Abstract
In an effort to further our understanding of lung cancer biology and to identify new candidate biomarkers to be used in the management of lung cancer, we need to probe these tissues and biological fluids with tools that address the biology of lung cancer directly at the protein level. Proteins are responsible of the function and phenotype of cells. Cancer cells express proteins that distinguish them from normal cells. Proteomics is defined as the study of the proteome, the complete set of proteins produced by a species, using the technologies of large-scale protein separation and identification. As a result, new technologies are being developed to allow the rapid and systematic analysis of thousands of proteins. The analytical advantages of mass spectrometry (MS), including sensitivity and high-throughput, promise to make it a mainstay of novel biomarker discovery to differentiate cancer from normal cells and to predict individuals likely to develop or recur with lung cancer. In this review, we summarize the progress made in clinical proteomics as it applies to the management of lung cancer. We will focus our discussion on how MS approaches may advance the areas of early detection, response to therapy, and prognostic evaluation.
Keywords: proteome, translational research, biomarkers
Lung cancer is the leading cause of cancer-related death worldwide among both males and females, with more than 1 million deaths annually (1). Non–small cell lung cancer (NSCLC) accounts for about 80% of all lung cancers. Although advances have been made in diagnosis and treatment strategies in the last decade, the prognosis of NSCLC patients is poor, with a 5-year overall survival of 15 to 20% (2). This is mainly due to a lack of early diagnosis tools, with more than 60% of the patients diagnosed with advanced or metastatic disease (3) and therefore not eligible for a curative surgical resection. Lung cancer is often suspected on the basis of abnormal chest imaging and/or nonspecific symptoms. Bronchoscopy, with cytopathologic examination of bronchoalveolar lavage, endobronchial brushings and biopsies obtained from the suspect area, is in general used as an initial diagnostic tool. However, while this procedure is 100% specific for lung cancer, the sensitivity is low and ranges from 30% for small peripheral lesions to 80% for central endobronchial tumors (4). More invasive and expensive diagnostic tests are often required, delaying the diagnosis and the subsequent treatment initiation. Surgical resection offers the best chance for cure. For patients undergoing surgery, the long-term prognosis remains disappointing, with a 5-year overall survival of 50% only (5). Recent studies showed that survival of surgically resected patients with NSCLC might be improved by systemic platinum-based adjuvant chemotherapy (6, 7), but which patients might benefit from this treatment cannot be determined accurately.
To improve lung cancer management and survival, there is a great need to develop screening and early diagnosis strategies that are sensitive, specific, and noninvasive; tools predicting prognosis to optimize treatment and avoid overtreatment; and tools identifying potential therapeutic targets. During the last 10 years, genomic and proteomic approaches have been used for these purposes. While epigenetic and genetic alterations are driving carcinogenesis and genomic studies have provided valuable information on lung cancer molecular biology (8), a proteomic approach opens a new window into the pathogenesis of lung cancer. In favor of this new approach are two main arguments. First, the phenotype of a cell is determined by proteins and cannot be predicted by genomics alone. Indeed, protein expression levels are poorly correlated to messenger RNA expression levels (9); and post-translational modifications such as phosphorylation, glycosylation, and proteolytic processing, which are common events, have the potential to significantly modify protein functions and characteristics of the cell or tissue where the protein is expressed. Second, while genetics require DNA extraction from tumor cells that are not easily obtained by noninvasive methods, proteomics do not necessarily need a direct access to tumor cells. Proteins can easily and noninvasively be obtained from various sources such as blood and exhaled breath condensate (EBC).
In this review, we summarize recent applications of mass spectrometry (MS) to proteomic profiling of lung cancer. We discuss the challenges of this approach, its limitations, and the potential applications to the management of lung cancer.
METHODOLOGICAL CONSIDERATIONS
A mass spectrometer analyzes proteins after their conversion to gaseous ions, based on their mass-to-charge ratio (m/z). It is made of three basic elements: an ion source charging proteins and converting them to gaseous ions, a mass analyzer separating them as a function of their m/z ratios, and a detector capturing the ionized proteins after separation. Two methods of ionization are routinely used: matrix-assisted laser desorption ionization (MALDI) uses a laser to desorb and ionize proteins from the solid phase to the gaseous phase, and electrospray ionization (ESI) ionizes and vaporizes proteins from liquid solutions (Table 1). The most frequently used mass analyzers are time-of-flights (TOF), quadruple ion traps, linear ion traps, obitraps, and Fourier-transform ion cyclotron resonance (FT-ICR) cells. The combination of the same or different mass analyzers allows one to select and fragment ions of interest to determine their structures or in the case of peptides their sequence. This analytical approach is referred to as tandem mass spectrometry or MS/MS. Mass spectrometers not only identify protein sequences, but can also detect post-translational modifications such as acetylations and phosphorylations. Overall, MS can detect significant changes in protein profiles associated with clinical features, such as the development of neoplasia, histology, response to chemotherapy, and prognosis.
