Abstract
This review focuses on recent research using genomics to examine lung carcinogenesis, histologic differentiation, and progression.
Keywords: adenocarcinoma, invasion, genomics, microarray, development
Lung cancer is the leading cause of cancer death in both men and women in the United States, despite its incidence being less than that of prostate cancer in men and breast cancer in women. With 166,000 deaths expected in 2008, the sum total of lung cancer deaths exceeds those of prostate, breast, and colon cancer combined (1). Prostate, breast, and colorectal carcinoma have all demonstrated significant improvements in 5-year survival over time and are currently 99, 88, and 64%, respectively. In contrast, the 5-year survival rate for lung cancer has remained relatively stable at 15%.
There are several potential explanations for the disparity between lung cancer survival and that of other common tumors, including late detection and histologic heterogeneity. Currently, over 75% of new lung cancer diagnoses are made in patients presenting with distant or regional metastatic disease (2). Histologically, prostate, breast, and colorectal carcinomas are uniformly adenocarcinoma and treatment is primarily determined by clinical stage, at times modified by results of molecular assays (3, 4). In contrast, only 30% of lung carcinoma is adenocarcinoma. Yet for the most part, until recently, non–small cell lung cancers (squamous, large cell, adenocarcinoma) were treated similarly, regardless of the biological heterogeneity associated with histology. It is likely that poor historical lung cancer response rates may in part be attributable to a relatively homogenous approach to a heterogeneous disease. Ongoing efforts are underway to identify clinically relevant biological properties of tumors that will facilitate individualized lung cancer treatment.
This research takes advantage of technical advances that allow rapid high-throughput assays to interrogate the genome (mRNA, microRNA, copy number, mutation analyses), proteome, and epigenome. In this review, we will focus on transcriptional profiling studies that have advanced our understanding of malignant transformation of lung epithelial cells and of lung cancer differentiation and progression. These advances have direct relevance to the goals of early detection, accurate prognostic assessment, and targeted therapy.
FIELD CARCINOGENESIS
Fifteen percent of lifetime smokers develop lung cancer, but 10% of lung cancers occur in never-smokers (5). In nonsmokers, exposure to secondhand smoke or to other lung carcinogens such as radon, asbestos, arsenic, or air pollution may be contributory. In both smokers and nonsmokers, genetic polymorphisms in genes associated with carcinogen metabolism, DNA damage repair, and cell cycle control may influence lung cancer susceptibility and modify the injury response associated with exposure to the dozens of carcinogens contained in tobacco smoke (6, 7). While the heritable component of lung cancer risk due to these genetic polymorphisms is most notable in early-onset cases occurring with less total smoke exposure, due to the limited number of genes studied, the overall attributable risk remains small even in this group of patients (7, 8). Recently, three genome-wide association studies identified a locus on chromosome 15q associated with lung cancer (9–11). The common locus variant near rs1051730 contains three genes encoding subunits of the nicotinic acetylcholine receptor (nAChR), CHRNA3, CHRNA5, and CHRNB4. The nAChR holoreceptor is a pentamer surrounding a gated channel responsive to ligands such as acetylcholine, nicotine, or its highly carcinogenic derivative 4(methylnitrosamine)-1-(3-pyridyl)-1-butanone (12). It is unclear whether the gene variant mediates susceptibility predominantly through influencing smoking behavior and nicotine dependence or through direct effects on lung tumor growth and suppression of apoptosis (13).
Because of tobacco smoke exposure, the bronchial epithelium of smokers is subject to field cancerization with alterations of airway cell DNA. Such alterations bring about oncogene activation, tumor suppressor gene silencing, and widespread loss of heterozygosity, all of which drive distinct gene expression signatures. While it is unclear which molecular alterations are required for neoplasia, in some instances, field carcinogenesis leads to malignant transformation of lung cancer progenitor cells. This malignant transformation may also require contributions from other systemic cell types such as bone marrow–derived stem cells (14, 15).
Microarray expression profiling has provided important information about cigarette smoke–associated lung carcinogenesis. Recently, we and others used gene expression profiling to identify gene signatures associated with cigarette smoking and lung adenocarcinoma in smokers and nonsmokers (16–20). Together, these results suggest that distinct cell transformation pathways and stromal tissue responses are involved in adenocarcinoma arising in smokers and nonsmokers (21). Observational studies using microarray analyses to profile bronchial epithelial cells obtained from bronchoscopic brushings of healthy smokers and nonsmokers identified overexpression of antioxidant genes and genes associated with xenobiotic metabolism by cells obtained from smokers (22, 23). However, these changes appeared to be potentially reversible when evaluated in a comparison of current versus former smokers. This transient genomic response to injury has been confirmed by in vitro studies using human bronchial epithelial cells exposed to cigarette smoke condensate and by in vivo rodent models of cigarette smoke exposure (24).
