE complex formation. We also observed a difference in the growth rates of wild-type cells in liquid media buffered at different pH. At pH 7.5, the growth rate of wild-type cells was much lower than that of cells grown in media buffered at pH 6.0 and no growth was observed in liquid media buffered at pH above 7.5 (data not shown).
Figure 5. Suppressing conditions are associated with decreased SNARE complex formation.
Cells grown under alkaline conditions have reduced levels of SNARE complexes. HA-SEC9 cells were grown in rich media buffered to pH 6.0 or pH 7.5. Lysates from these cells were prepared and levels of SNARE complex formation assayed by immunoprecipitation with antibodies against HA-Sec9p and subsequent western blotting for Snc1/2p and Sso1/2p. Input lanes (I) represent 2% of the lysate that was immunoprecipitated and loaded in the pellet lanes (P). The relative amount of Sso1/2p and Snc1/2p that is co-immunoprecipitated with HA-Sec9p is reduced in lysates from cells grown at pH 7.5 relative to the amount of precipitated SNAREs from cells grown at pH 6.0.
Slow growth conditions suppress the temperature-sensitivity of sec9 mutations
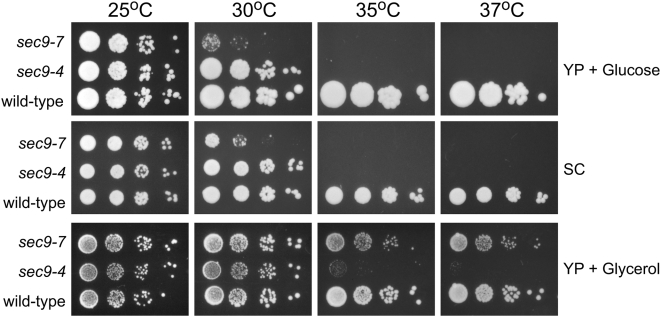
As noted above, cell growth was inhibited in liquid culture under alkaline conditions. In addition, disruption of V-ATPase subunit genes also caused growth defects when cells were grown on un-buffered media (Figure 1 and data not shown). This led us to hypothesize that the observed suppression of sec9 might be the result of culture conditions that decrease growth. Slower cell growth would be expected to reduce the rate of secretory vesicle formation and thereby lower the requirement for SNARE function. To test this hypothesis, we asked whether reducing overall growth rates by another means would also result in suppression of sec9. Yeast are unable to ferment glycerol, thus the growth rate on media containing glycerol, as the sole carbon source, is much slower than the growth on media that contains glucose. Both sec9-4 and sec9-7 mutant cells showed growth defects at 30°C and failed to grow at 35°C or higher on YP+glucose media. In contrast, both cell types grew and formed individual colonies at 35°C and 37°C when plated on YP+glycerol media (Figure 6). There was no difference in the pH of these two types of media. This result demonstrates that the overall growth conditions can influence the phenotype of sec9 mutants. When compared to wild-type cells, sec9-7 mutant cells grew better at elevated temperatures than sec9-4 mutant cells. It was particularly striking that the colony size of sec9-7 mutants was similar to the size of wild-type colonies at 37°C on YP+glycerol media.
Figure 6. Decreased growth suppresses the temperature-sensitivity of sec9 alleles.
Serial dilutions of sec9-7, sec9-4, or wild-type cell suspensions were spotted onto rich media containing glucose (YP+glucose), synthetic complete media (SC) or rich media with glycerol (YP+glycerol) and incubated at the indicated temperature for 3 days (YP+glucose and SC) or 5 days (YP+glycerol). On YP+glucose, sec9 mutants are unable to grow at temperatures above 35°C and show decreased growth at 30°C, relative to wild-type cells. Compared to YP+glucose, the growth of wild-type cells is decreased when plated on SC or YP+glycerol and under these conditions both sec9-4 and sec9-7 cells show increased growth at semi- and non-permissive temperatures. The relative increase in fitness associated with growth on YP+glycerol is higher for sec9-7 mutants compared to sec9-4 mutant cells.
Discussion
We designed a genetic screen to identify proteins that influence SNARE complex formation by isolating suppressors of mutations defective in SNARE complex formation. Our hypothesis was that sec9 suppressing conditions (either by extragenic suppressors or changes in culture media) would be due to an increase in SNARE complex formation. Contrary to our hypothesis, we found that suppressing conditions led to a decrease in SNARE complex formation as assayed by co-immunoprecipitation. In addition, we observed suppression of two different alleles of sec9 that have intrinsic differences in their ability to form SNARE complexes. As an alternative test of whether suppression is linked to increased SNARE complex formation, we examined if genetic enhancement of SNARE complex formation results in suppression of sec9 mutations. Overexpression of SRO7 results in increased SNARE complex levels, but does not suppress sec9 mutations. Together these results exclude the possibility that suppression is due to increased SNARE complex formation.