TABLE 1.
MASS SPECTROMETRY–BASED PROTEOMICS TECHNOLOGY PLATFORMS
| MALDI MS | Two-Dimensional Gel | Tandem MS | MRM MS/MS | |
|---|---|---|---|---|
| Purpose | Profiling | Profiling and ID | Profiling and ID | Quantitation |
| Sample preparation | ± Depletion, fractionation | ± Depletion, fractionation | Digestion | Digestion |
| Analytes | MS Peaks | Spots | MS-MS Spectra | Peak areas |
| Representative of | Proteins, peptides | Proteins | Peptide sequences | Protein amounts |
| Depth of analysis | 100–300 peaks | 100–1,000 proteins | 500–3,000 proteins | Single target protein |
| Analysis compare | Patterns | Patterns | Inventories | Amounts |
| Protein identification | Difficult | Readily by MS | High-throughput | Quantitative |
| Information | Correlation to biology | Link to biology | Link to biology | Verification |
Definition of abbreviations: MALDI = matrix-assisted laser desorption/ionization; MRM = multiple reaction monitoring; MS = mass spectrometry.
Matrix-assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry
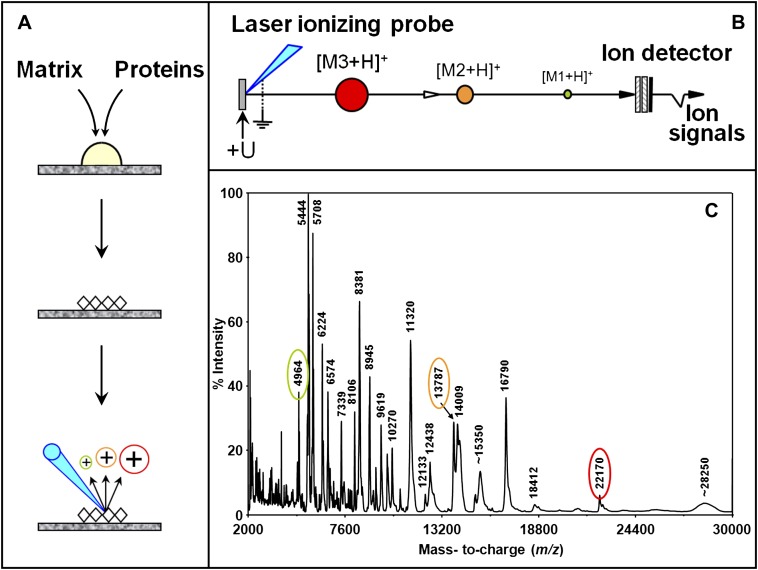
Among proteomic technologies, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) is a technique that has allowed rapid progress in cancer biology. It is a simple and high-throughput technique that analyzes with high sensitivity and specificity intact proteins expressed in complex biological mixtures, such as serum, urine, and tissues. This technique requires the sample co-crystallization with a matrix that absorbs laser energy and subsequently ejects and ionizes molecules via a proton transfer mechanism into the gas phase, forming ions with the general formula [M+H]+ (Figure 1). Ions are then accelerated in the ion source by a fixed potential difference and travel a fixed-length field-free distance before reaching the detector at a speed inversely proportional to their m/z ratios (lighter ions are faster to reach the detector than the heavier ions for a same charge). The time taken by each ion to hit the detector creates a signal, which indicates m/z ratio in the x-axis and ion intensity in the y-axis. Because the MALDI process essentially favors the production of singly charged molecular ions, it allows the analysis of complex protein mixtures without fractionation (10). Intact proteins with molecular weights (MW) 1 to over 200 kD can be determined with high accuracy. Several characteristics of MALDI-TOF MS make it a widely used technique for the analysis of complex biological samples (such as tissues, whole cells, laser-captured microdissected cells, blood, serum, urine) with high mass accuracy (far better than any gel system), high-throughput capability (sample analysis in seconds), small required sample size (possible analysis of just a few cells) and higher tolerance for salts, buffers, or biological contaminants. When used in combination with surface chromatography, this method is also known as surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS). It uses chromatographic chip arrays to selectively bind subsets of proteins from complex samples. The surfaces can be washed to remove nonspecifically bound proteins and substances that can interfere with the ionization process. Then, matrix solution is applied to the array binding the proteins and MALDI-TOF MS is performed.
Figure 1.