In addition, a field defect in smokers closely associated with lung cancer was reported by Spira and colleagues, who acquired gene expression signatures from nonmalignant bronchial cell brushings in a large number of smokers with and without lung cancer and observed that signatures from uninvolved cells in patients with cancer were distinct from those obtained from healthy smokers (25). This suggests that an epithelial “field defect” typically associated with cigarette smoke exposure may be specific for cancer. If validated prospectively, molecular testing of epithelial cells acquired throughout the aerodigestive tract may provide useful information for risk assessment.
GENOMICS OF LUNG CANCER DIFFERENTIATION AND PROGRESSION
Lung Developmental Pathways in Lung Cancer
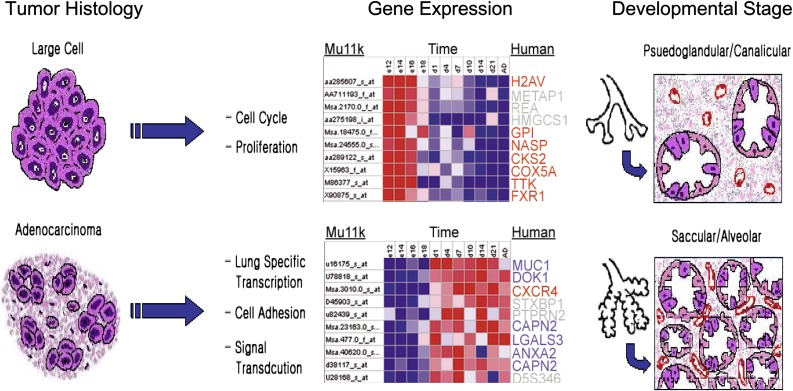
Current paradigms suggest that lung carcinomas arise from pluripotent stem and progenitor cells capable of differentiation into one or several histologic cell types. These paradigms suggest that lung tumor cell ontology is determined by the consequences of gene transcriptional activation and/or repression events that recapitulate embryonic lung development (26, 27). The hypothesis that lung cancer arises from aberrant expression of genes involved in lung development is supported by gene expression studies demonstrating similarities between signatures obtained from human lung tumors and signatures characteristic of normal lung development (Figure 1). In an analysis of 32 non–small cell lung carcinoma (NSCLC) specimens and 7 normal specimens, unsupervised hierarchical analysis segregated tumors on the basis of histologic type and differentiation (28). Supervised clustering analysis of tumors identified numerous genes with known important function in embryonic lung development. Comparison of human lung tumor histology classifiers with genes temporally activated during mouse lung development reveals that genes expressed by large cell carcinoma (LCC) are similarly expressed during the early pseudoglandular and canalicular stages of lung development, while those expressed by adenocarcinoma mirror those expressed during the later terminal sac and alveolar stages. In addition to highlighting the expression of proliferation-associated genes by LCC and of differentiation-associated genes by adenocarcinoma, these results suggest a recapitulation of developmentally regulated pathways in lung tumors.
Figure 1.
Comparison of gene expression of human lung tumors with genes temporally expressed during lung development suggests a recapitulation of developmentally regulated pathways. Nearest neighbor analysis was used to classify and rank murine (Mu11k) gene markers associated with two groups associated with lung development stage. Each row in the heat map (middle panel) shows gene expression over time for murine genes. Lung development stages represented by Group 1 (Days E12–E18, top panel) and Group 2 (Postnatal Day 1–Adult, lower panel) highly expressed genes are pseudoglandular/canalicular and terminal sac/alveolar, respectively. The corresponding human histology gene orthologs for large cell carcinoma are indicated in red, adenocarcinoma in blue. Specifically, genes expressed by large cell carcinoma mirror those expressed during the early pseudoglandular and canalicular stages of lung development, while those expressed by adenocarcinoma are predominantly expressed during the later terminal sac and alveolar stages.
Similar observations were reported by Bonner and colleagues, who compared gene expression profiles of human lung tumors with those obtained from a murine model of lung adenocarcinoma (29). Liu and colleagues (30) found that genes associated with early lung development were more often expressed by small cell lung carcinoma (SCLC) and by tumors associated with poor prognosis. In addition, Glinsky and colleagues reported that a gene signature of “stemness” derived from BMI-1–regulated genes in normal stem cells is associated with metastasis and survival in several tumor types, including NSCLC (31). Taken together, these observations suggest that poor differentiation is linked to molecular parameters of early development representing lung stem and progenitor cell programs, and that gene signatures of these phenotypes are important for lung cancer differentiation, progression, and clinical outcome.