The results of our screen revealed that disruption of the yeast V-ATPase is able to suppress sec9 mutations. This was initially intriguing, as V-ATPase subunits physically interact with SNAREs and the V-ATPase has been implicated in membrane fusion in other genetic model systems. In these studies, loss-of-function mutations in V-ATPase subunits produce phenotypes that are attributable to defects in membrane fusion independent of acidification [15]–[17]. In each of these cases, the loss of the V-ATPase results in defects in membrane fusion, suggesting a facilitatory role for the V-ATPase in membrane fusion. However, our results indicate that loss of V-ATPase function suppresses two different sec9 mutations. This is inconsistent with a facilitatory role for the V-ATPase in secretory vesicle fusion, which predicts that disruption of the V-ATPase would enhance the phenotype of mutations in sec9. Although our results establish a genetic interaction between the V-ATPase and SNAREs, this interaction is likely a secondary consequence of the growth defect of V-ATPase subunit mutations.
The results presented here suggest that a function of Sec9p and SNARE complex formation is influenced by the physiological state of the cell. One common feature of the different suppressing conditions is that they all confer a reduction in the growth rate of wild-type cells, while mutant backgrounds that do not affect growth (pep4Δ prb1Δ) fail to suppress sec9 mutations. Our interpretation of these results is that mutations in SEC9 affect a constitutive function, but that the temperature-sensitive phenotype is a manifestation of greater cellular requirement for this absent function under the increased physiological demands of higher temperature. This is consistent with previous observations that the level of SNARE complexes can be controlled by the availability of Sec9p, suggesting that Sec9p is limiting for SNARE complex formation [37]. Suppressing conditions decrease physiological demands by slowing the growth rate and thereby bring the cells back to normal homeostasis at elevated temperatures. Because Sec9-4p and Sec9-7p have differing propensities to form SNARE complexes, this constitutive function of SEC9 may be independent of SNARE complex formation. Further characterization of the sec9 suppression phenotype and molecular genetic studies could be utilized to identify this novel function.
Materials and Methods
Yeast Culture Conditions
Standard techniques and media were used for cell growth, strain construction, and transformation [38], [39]. Buffered media was prepared by the addition of 50 mM MES, 50 mM MOPS and adjusting to the indicated pH.
Isolation and identification of suppressor mutations
Cells harboring the sec9-4 allele were plated on YPD media and incubated at 30°C. Colonies that formed were selected and back-crossed with wild-type cells and the resulting diploids were subject to meiotic analysis to verify that the suppressing mutations were extragenic and characterization of secondary phenotypes associated with the suppressing mutations, in this case cold-sensitivity and poor growth on media containing glycerol. Once a selectable secondary phenotype was determined, cells containing the suppressor mutation were transformed with a genomic library in YCp50 and transformants were screened for rescue of the secondary phenotype. Plasmids conferring rescue were recovered from yeast and analyzed by restriction mapping and sequencing.
Yeast Molecular Genetics
Deletion of VMA1 and VMA16 was by PCR-mediated gene disruption [40]. For disruption of VMA1 a HIS3 deletion cassette was generated by PCR using (ATTCTTAGAGTTAAAAAGCAAATAGAGAAGAAAAGAAACACGGATCCCCGGGTTAATTAA) and (CATCTAACAAATATACCAGAAGATAAATGCTACATATATCGAATTCGAGCTCGTTTAAAC) while the deletion cassette for VMA16 was generated with (GGAAGGCGAATAAAATACAGGAGCTAGAGCGTGTAAGATACGGATCCCCGGCTTAATTAA) and (TAGCTCGTAAAAACGGAAAAGAAAAGCCTGGTTTGAGCGCGAATTCGAGCTCGTTTAAAC) using pFA6-HIS5MX as template.
HA-Sec9p was generated by transformation of a SEC9::HIS3:PGAL1-HA 3 -SEC9 cassette into diploid MATa/MATα his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 ura3-52/ura3-52 cells. The transformation amplicon was generated by PCR using (TTGATCCTGCTTTTAACAATTTCGAATCGTTTGCGTAATTGAATTCGAGCTCGTTTA AAC) and (CCTCCTCTGGAGGCTTAATCTTAAAAAATTTCTTTAATCCGCACTGACGAGCGTAATCTG) using pFA6a-HIS3MX6-PGAL1-HA3 as a template, resulting in a transformation cassette directed in-frame to the N-terminus of Sec9p. Diploid transformants were sporulated and subjected to meiotic segregation analysis. Tetrads exhibited a 2∶2 segregation pattern for histidine prototrophy that was linked with the inability to grow on YPD media. Histidine prototrophic spores were tested for production of HA-Sec9p by western blotting using antibodies against HA.