Principle of matrix-assisted laser desorption/ionization (MALDI-TOF MS) time-of-flight mass spectrometry. (A) Analyte molecules are first mixed with matrix molecules in a ratio of about 1 to 5,000. Upon solvent evaporation, matrix-analyte co-crystals form. In the ion source of the instrument, irradiation of these with a brief laser pulse initiates the desorption-ionization events. (B) The newly formed molecular ions are accelerated in the source of the instrument by a constant potential difference, giving all of the ions of the same charge the same energy. Ions are therefore separated in time according to their mass as they travel the length of the time-of-flight. (C) MALDI-TOF mass spectrum obtained in the positive ionization mode from a complex protein mixture. By a large majority, essentially singly charged ions (z = 1) are observed. Reprinted by permission from Reference 131.
MALDI MS is a very sensitive analytical technique, particularly for peptides and proteins. The amount of sample necessary for analysis ranges from a few fentomoles for peptides to a few picomoles for higher MW proteins (∼> 50 kD) deposited on target. As with every other ionization method, ion suppression effects occur with MALDI. These happen in the gas phase during the desorption/ionization step and occur when molecules with different proton affinities compete for the ionizing protons.
Liquid Chromatography Tandem Mass Spectrometry
Powerful for the separation and identification of peptides and proteins in a complex mixture, this technique directly couples liquid chromatography (HPLC) with ESI MS and has had a profound impact on tumor protein profiling (11).
With electrospray ionization, analyte samples are directly analyzed from solution. Ions are formed by an electrospray process by pushing the analyte solution through a thin needle biased at positive voltage. A continuous spray forms at the tip of the needle. The spray process forms very small droplets that progressively desolvate liberating ions. One of the fundamental properties of electrospray-produced peptide and protein ions is that they carry multiple charges. The MW of the analyte is then obtained by deconvoluting the signal distribution. One limitation of ESI MS is that because each sample molecule generates a distribution of ions, it becomes increasingly difficult to analyze and deconvolute overlapping signal distributions from complex mixtures. However, electrospray ionization is performed from a liquid sample, and liquid-based chromatographic separation systems such as reverse-phase HPLC can be directly coupled to the mass spectrometers (Figure 2) for mass determination (liquid chromatography [LC]-MS) and peptide sequence analyses (LC-MS/MS).
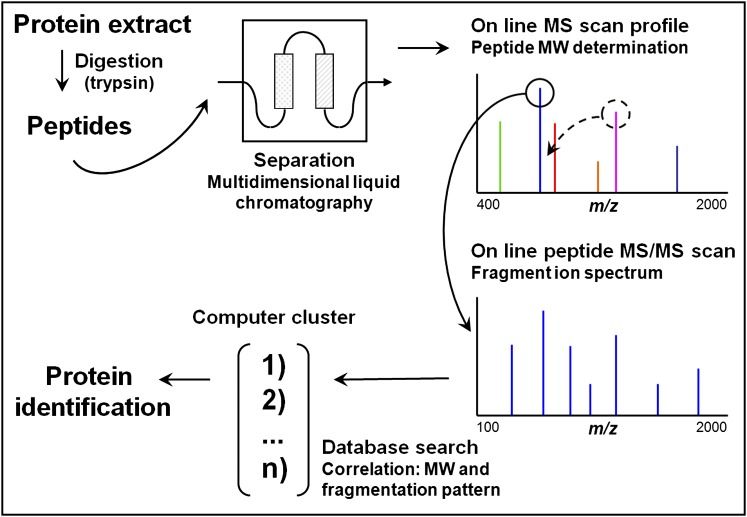
Figure 2.
Bottom up or shotgun protein identification by liquid chromatography (LC)-mass spectrometry (MS)/MS from a complex protein mixture. The protein mixture is first digested (by trypsin) and the resulting peptides are separated by multidimensional liquid chromatography (typically strong cation exchange followed by reverse-phase separation) coupled online to the mass spectrometer. As they elute, the m/z ratios of the peptides are first determined followed by one or several MS/MS scans from the most abundant peptide signals. This cycle is repeated until all of the peptides have eluted from the chromatography column. For each precursor peptide selected for MS/MS, peptides of similar nominal mass are extracted from sequence databases and predicted fragmentation patterns are derived in silico. These patterns are then compared with the experimental fragmentation spectrum to generate correlation scores. Positive identification of a protein is based on the observation of two or more peptides issued from its sequence. Modified by permission from Reference 131.