Neuroendocrine Tumors
One of the first studies using gene expression profiling to examine lung cancer histologic diversity compared carcinoids and small cell carcinoma tumors (32). Despite sharing neuroendocrine differentiation, the molecular profiles suggested different cells of origin for these tumor types. This molecular distinction was supported by a subsequent study from Bhattacharjee and colleagues (33) showing two distinct clusters of small cell carcinoma and carcinoid tumors, with small cell carcinoma characterized by frequent expression of proliferation markers such as MCM2, PCNA, MCM6, and thymidylate synthase. This was later confirmed by Virtanen and coworkers (34), who identified a carcinoid subgroup distinct from high-grade neuroendocrine carcinomas. Interestingly, large cell neuroendocrine carcinoma, a high-grade neuroendocrine carcinoma, was not readily distinguishable from small cell carcinoma by gene expression profiling. This observation suggested that despite being classified as an NSCLC subtype, large cell neuroendocrine carcinoma bears more similarity to small cell carcinoma than to other NSCLC subtypesn and illustrates how gene expression profiling can explain similar clinical features observed in histologically distinct tumors.
Squamous Cell Carcinoma
Researchers have examined gene expression profiles of squamous cell carcinomas (SCC) of the lung to address three major areas of concern: (1) to account for the histologic heterogeneity of NSCLC and to differentiate squamous cell carcinoma from other subtypes, (2) to identify prognostic factors specific to squamous cell carcinoma, and (3) to aid in distinguishing primary pulmonary SCC from metastatic SCC of the head and neck (HNSCC).
Several studies have identified gene expression profiles that distinguish pulmonary adenocarcinoma from pulmonary SCC (35–38). Notably, a shared set of genes encoding cytokeratins and cell adhesion proteins consistently distinguish squamous cell carcinoma from adenocarcinoma. These classifiers include cytokeratin 14 and components of desmosomes, hemi-desmosomes, and gap junctions, such as integrins ITGB4, desmocollin, and desmoplakin.
A consistent finding of unsupervised clustering analysis in pulmonary SCC is the generation of two clusters not attributable to histology, differentiation, or stage (39, 40). Rather, these clusters were enriched in prognosis associated genes encoding proteins with function in the cell cycle and proliferation. Larson and colleagues (41) compared gene expression profiles of node-negative SCC tumors (N0) with signatures obtained from tumors with ipsalateral hilar or peribronchial nodal involvement (N1) via direct nodal invasion and with signatures obtained from N1 tumors with regional lymph node metastases via lymphatics, and found that N1 tumors with direct lymphatic invasion were similar to N0 tumors and distinct from N1 tumors with lymphatic spread, both in terms of molecular features and prognosis.
Although immunohistochemistry panels are effective in determining a site of primary origin for adenocarcinoma, equally effective panels do not exist for squamous cell carcinoma. Because primary SCC of the lung must be distinguished from metastatic HNSCC, this poses a diagnostic dilemma in patients with a history of HNSCC who subsequently develop a pulmonary nodule. Toward this end, Talbot and colleagues (42) developed a 500-gene classifier that distinguished pulmonary SCC from SCC of the tongue. In addition, Vachani and colleagues (43) used a training set of 18 HNSCC and 10 pulmonary squamous cell carcinomas to develop a 10-gene profile to distinguish these two tumor types. This classifier was validated on 122 subjects from previously published independent datasets, with 96% accuracy. Using real-time PCR, they correctly classified 12 independent samples as either of head and neck or pulmonary origin. These studies did not use microdissection; thus, it is possible that the signal from the host tissue may confound interpretation of gene signatures of tumor origin. However, if validated by others using microdissected specimens, these genomic classifiers hold promise for addressing important clinical staging issues in squamous cell carcinoma.
Adenocarcinoma
Somatic DNA alterations including mutations, amplifications, deletions, and translocations are common in tumors and are required for the activation of oncogenes and the inactivation of tumor suppressor genes that drive carcinogenesis. Large-scale efforts to systematically characterize these alterations in lung cancer and other tumors (44, 45) have provided insights into DNA structural alterations in non–small cell lung cancer. Adenocarcinoma, the most frequent histologic subtype of NSCLC, is characterized by multiple somatic DNA alterations. Recently, Weir and colleagues comprehensively examined DNA copy number alterations in 371 lung adenocarcinoma specimens (46) and found that the top focal regions of amplification and deletion included 14q13.3, 12q15, 8q24.21, 7p11.2, and 8q21.13. These results confirmed amplifications and deletions reported in other smaller studies, and also identified novel alterations such as amplification of the transcription factor TTF-1 on chromosome 14q13.3. TTF-1 encodes thyroid transcription factor 1, a member of the Nk-2 homeobox family that binds to and activates the promoter of thyroid- and lung-specific genes. Interestingly, other groups using similar approaches also identified TTF-1 amplification as a frequent lung adenocarcinoma alteration, thus providing independent validation of this finding (47, 48). Together, these observations link lung tumor differentiation states and histologic grade with epithelial cell developmental pathways (28, 49), and suggest that these DNA amplification events are important in mediating lung cancer initiation, differentiation, and progression.