SRO7-URA3-2μ was generated by amplifying SRO7 using (GGACTAGTCCTGAAGCTAATCCTTAACAGCGG) and (CGGGATCCACGTCTCAAAACAATTGGGCC) on a yeast genomic DNA template with Expand High-Fidelity polymerase, digesting with BamHI and SpeI, and ligation into the BamHI-SpeI sites of pRS426. This construct was verified by restriction digest.
In vivo SNARE complex formation assays
Immunoprecipitation experiments were performed as previously described [41], [42] with minor modifications. Cells inoculated from saturated cultures were grown overnight in selective media at 25°C to an OD600 of ∼0.4–1.0. Cells were harvested and washed in ice-cold TAF buffer (20 mM Tris-Cl pH 7.5, 20 mM NaN3, 20 mM NaF) and suspended in ice-cold IP buffer (50 mM HEPES, pH 7.4, 150 mM KCl, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, 2× Complete protease inhibitor (Roche)). Crude cell lysates were generated by beating with 0.5 mm zirconia/silica beads for 2 cycles of 4 minutes each in a Mini-beadbeater 8 at 4°C (Biospec). Lysates were cleared by centrifugation for 20 minutes at 15,000 rpm in a microcentrifuge and the supernatants were assayed for protein concentration by Bradford assay using BSA as a standard. Cleared lysates were diluted to an equal protein concentration and equal volume aliquots were then pre-cleared with ∼80 ul of equilibrated IgG-Sepharose beads by rocking samples for 30 minutes at 4°C, spinning down the beads (30 s at 1000×g) and keeping the supernatant. Epitopes were bound by adding 10 µl of anti-HA antibody (Covance) and incubated with rocking overnight at 4°C. Immuno-complexes were precipitated by the addition of ∼20 µl IgG-Sepharose beads and incubation for 2 hours, followed by spinning the beads down (1000×g, 30 s) and washing 5 times with ice-cold IP buffer. Proteins were elutated from the beads by boiling in 1× SDS-sample buffer. Precipitates and input samples were subject to SDS-PAGE, transferring to nitrocellulose membranes and Western blotting with antibodies against Sso1/2p or Snc1/2p. Assay of percent SNARE immunoprecipitation (Figure 4b) was determined using the Odyssey Infrared Imaging System and comparing the integrated intensity of bands in pellet lanes to bands present in samples derived from 5% of the lysate used for immunoprecipitation.
Acknowledgments
We thank Christian Dimaano and Markus Babst for kindly providing pep4Δ and prb1Δ mutations and the laboratories of Susan Ferro-Novick, Steven M. Strittmatter, and Marc Hammarlund for providing laboratory space and equipment.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grants from the National Institute of Health (GM35370 and GM082861) to P.N. and a Postdoctoral Fellowship (PF-07-037-01-CSM) from the American Cancer Society to D.W. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Field C, Schekman R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. J Cell Biol. 1980;86:123–128. doi: 10.1083/jcb.86.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 4.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 5.Gerst JE. SNAREs and SNARE regulators in membrane fusion and exocytosis. Cell Mol Life Sci. 1999;55:707–734. doi: 10.1007/s000180050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 7.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 8.Brennwald P, Kearns B, Champion K, Keränen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 9.Rossi G, Salminen A, Rice LM, Brünger AT, Brennwald P. Analysis of a yeast SNARE complex reveals remarkable similarity to the neuronal SNARE complex and a novel function for the C terminus of the SNAP-25 homolog, Sec9. J Biol Chem. 1997;272:16610–16617. doi: 10.1074/jbc.272.26.16610. [DOI] [PubMed] [Google Scholar]
- 10.Aalto MK, Ronne H, Keränen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Protopopov V, Govindan B, Novick P, Gerst JE. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- 12.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayer MJ, Reese C, Buhler S, Peters C, Mayer A. Vacuole membrane fusion: V0 functions after trans-SNARE pairing and is coupled to the Ca2+-releasing channel. J Cell Biol. 2003;162:211–222. doi: 10.1083/jcb.200212004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters C, Bayer MJ, Bühler S, Andersen JS, Mann M, Mayer A. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–588. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- 15.Hiesinger PR, Fayyazuddin A, Mehta SQ, Rosenmund T, Schulze KL, et al. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 2005;121:607–620. doi: 10.1016/j.cell.2005.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liégeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol. 2006;173:949–961. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peri F, Nüsslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 18.Galli T, McPherson PS, De Camilli P. The V0 sector of the V-ATPase, synaptobrevin, and synaptophysin are associated on synaptic vesicles in a Triton X-100-resistant, freeze thawing sensitive complex. J Biol Chem. 1996;271:2193. doi: 10.1074/jbc.271.4.2193. [DOI] [PubMed] [Google Scholar]