A fully automated LC-MS platform follows a “bottom-up” analytical approach (as opposed to a “top down” approach, in which intact proteins are ionized and fragmented to peptidic fragments). In the bottom-up (also referred to as “shotgun”) proteomic approach, proteins are first digested with site-specific proteases, and the resulting peptides are separated by LC and analyzed online by fast cycles of ESI MS and MS/MS (Figure 2). Cycles consist of an initial MS scan in which charged peptides are measured according to their m/z ratios. The most abundant of these are then sequentially selected for MS/MS analyses. The resulting fragment ions are then analyzed in a second MS scan according to their m/z ratios. Based on our understanding of the fragments produced in the collision cell and their precise MW, peptide sequences can be deduced. Through comparisons with predicted sequences of the same nominal mass in gene and protein databases, peptides are identified and proteins from which they came are deduced. However, with extremely complex protein mixtures, confident and reproducible identification by MS/MS sequencing becomes difficult. Also, the high-abundance proteins may obscure the low-abundance ones. To overcome these problems, different separation methods are combined with MS analysis, such as size exclusion, anion exchange, strong cation exchange, isoelectric focusing, and reverse phase chromatography. In particular, multidimensional protein identification technology (MudPIT), a combination of strong cation exchange and reverse phase columns, can be adapted to a shotgun MS proteomic platform (12–14), taking advantage of ion exchange and reverse phase separations, data-independent scanning (15), and a reduced total analysis time.
Commonly, two types of ESI mass analyzers are used for high-throughput proteomic analyses, namely ion traps (16, 17) and Fourier-transformed mass analyzers (FTMS) (18, 19). These physically “trap” ions in their centers. Once the ions are cooled in the center of the trap, they can be sequentially ejected from the trap according to their m/z ratios and then detected. In modern ion trap instruments such as high-capacity or linear traps, the sequence of events from MS to MS/MS measurements is very fast (in the ∼0.1- to 0.2-s time frame). These instruments are ideal for high throughput proteomic analyses in the LC-MS configuration (17, 20). Although ion traps offer very high-throughput and sensitivity, they lack mass resolution and accuracy (typically above 100 ppm). FTMS instruments currently provide the highest mass measurement accuracy available for structural characterization of peptides, proteins, and other biomolecules (18, 21, 22). FTMS instruments consist of an ion source, some ion optics to transfer the ions into the magnetic field, and the ion cyclotron resonance (ICR) cell or Penning trap (18, 21, 22). Ions are trapped in the ICR by a magnetic field, excited by a resonant excitation pulse, and the resulting changes in image charge around detector electrodes are digitized by fast Fourier Transform and stored in computer memory. One of the advantages of FTMS is the very high resolving power, providing mass accuracy routinely better than 1 ppm. LC-MS (and LC-MS/MS) can be performed using FTMS, but the duty cycle limits the abilities of the instrument to effectively perform a fast scan of the LC run and subsequently perform MS/MS measurements. To partially circumvent this limitation, hybrid ion trap/FTMS instruments have been developed. In this case, peptides from the LC run are selected, fragmented, and the resulting ions are analyzed in the ion trap while the ICR cell is used to accurately measure the MW of the parent ions (thereby achieving higher cycle rates) (23). The newly developed ion trap/orbitrap mass spectrometers (24) are capable of performing similar measurements (25–27).
Imaging Mass Spectrometry
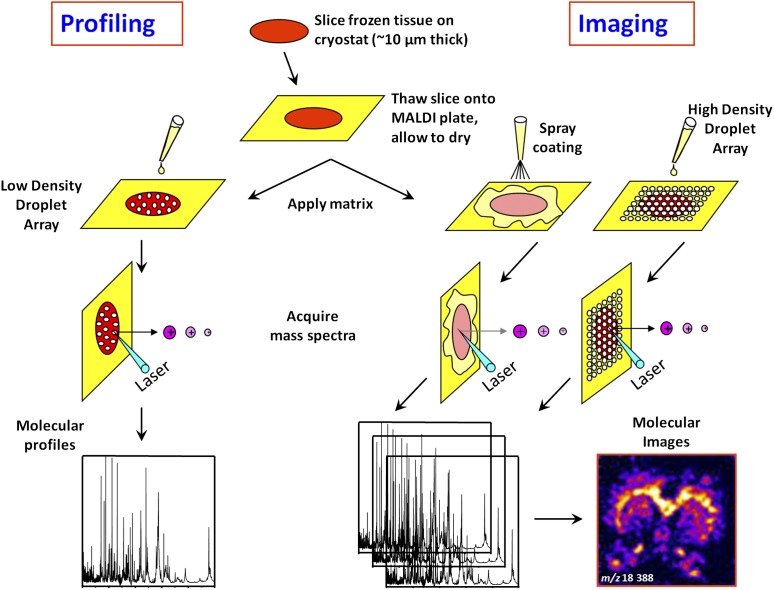
MALDI MS profiling/imaging is a technology used for direct mapping and high-resolution imaging of biomolecules present in tissue sections (28, 29). Frozen tissue sections approximately 10 μm thick are mounted on conductive target plates (indium-tin oxide–coated glass slides) (Figure 3). Matrix is then homogeneously deposited in a manner to avoid delocalization of the analytes contained within the sections. Several strategies can be used including spray-coating and automated printing of high-density droplet arrays. Individual spectra are acquired from the entire surface of the section with a fixed spatial resolution (50–300 μm). Each spectrum contains unique proteomic information representative of the underlying histology. From a single raster of a section, when integrating the intensities of specific m/z signals and plotting these as a function of their spatial coordinates, hundreds of ion (or protein) images can be visualized (Figure 3) (30). This gives us the tissue distribution of protein expression. However, it has several advantages upon immuno-histochemistry (IHC): it does not require the use of antibodies, can map the expression of hundreds of proteins in a single section, and it can detect post-translational modifications (31, 32). This technique shows potential for biomarker tissue localization, understanding of the molecular complexity of tumor tissues, and assessment of surgical margins in resected tumors (33, 34). Also, by imaging drugs and their metabolites, it allows direct analysis of tissue pharmacokinetics and drug metabolism (35), suggesting a possible application in assessment of response to therapy (36).