Lung Adenocarcinoma Invasion
Lung adenocarcinoma, the most frequent histologic type of NSCLC, is heterogeneous. Histologically, subclassification of adenocarcinoma is based upon World Health Organization (WHO) criteria, determined predominantly by cell morphology and growth pattern, along a spectrum including noninvasive bronchioloalveolar carcinoma (BAC), adenocarcinoma with mixed subtypes (AC-Mixed), and pure invasive adenocarcinoma (IAC) (50). Lung cancer metastasis represents the final step of a complex sequence composed of invasion (loss of cell–cell adhesion, increased cell motility, and basement membrane degradation), vascular intravasation and extravasation, establishment of a metastatic niche, and angiogenesis. Recent research has focused on characterizing the molecular mechanisms and clinical implications of adenocarcinoma invasion, the initial step of metastasis.
Paralleling malignancies in other organs, such as breast and cervix, where tumors are defined as noninvasive (in situ carcinoma), microinvasive (microscopic invasion), or as invasive carcinomas, the extent of the invasive component seen in lung adenocarcinoma is associated with clinical outcomes. Notably, this is most relevant in the lung for adenocarcinoma, because the noninvasive component is generally minimal in squamous cell carcinoma and often absent in large cell carcinoma. The clinical importance of lung adenocarcinoma invasion is supported by several recent studies (51–56) indicating that the risk of death in noninvasive BAC tumors is significantly lower than that of pure invasive tumors and in tumors with less than 0.6 cm of fibrosis or linear invasion. These results suggest that the prognosis of BAC is favorable, and suggest a similarly favorable prognosis for the subset of AC-Mixed subtype tumors with limited invasion (<6 mm).
We (57) and others (58–60) have used microarray gene expression profiling of lung adenocarcinoma tumors to identify signatures associated with histologic subtype and invasion. The results of unsupervised analyses are striking in that in each instance, tumors segregate into three major clades composed predominantly of BAC, AC-Mixed subtype, and pure invasive tumors, providing biological plausibility for the notion that these adenocarcinoma subtypes are distinct tumors. Taken together with the clinical data indicating distinct prognoses for each subtype, these studies have motivated consideration of revising the WHO lung adenocarcinoma classification scheme to reinforce the designation of purely noninvasive tumors and to create a designation for minimally invasive tumors.
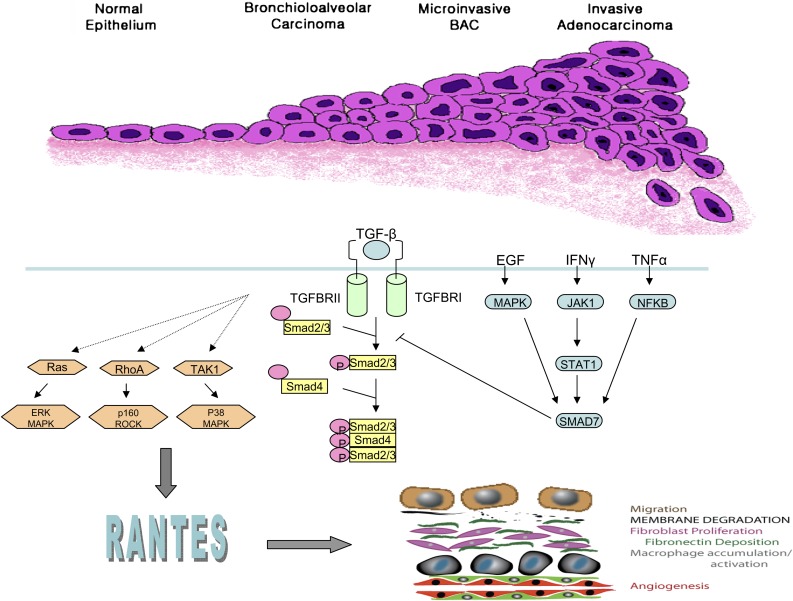
To identify the molecular pathways important in mediating the acquisition of invasion by lung adenocarcinoma, we performed supervised analysis of mRNA microarray data to identify genes differentially expressed in noninvasive BAC and in AC-mixed type tumors. Among the genes differentially expressed in the progression from BAC to invasive tumors was the type II transforming growth factor β receptor (TGFβRII), which was less highly expressed by AC-Mixed and solid invasive tumors compared with BAC. This finding, which suggested that TGFβRII repression was required for lung adenocarcinoma invasion, is supported by genetic models combining targeted deletion of TGFβRII with other oncogenic events such as Apc mutation in colon tumors and Kras mutations in pancreatic and oropharyngeal carcinomas (61–63). The phenotypes of these TGFβRII-deficient cancer models clearly demonstrate the importance of TGF-β signaling in tumor invasion, yet the downstream signaling mechanisms are undefined.