- 19.Lehman K, Rossi G, Adamo JE, Brennwald P. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J Cell Biol. 1999;146:125–140. doi: 10.1083/jcb.146.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagami M, Toh-e A, Matsui Y. Sro7p, a Saccharomyces cerevisiae counterpart of the tumor suppressor l(2)gl protein, is related to myosins in function. Genetics. 1998;149:1717–1727. doi: 10.1093/genetics/149.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsui Y, Toh-E A. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol Cell Biol. 1992;12:5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gangar A, Rossi G, Andreeva A, Hales R, Brennwald P. Structurally conserved interaction of Lgl family with SNAREs is critical to their cellular function. Curr Biol. 2005;15:1136–1142. doi: 10.1016/j.cub.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 23.Hattendorf DA, Andreeva A, Gangar A, Brennwald PJ, Weis WI. Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature. 2007;446:567–571. doi: 10.1038/nature05635. [DOI] [PubMed] [Google Scholar]
- 24.Grosshans BL, Andreeva A, Gangar A, Niessen S, Yates JR, et al. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J Cell Biol. 2006;172:55–66. doi: 10.1083/jcb.200510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheviet S, Bezzi P, Ivarsson R, Renström E, Viertl D, et al. Tomosyn-1 is involved in a post-docking event required for pancreatic beta-cell exocytosis. J Cell Sci. 2006;119:2912–2920. doi: 10.1242/jcs.03037. [DOI] [PubMed] [Google Scholar]
- 26.Gracheva EO, Burdina AO, Holgado AM, Berthelot-Grosjean M, Ackley BD, et al. Tomosyn Inhibits Synaptic Vesicle Priming in Caenorhabditis elegans. PLoS Biol. 2006;4:e261. doi: 10.1371/journal.pbio.0040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prelich G. Suppression mechanisms: themes from variations. Trends Genet. 1999;15:261–266. doi: 10.1016/s0168-9525(99)01749-7. [DOI] [PubMed] [Google Scholar]
- 28.Hirata R, Ohsumk Y, Nakano A, Kawasaki H, Suzuki K, Anraku Y. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 29.Hirata R, Graham LA, Takatsuki A, Stevens TH, Anraku Y. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J Biol Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.8.4795. [DOI] [PubMed] [Google Scholar]
- 30.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 31.Sørensen SO, van den Hazel HB, Kielland-Brandt MC, Winther JR. pH-dependent processing of yeast procarboxypeptidase Y by proteinase A in vivo and in vitro. Eur J Biochem. 1994;220:19–27. doi: 10.1111/j.1432-1033.1994.tb18594.x. [DOI] [PubMed] [Google Scholar]
- 32.Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nebes VL, Jones EW. Activation of the proteinase B precursor of the yeast Saccharomyces cerevisiae by autocatalysis and by an internal sequence. J Biol Chem. 1991;266:22851–22857. [PubMed] [Google Scholar]
- 34.Zubenko GS, Park FJ, Jones EW. Mutations in PEP4 locus of Saccharomyces cerevisiae block final step in maturation of two vacuolar hydrolases. Proc Natl Acad Sci U S A. 1983;80:510–514. doi: 10.1073/pnas.80.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Munoz GA, Kane PM. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem. 2008;283:20309–20319. doi: 10.1074/jbc.M710470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munson M, Chen X, Cocina AE, Schultz SM, Hughson FM. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat Struct Biol. 2000;7:894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- 37.Munson M, Hughson FM. Conformational regulation of SNARE assembly and disassembly in vivo. J Biol Chem. 2002;277:9375–9381. doi: 10.1074/jbc.M111729200. [DOI] [PubMed] [Google Scholar]
- 38.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 40.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 41.Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grote E, Novick PJ. Promiscuity in Rab-SNARE interactions. Mol Biol Cell. 1999;10:4149–4161. doi: 10.1091/mbc.10.12.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]