Figure 3.
Scheme outlining the different steps involved in profiling and imaging mass spectrometry of tissue samples. Sections are cut (∼10 μm thick) from fresh frozen biopsies and mounted on a conductive target plate. In the “Profiling” experiment, droplets of matrix are deposited on the sections at discrete locations of interest defined by histology. Protein profiles are then acquired. In the “Imaging” experiment, matrix is deposited in a homogeneous manner on the sections (spray or droplet array deposition), minimizing analyte delocalization. The entire surface of the sections is then systematically analyzed by moving the sample with a fixed step (ranging between 50 and 300 μm) defining the imaging resolution. At each grid coordinate, a protein profile is acquired. Protein expression maps (or images) can be reconstructed for every m/z value detected by integrating the corresponding signal intensities and plotting these as a function of sampling coordinates.
Quantitative Proteomics
Several MS-based approaches have been developed that allow for the relative or absolute quantification of proteins. Most protocols involve the use of stable isotopes to differentially label proteins or peptides before mixing samples for multiplexing onto the same analytical run (37–44). This negates instrumental variations and enables direct quantification of the same m/z values between the different isotopic labelings. These in vitro labeling strategies are, however, susceptible to technical variation introduced during the protein/peptide labeling and enrichment steps, and therefore require replicate analyses. One well-established technique is referred to as isotope-coded affinity tagging (ICAT), which employs stable heavy and light isotope affinity tags that are reactive toward cysteine residues (38). The tagging is performed on the intact protein before enzymatic digestion whereby the sample and reference protein extracts are tagged with either the light or the heavy tag, respectively. The extracts are then mixed and digested with a protease. The ICAT tags typically contain a biotin group that allows the separation of the tagged peptides using a streptavidin affinity purification step. The tagged peptides are then analyzed by LC-MS and quantification is performed by MS by monitoring the intensity of peptide pairs that are separated by the mass difference expected between the light and heavy tags. Recent “label-free” variations for quantitative LC-MS/MS strategies rely on peak intensity measurements of peptides detected by MS (45–47) or on the number of ions per protein detected in a mass spectrometric experiment (48).
Difference Gel Electrophoresis (DIGE) technology adds an essential quantitative component to two-dimensional gel-based proteomics to a level whereby even subtle changes in protein abundance and charge-altering post-translation modifications (such as acetylation and phosphorylation, among others) can be monitored from multiple experimental conditions with statistical confidence (49–53). DIGE overcomes many of the limitations commonly associated with two-dimensional gels, such as analytical (gel-to-gel) variation and limited dynamic range that can severely hamper a quantitative differential-display study. This is accomplished by multiplexing samples that have been pre-labeled with spectrally resolvable fluorescent dyes (Cy2, Cy3, and Cy5) into the same analytical run (2D gel), as this removes gel-to-gel variation from the quantitative measurements made for each resolved protein between the three dye excitation/emission spectral channels (54, 55). Although direct quantification is performed between the Cy dye channels within a gel without interference from gel-to-gel variation, this is not performed between the two individual samples co-resolved in the same gel. Rather, the Cy3:Cy2 and Cy5:Cy2 ratios for each protein are normalized across all of the gels in a large experiment, using the Cy2 signals for separate normalization of each protein under survey. This also allows for replicate samples from multiple conditions to be inter-compared using univariate statistical analyses (Student's t test, ANOVA) as well as multivariable statistical analyses (principle component analysis, hierarchal cluster analysis) (56, 57).