We used a tumor cell invasion system and microarray analysis to identify and characterize downstream mediators of TGF-β signaling important for lung adenocarcinoma invasion (57). Among potential mediators identified was the chemokine CCL5 (RANTES [regulated on activation, normal T cell expressed, and presumably secreted]), which is up-regulated by invasive tumors and TGFβRII knockdown cells. RANTES is involved in immunoregulatory and inflammatory processes and is transcribed and secreted not only by T cells, other inflammatory cells, and stromal cells, but also by tumor cells and normal bronchial epithelium. RANTES is a ligand for chemokine receptors CCR1, CCR3, CCR4, and CCR5, which are expressed on epithelial cells, macrophages, lymphocytes, dendritic cells, and stromal cells (64–67). Inhibition of RANTES signaling was found to be associated with abrogation of tumor invasion, suggesting that RANTES is required for invasion in TGFβRII repressed lung adenocarcinoma cells (Figure 2). The clinical significance of this pathway is further supported by the finding that tumor expression of both RANTES and CCR5 by a large panel of lung adenocarcinoma is associated with patient survival. These studies illustrate how information gained from global expression profiling of tumors can be used to identify key pathways and genes mediating tumor growth, invasion, and metastasis.
Figure 2.
Classification of lung adenocarcinoma reflects progression along a spectrum of invasiveness spanning epithelial atypia, bronchioloalveolar adenocarcinoma (BAC), BAC with foci of microinvasion, and invasive adenocarcinoma. In cells with repression of TGFβ-RII (lower panel), signaling through the Smad pathway is abrogated (right). Invasiveness is mediated through noncanonical signaling pathways that require RANTES (left).
Gene Expression Predictors of Lung Cancer Survival
Proof of concept that tumor mRNA profiling provides clinically significant information regarding patient outcome after resection has been established. The promise of this approach to segregate tumors and to direct patient treatment based on these signatures is being prospectively tested in two breast cancer clinical trials. For NSCLC, several predictors have been developed, which for the most part are based on methodologically sound approaches that include independent validation. As indicated in Table 1, the results of these studies are heterogeneous both in terms of the number of genes in the predictors and in the specific genes included in each signature. This heterogeneity is expected, given differences in study design, assay platform, tumor histology, and patient selection. Subsequently, Shedden and colleagues published the results of a large, multicenter, blinded evaluation of several genomic signatures of prognosis in 442 adenocarcinomas (68). The addition of clinical covariates enhanced the performance of novel signatures, which performed slightly better than previously published signatures (69, 70). None of the signatures performed significantly better than the others. This landmark study is a model for the meticulous handling of challenges inherent in translational cancer genomic studies, and for its vast data repository of clinical and pathologically annotated data that now is available for testing of new signatures and hypotheses. It remains unclear which genomic predictor of prognosis is best or whether specific genes or entire signatures are most important in predicting outcome. Independent prospective evaluation of the predictive accuracy of these signatures, prospective clinical trials, and application to small biopsy specimens (71) are appropriate and will be required to extend this important area of research.
TABLE 1.
RECENT GENE EXPRESSION PREDICTORS OF NON–SMALL CELL LUNG CANCER OUTCOME
| Classifier | Histology | Genes | Outcome | Platform |
|---|---|---|---|---|
| Duke Metagene (69) | NSCLC, Stages I, II, III | 79 | Recurrence | Affymetrix U133A |
| NCI Cytokine (72) | Adenocarcinoma, Stages I, II, III, IV | 15 | Lymph node metastasis and survival | RT-PCR |
| Milan 10 gene (73) | Adenocarcinoma, Stage I | E2F1, E2F4, HOXB7, HSPG2, MCM6, NUDCD1, RRM2, SERPINB5, SF3B1, SCGB3A1 | Overall Survival | RT-PCR |
| Taiwan 5 gene (70) | NSCLC Stages I, II, III | DUSP6, ERBB3, MMD, STAT1, LCK | Overall and Relapse Free Survival | RT-PCR |
| Toronto 3 gene (74) | NSCLC Stages I, II, III | CCR7, HIF1A, STX1A | Overall Survival | RT-PCR |
Definition of abbreviation: NSCLC = non–small cell lung cancer.
Gene expression profiling studies of lung tumors not only provide important information regarding prognosis and survival, but also help to identify potential therapeutic targets by offering key insights into how genetic alterations affect lung tumorigensis and metastasis. As medicine becomes increasingly reliant on genomic information, these studies hold much promise for the diagnosis and treatment of lung cancer.
Supported by the NIH (1RO1CA120174 to C.A.P.), the American Cancer Society (RSG-CNE-108857), Joan's Legacy Foundation, and by the Flight Attendants Medical Research Institute.