Multiple reaction monitoring (MRM) MS approaches are fast becoming well adapted to monitor and quantify very specific protein targets within complex mixtures (58, 59). In this case, a specific tryptic peptide is selected as a stoichiometric representative of the protein from which it is cleaved. Most MRM assays use electrospray ionization followed by two stages of mass selection: the first stage selects the mass of the intact peptide and, after fragmentation by collision, the second stage selects a specific fragment of the peptide. The two mass filters produce a very specific and sensitive response that can be used to detect and integrate a peptide peak in a one- or multi-dimensional chromatographic separation (LC-MS). This approach usually provides absolute structural specificity for the peptide, and in combination with a stable isotope-labeled internal standard of the same target peptide, it can provide absolute quantitation of peptide concentration over a range of four orders of magnitude (58–60). MRM measurements can be multiplexed to monitor several tens of targeted MS/MS transition from a single LC-MS run (4).
APPLICATIONS TO LUNG CANCER–RELATED BIOSPECIMENS
Lung Tissues
MS techniques have been used after different methods of separation to identify specific proteins from lung cancer tissues. Using two-dimensional gel electrophoresis combined with MS, Oh and coworkers (61) constructed a database containing protein expression data on more than 1,000 lung cancer tissues, intended to facilitate development of novel classifications for lung cancer and identification of novel markers for early diagnosis. Chen and colleagues (9) used two-dimensional gel electrophoresis to compare protein expression levels of 93 lung adenocarcinomas (ADC) to 10 uninvolved lung samples, and performed MALDI MS or peptide sequencing to identify 9 enzyme proteins significantly increased in lung ADC. Using two-dimensional gel electrophoresis with subsequent analysis by MALDI and ESI MS, Bergman and coworkers (62) identified overexpressed truncated forms of cytokeratins 6D and 8, and of cathepsin D as markers of tumor proliferation. This was confirmed by Gharib and colleagues (63); moreover, two isoforms of CK7, one of CK8, and one of CK19 were associated with survival. Performing two-dimensional gel electophoresis, MS and microarray analysis on 90 lung ADC tissues, Chen and coworkers (64) identified PGK1 as a survival predictor in stage I lung cancer. This was confirmed in an independent validation set of 117 ADC and squamous cell carcinomas (SCC) using tissue microarrays (TMA). Alfonso and colleagues (65) analyzed 12 surgically resected lung cancers with two-dimensional gel combined to MALDI MS and identified several proteins previously reported (such as annexin II, cathepsin D, HSP27, stathmin, MnSOD), confirming the validity of this technique in the identification of candidate biomarkers. Using MALDI-TOF MS to compare 10 NSCLC to 10 normal lung tissue lysates, Campa and coworkers (66) identified two overexpressed proteins in lung cancer specimens: the macrophage migration inhibitory factor (MMIF) and the cyclophilin A. These proteins were not found to be prognostic factors of disease based on a lung cancer TMA (67).
With recent technological advances, MS is now not only used to identify specific proteins or fragments, but also to characterize protein profiles that are associated with tumor characteristics and behavior. Our group used MALDI-TOF MS to profile 79 NSCLC and 14 normal lung frozen tissue sections, then selected differentially expressed MS signals (68) and defined a class prediction model using established methods (69). We identified protein signals that allowed the classification of lung tumors by histology, the distinction of primary tumors from metastases, and the identification of nodal involvement with 75% accuracy. We also identified a 15-signal signature classifying patients into good and poor prognosis groups. Recently we tested these proteins by IHC on formalin-fixed paraffin-embedded (FFPE) tissues from patients with NSCLC, and showed that the combined IHC scores of calmodulin, thymosin β4, and thymosin β10 were correlated to survival (70). We also found that cofilin-1 was correlated to a better outcome when the IHC score was low in patients without lymph node involvement or when the score was high in patients with positive nodes. To elucidate the biology of lung tumor development, we performed MALDI-TOF MS on 25 normal lung, 29 normal bronchial epithelium, 20 preinvasive, and 36 invasive lung cancer tissues from 53 patients (71). We found specific protein expression patterns classifying bronchial and alveolar tissue with normal histology from preinvasive bronchial lesions and invasive lung cancers with 90% accuracy.
To identify new prognostic factors for patients with NSCLC who had been surgically resected, Yanagisawa and coworkers (72) analyzed protein profiles of 174 NSCLC tumors and 27 normal lung tissues with MALDI MS. In the training set (116 NSCLC and 20 normal tissues), when comparing MS signals of patients with high risk of recurrence (who died within 5 years of surgery because of relapse) to those with low risk of recurrence (alive without any sign of relapse after a median follow-up of 89 months), 25 signals were found to be differentially expressed, associated with both relapse-free and overall survivals. In the independent validation set (58 NSCLC and 7 normal tissues), the signature was also significantly associated with relapse-free survival and overall survival among patients with stage I disease. For the other stages, only the association with overall survival was statistically significant. This 25-signal signature distinguished patients with NSCLC with good prognosis from those with poor prognosis better than the prognostic factors currently used in clinic, such as histology and TNM classification. By detecting which patients are likely to relapse after surgery, this 25-signal signature may help to decide which patients will benefit from systemic adjuvant therapy. The authors also identified approximately half of the proteins present in the signature and showed that they are involved in cell migration, cell death, cell cycle, protein metabolism, and transcription. These proteins included ribosomal protein L26-like 1, acylphosphatase, and phosphoprotein enriched in astrocytes 15. A better understanding of their role in NSCLC tumorigenesis and progression may lead to improved treatments.