Conflict of Interest Statement: A.C.B. is a co-inventor involved in a patent application “Gene expression profiles correlating with histology and prognosis” measuring gene expression of a set of genes with positive correlation with histologic subtype and prognosis. To date, no financial benefit has been derived from this application. R.L.T. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.A.P. received $125,000 in 2006–2007 from Philip Research, NA as research grants and is co-inventor for a pending patent filed by Columbia University for expression profiles correlating with histology and prognosis.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin 2008;58:71–96. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin 2006;56:106–130. [DOI] [PubMed] [Google Scholar]
- 3.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–2826. [DOI] [PubMed] [Google Scholar]
- 4.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, et al. Trastuzumab plus adjuvant chemotherapy for operable her2-positive breast cancer. N Engl J Med 2005;353:1673–1684. [DOI] [PubMed] [Google Scholar]
- 5.Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: The role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev 2003;12:689–698. [PubMed] [Google Scholar]
- 6.Hecht SS. Cigarette smoking and lung cancer: chemical mechanisms and approaches to prevention. Lancet Oncol 2002;3:461–469. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz AG, Prysak GM, Bock CH, Cote ML. The molecular epidemiology of lung cancer. Carcinogenesis 2007;28:507–518. [DOI] [PubMed] [Google Scholar]
- 8.Vineis P, Anttila S, Benhamou S, Spinola M, Hirvonen A, Kiyohara C, Garte SJ, Puntoni R, Rannug A, Strange RC, et al. Evidence of gene gene interactions in lung carcinogenesis in a large pooled analysis. Carcinogenesis 2007;28:1902–1905. [DOI] [PubMed] [Google Scholar]
- 9.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag snps identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008;40:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008;452:633–637. [DOI] [PubMed] [Google Scholar]
- 11.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452:638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuller HM, Orloff M. Tobacco-specific carcinogenic nitrosamines: ligands for nicotinic acetylcholine receptors in human lung cancer cells. Biochem Pharmacol 1998;55:1377–1384. [DOI] [PubMed] [Google Scholar]
- 13.Lam DC, Girard L, Ramirez R, Chau WS, Suen WS, Sheridan S, Tin VP, Chung LP, Wong MP, Shay JW, et al. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res 2007;67:4638–4647. [DOI] [PubMed] [Google Scholar]
- 14.Houghton J, Stoicov C, Nomura S, Rogers AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR, Wang TC. Gastric cancer originating from bone marrow-derived cells. Science 2004;306:1568–1571. [DOI] [PubMed] [Google Scholar]
- 15.Avital I, Moreira AL, Klimstra DS, Leversha M, Papadopoulos EB, Brennan M, Downey RJ. Donor-derived human bone marrow cells contribute to solid organ cancers developing after bone marrow transplantation. Stem Cells 2007;25:2903–2909. [DOI] [PubMed] [Google Scholar]
- 16.Powell CA, Spira A, Derti A, DeLisi C, Liu G, Borczuk A, Busch S, Sahasrabudhe S, Chen Y, Sugarbaker D, et al. Gene expression in lung adenocarcinomas of smokers and nonsmokers. Am J Respir Cell Mol Biol 2003;29:157–162. [DOI] [PubMed] [Google Scholar]
- 17.Woenckhaus M, Klein-Hitpass L, Grepmeier U, Merk J, Pfeifer M, Wild P, Bettstetter M, Wuensch P, Blaszyk H, Hartmann A, et al. Smoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancers. J Pathol 2006;210:192–204. [DOI] [PubMed] [Google Scholar]
- 18.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One 2008;3:e1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura K, Bowman ED, Simon R, Peng AC, Robles AI, Jones RT, Katagiri T, He P, Mizukami H, Charboneau L, et al. Laser capture microdissection and microarray expression analysis of lung adenocarcinoma reveals tobacco smoking- and prognosis-related molecular profiles. Cancer Res 2002;62:3244–3250. [PubMed] [Google Scholar]
- 20.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339–346. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol 2007;25:561–570. [DOI] [PubMed] [Google Scholar]
- 22.Hackett NR, Heguy A, Harvey BG, O'Connor TP, Luettich K, Flieder DB, Kaplan R, Crystal RG. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol 2003;29:331–343. [DOI] [PubMed] [Google Scholar]
- 23.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA 2004;101:10143–10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebel S, Gerstmayer B, Bosio A, Haussmann H-J, Miert EV, Muller T. Gene expression profiling in respiratory tissues from rats exposed to mainstream cigarette smoke. Carcinogenesis 2004;25:169–178. [DOI] [PubMed] [Google Scholar]
- 25.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med 2007;13:361–366. [DOI] [PubMed] [Google Scholar]
- 26.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature 2001;414:105–111. [DOI] [PubMed] [Google Scholar]
- 27.Heguy A, O'Connor TP, Luettich K, Worgall S, Cieciuch A, Harvey B-G, Hackett NR, Crystal RG. Gene expression profiling of human alveolar macrophages of phenotypically normal smokers and nonsmokers reveals a previously unrecognized subset of genes modulated by cigarette smoking. J Mol Med 2006;84:318. [DOI] [PubMed] [Google Scholar]
- 28.Borczuk AC, Gorenstein L, Walter KL, Assaad AA, Wang L, Powell CA. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol 2003;163:1949–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonner AE, Lemon WJ, Devereux TR, Lubet RA, You M. Molecular profiling of mouse lung tumors: association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene 2004;23:1166–1176. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Kho AT, Kohane IS, Sun Y. Predicting survival within the lung cancer histopathological hierarchy using a multi-scale genomic model of development. PLoS Med 2006;3:e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 2005;115:1503–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anbazhagan R, Tihan T, Bornman DM, Johnston JC, Saltz JH, Weigering A, Piantadosi S, Gabrielson E. Classification of small cell lung cancer and pulmonary carcinoid by gene expression profiles. Cancer Res 1999;59:5119–5122. [PubMed] [Google Scholar]
- 33.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mrna expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 2001;98:13790–13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Virtanen C, Ishikawa Y, Honjoh D, Kimura M, Shimane M, Miyoshi T, Nomura H, Jones MH. Integrated classification of lung tumors and cell lines by expression profiling. Proc Natl Acad Sci USA 2002;99:12357–12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boelens MC, van den Berg A, Vogelzang I, Wesseling J, Postma DS, Timens W, Groen HJ. Differential expression and distribution of epithelial adhesion molecules in non-small cell lung cancer and normal bronchus. J Clin Pathol 2007;60:608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kettunen E, Anttila S, Seppanen JK, Karjalainen A, Edgren H, Lindstrom I, Salovaara R, Nissen AM, Salo J, Mattson K, et al. Differentially expressed genes in nonsmall cell lung cancer: expression profiling of cancer-related genes in squamous cell lung cancer. Cancer Genet Cytogenet 2004;149:98–106. [DOI] [PubMed] [Google Scholar]
- 37.Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC, Schnabel P, Warth A, Poustka A, Sultmann H, et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer 2009;63:32–38. [DOI] [PubMed] [Google Scholar]
- 38.Wang T, Hopkins D, Schmidt C, Silva S, Houghton R, Takita H, Repasky E, Reed SG. Identification of genes differentially over-expressed in lung squamous cell carcinoma using combination of cDNA subtraction and microarray analysis. Oncogene 2000;19:1519–1528. [DOI] [PubMed] [Google Scholar]
- 39.Inamura K, Fujiwara T, Hoshida Y, Isagawa T, Jones MH, Virtanen C, Shimane M, Satoh Y, Okumura S, Nakagawa K, et al. Two subclasses of lung squamous cell carcinoma with different gene expression profiles and prognosis identified by hierarchical clustering and non-negative matrix factorization. Oncogene 2005;24:7105–7113. [DOI] [PubMed] [Google Scholar]
- 40.Larsen JE, Pavey SJ, Passmore LH, Bowman R, Clarke BE, Hayward NK, Fong KM. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis 2007;28:760–766. [DOI] [PubMed] [Google Scholar]
- 41.Larsen JE, Pavey SJ, Bowman R, Yang IA, Clarke BE, Colosimo ML, Hayward NK, Fong KM. Gene expression of lung squamous cell carcinoma reflects mode of lymph node involvement. Eur Respir J 2007;30:21–25. [DOI] [PubMed] [Google Scholar]
- 42.Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, Oc P, Socci ND, Ngai I, Carlson D, Ghossein R, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res 2005;65:3063–3071. [DOI] [PubMed] [Google Scholar]
- 43.Vachani A, Nebozhyn M, Singhal S, Alila L, Wakeam E, Muschel R, Powell CA, Gaffney P, Singh B, Brose MS, et al. A 10-gene classifier for distinguishing head and neck squamous cell carcinoma and lung squamous cell carcinoma. Clin Cancer Res 2007;13:2905–2915. [DOI] [PubMed] [Google Scholar]
- 44.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature 2007;446:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins FS, Barker AD. Mapping the cancer genome: pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am 2007;296:50–57. [PubMed] [Google Scholar]
- 46.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA 2007;104:16663–16668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator ttf-1. Cancer Res 2007;67:6007–6011. [DOI] [PubMed] [Google Scholar]
- 49.Borczuk AC, Powell CA. Expression profiling and lung cancer development. Proc Am Thorac Soc 2007;4:127–132. [DOI] [PubMed] [Google Scholar]
- 50.Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new world health organization classification of lung tumours. Eur Respir J 2001;18:1059–1068. [DOI] [PubMed] [Google Scholar]
- 51.Maeshima AM, Niki T, Maeshima A, Yamada T, Kondo H, Matsuno Y. Modified scar grade: a prognostic indicator in small peripheral lung adenocarcinoma. Cancer 2002;95:2546–2554. [DOI] [PubMed] [Google Scholar]