Two recent improvements of this technology include its application to cytologic preparation and to formalin fixed paraffin embedded (FFPE) tissue sections. Amann and colleagues developed a technique allowing the use of fine-needle aspiration (FNA) samples for MALDI-TOF MS analysis (73). FNA is commonly used in the management of lung nodules by providing cells in suspension for cytologic analysis, but the samples are usually highly contaminated with blood and diverse debris. Cells collected by FNA were centrifuged onto indium-tin oxide–coated slides, fixed, stained with cresyl violet for microscopic analysis, and clumps of cancer cells were selectively spotted with matrix and analyzed by MALDI-TOF MS. High-quality, specific, and highly reproducible protein profiles were obtained and allowed classification of cancerous preparations from controls. Second, Groseclose and coworkers performed high-throughput analysis of the protein content of FFPE tissue microarrays, using MALDI imaging MS after on-tissue tryptic digestion to select and identify a much larger number of proteins, and successfully distinguished the different lung cancer histologies based on their proteomic profiles (74). This high-throughput approach may be especially valuable in efforts to correlate cancer biology to clinical information.
MS analysis of lung cancer tissues still requires invasive approaches for sample collection, providing a rationale to investigate its application in biospecimens such as blood and EBC that would be better suited for clinical application.
Blood
Blood proteome analysis assumes that tissue perfusion of tumors or host responses contribute to modification of circulating proteins or peptides. Because it is noninvasive, easy, fast, and amenable to repetitive measurements over time, this approach appears very appealing to researchers addressing the discovery of biomarkers, potentially allowing early diagnosis of cancer, monitoring of disease status, development of targeted therapies, evaluation of response to therapy, and survival. It may improve our diagnostic accuracy and decrease the number of thoracotomies currently required for pathologic evidence of malignant cells. Various serum biomarkers have already been investigated in lung cancer, but have not been proven useful in clinical practice because of their limited sensitivity and/or specificity. For example, carcinoembryonic antigen (CEA) displayed 95% specificity but only 26 to 33% sensitivity in the diagnosis of lung cancer (75–77). Also, most of these markers have a better sensitivity in advanced stages of lung cancers and are not useful for early diagnosis or screening.
Several studies using MALDI-based approaches reported serum protein expression profiles that distinguish patients with various cancers from control subjects (78–85). In one of these studies, using MALDI-TOF MS, Sidransky and colleagues (78) identified a serum protein profile in patients with head and neck cancer achieving 73% sensitivity and 90% specificity. When applied to the serum of patients with lung cancer, these profiles discriminated lung SCC with 52%, ADC with 34%, and large cell carcinoma with 40% sensitivity. However, the study was not designed to address whether this protein profile can discriminate patients with lung cancer from control subjects.
In an effort to discover serum biomarkers to improve lung cancer management, Patz and coworkers (86) used two-dimensional difference gel electrophoresis (DIGE) and MALDI-TOF MS. They identified three differentially expressed serum proteins by two-dimensional DIGE (transferrin, retinol-binding protein [RBP], and haptoglobin) and one by MALDI-TOF MS (α1-antitrypsin). They assayed these four proteins as well as two others previously known to be cancer associated (SCC antigen, CEA) on a training set of sera from 100 patients (50 patients with lung cancer, 50 control subjects). Using a Classification and Regression Tree (CART) analysis, they found that four of these proteins (CEA, RBP, α1-antitrypsin, and SCC antigen) were able to distinguish lung cancer cases from control subjects with 89.3% sensitivity and 84.7% specificity in the training set. When applied to the independent validation set, these markers displayed 77.8% sensitivity and 75.4% specificity. Using the classification scheme produced by the CART analysis, the probability of lung cancer for each patient was determined based on the terminal node into which he or she fell. For patients assigned to three of the different terminal nodes, the probability of having cancer was 92% in the training set and 90% in the validation set. When used alone, none of these four markers had sufficient diagnostic power, but when combined, they appeared to have value in suggesting lung cancer diagnosis and may be helpful for clinical management at different levels.