- 52.Borczuk AC, Qian F, Eleazar J, Assad A, Kazeros A, Powell CA. Invasive size is an independent predictor of survival in pulmonary adenocarcinoma. Am J Surg Pathol (In press) [DOI] [PMC free article] [PubMed]
- 53.Rena O, Papalia E, Ruffini E, Casadio C, Filosso PL, Oliaro A, Maggi G. Stage I pure bronchioloalveolar carcinoma: recurrences, survival and comparison with adenocarcinoma of the lung. Eur J Cardiothorac Surg 2003;23:409–414. [DOI] [PubMed] [Google Scholar]
- 54.Sakurai H, Maeshima A, Watanabe S, Suzuki K, Tsuchiya R, Maeshima AM, Matsuno Y, Asamura H. Grade of stromal invasion in small adenocarcinoma of the lung: histopathological minimal invasion and prognosis. Am J Surg Pathol 2004;28:198–206. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki K, Yokose T, Yoshida J, Nishimura M, Takahashi K, Nagai K, Nishiwaki Y. Prognostic significance of the size of central fibrosis in peripheral adenocarcinoma of the lung. Ann Thorac Surg 2000;69:893–897. [DOI] [PubMed] [Google Scholar]
- 56.Yim J, Zhu LC, Chiriboga L, Watson HN, Goldberg JD, Moreira AL. Histologic features are important prognostic indicators in early stages lung adenocarcinomas. Mod Pathol 2007;20:233–241. [DOI] [PubMed] [Google Scholar]
- 57.Borczuk AC, Kim HK, Yegen HA, Friedman RA, Powell CA. Lung adenocarcinoma global profiling identifies type II transforming growth factor-beta receptor as a repressor of invasiveness. Am J Respir Crit Care Med 2005;172:729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K, Mitsudomi T, Takahashi T. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol 2006;24:1679–1688. [DOI] [PubMed] [Google Scholar]
- 59.Motoi N, Szoke J, Riely GJ, Seshan VE, Kris MG, Rusch VW, Gerald WL, Travis WD. Lung adenocarcinoma: modification of the 2004. WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. Am J Surg Pathol 2008;32:810–827. [DOI] [PubMed] [Google Scholar]
- 60.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816–824. [DOI] [PubMed] [Google Scholar]
- 61.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CV, Moses HL. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active kras expression. Genes Dev 2006;20:3147–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu SL, Herrington H, Reh D, Weber S, Bornstein S, Wang D, Li AG, Tang CF, Siddiqui Y, Nord J, et al. Loss of transforming growth factor-beta type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev 2006;20:1331–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Munoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, Sozmen EG, Madison BB, Pozzi A, Moon RT, et al. Transforming growth factor beta receptor type ii inactivation induces the malignant transformation of intestinal neoplasms initiated by apc mutation. Cancer Res 2006;66:9837–9844. [DOI] [PubMed] [Google Scholar]
- 64.Fraziano M, Cappelli G, Santucci M, Mariani F, Amicosante M, Casarini M, Giosue S, Bisetti A, Colizzi V. Expression of CCR5 is increased in human monocyte-derived macrophages and alveolar macrophages in the course of in vivo and in vitro mycobacterium tuberculosis infection. AIDS Res Hum Retroviruses 1999;15:869–874. [DOI] [PubMed] [Google Scholar]
- 65.Kunkel EJ, Boisvert J, Murphy K, Vierra MA, Genovese MC, Wardlaw AJ, Greenberg HB, Hodge MR, Wu L, Butcher EC, et al. Expression of the chemokine receptors CCR4, CCR5, and CXCR3 by human tissue-infiltrating lymphocytes. Am J Pathol 2002;160:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma B, Liu W, Homer RJ, Lee PJ, Coyle AJ, Lora JM, Lee CG, Elias JA. Role of CCR5 in the pathogenesis of IL-13-induced inflammation and remodeling. J Immunol 2006;176:4968–4978. [DOI] [PubMed] [Google Scholar]
- 67.van Deventer HW, O'Connor W Jr, Brickey WJ, Aris RM, Ting JP, Serody JS. C-C chemokine receptor 5 on stromal cells promotes pulmonary metastasis. Cancer Res 2005;65:3374–3379. [DOI] [PubMed] [Google Scholar]
- 68.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, Kratzke R, Watson MA, Kelley M, Ginsburg GS, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med 2006;355:570–580. [DOI] [PubMed] [Google Scholar]
- 70.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 2007;356:11–20. [DOI] [PubMed] [Google Scholar]
- 71.Borczuk AC, Shah L, Pearson GD, Walter KL, Wang L, Austin JH, Friedman RA, Powell CA. Molecular signatures in biopsy specimens of lung cancer. Am J Respir Crit Care Med 2004;170:167–174. [DOI] [PubMed] [Google Scholar]
- 72.Seike M, Yanaihara N, Bowman ED, Zanetti KA, Budhu A, Kumamoto K, Mechanic LE, Matsumoto S, Yokota J, Shibata T, et al. Use of a cytokine gene expression signature in lung adenocarcinoma and the surrounding tissue as a prognostic classifier. J Natl Cancer Inst 2007;99:1257–1269. [DOI] [PubMed] [Google Scholar]
- 73.Bianchi F, Nuciforo P, Vecchi M, Bernard L, Tizzoni L, Marchetti A, Buttitta F, Felicioni L, Nicassio F, Di Fiore PP. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest 2007;117:3436–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lau SK, Boutros PC, Pintilie M, Blackhall FH, Zhu C-Q, Strumpf D, Johnston MR, Darling G, Keshavjee S, Waddell TK, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol 2007;25:5562–5569. [DOI] [PubMed] [Google Scholar]