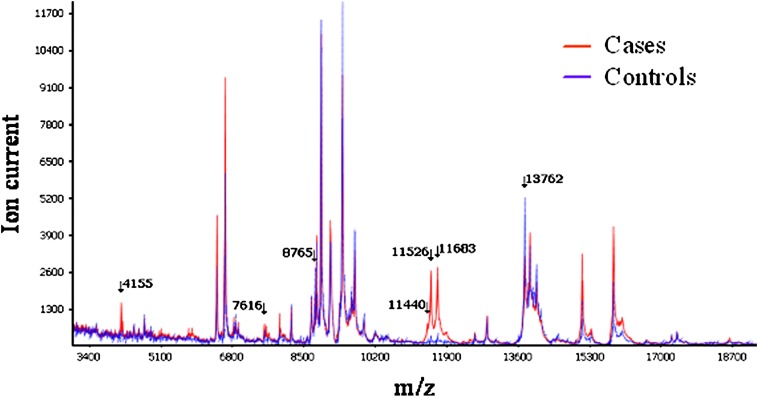
To demonstrate a role of noninvasive diagnosis of lung cancer, we used MALDI MS to analyze undepleted and unfractionated serum from a total of 288 patients with NSCLC and control subjects divided into training (92 cases, 92 controls) and test (50 cases, 56 controls) sets. In the training set, a seven-signal proteomic signature was defined (Figure 4) distinguishing lung cancer serum from matched controls with an overall accuracy of 78%, a sensitivity of 67.4%, and a specificity of 88.9%. In the test set, the signature reached an overall accuracy of 72.6%, a sensitivity of 58%, and a specificity of 85.7% (87). Because diagnosis of early-stage lung cancer is important, we searched for a protein signature discriminating stage I lung cancers from controls and found a six-signal signature reaching 70.8% sensitivity and 84.4% specificity in the training set (24 cases), and 57.1% sensitivity and 71.4% specificity in the test set (14 cases). With a multivariate logistic regression model applied on a total of 223 cases and controls, we showed that the serum signature was associated with lung cancer diagnosis independent of sex, smoking status, smoking pack-years, and C-reactive protein levels, and that it had the strongest association with lung cancer diagnosis. In this signature, three features displayed the highest discriminatory value. They were identified as a cluster of truncated forms of serum amyloid A (SAA), an acute phase protein secreted into circulation in several inflammatory diseases and cancers (88, 89), including lung cancer (67, 90, 91).
Figure 4.
MALDI MS serum spectra from individuals with lung cancer and matched control subjects. Average intensity of spectrum analysis between matched cases (red line) and controls (blue line) are presented. Arrows point to m/z values of discriminatory features. Reprinted by permission from Reference 132.
Han and colleagues (92) used SELDI-TOF MS to analyze the serum of 253 individuals split into a training set (89 NSCLC, 68 controls) and a validation set (62 NSCLC, 34 controls). From the proteomic spectra of serum samples obtained in the training set, using Biomarker Pattern software, they generated a classification tree with three different protein masses that effectively identified patients with lung cancer from controls with 94% accuracy, 91% sensitivity, and 97% specificity. When applied to the validation test, the classification tree allowed 89% sensitivity, 91% specificity, and 90% positive predictive value. The authors also used electrochemiluminescent immunoassay to detect CEA and Cyfra21-1 serum levels, and showed that the specificity and sensitivity of these biomarkers taken individually or in combination were significantly lower compared with the SELDI proteomic profile (42% sensitivity and 72% specificity for Cyfra21-1; 46% sensitivity and 76% specificity for CEA). Using SELDI-TOF MS on serum samples from 158 patients with lung cancer and 50 control subjects, Yang and coworkers (93) reported a five-signal protein signature distinguishing lung cancer cases from controls with 86.9% sensitivity and 80.0% specificity in the validation set.
To identify patients with NSCLC who are likely to benefit from treatment with epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs), Taguchi and colleagues (94) used MALDI MS on pretreatment serum of 302 patients treated with gefitinib or erlotinib, 139 of them assigned into a training set (from three cohorts) and 163 into a validation set (from two independent cohorts). Sera from 158 patients with NSCLC not treated with EGFR TKIs (from three cohorts) were also tested. Based on survival and time to progression after EGFR TKIs treatment, an algorithm based on eight MS signals was developed in the training set. The classification algorithm was then applied to the validation set, successfully identifying patients with improved outcome after EGFR TKIs treatment. Indeed, the median survival of patients in the predicted “good” and “poor” groups was 207 and 92 days, respectively, with a hazard ratio [HR] of death of 0.50 (0.24–0.78, 95% confidence interval [CI]), in the first cohort, and 306 and 107 days, respectively, with a HR of 0.41 (0.17–0.63, 95% CI) in the second cohort. The algorithm kept its predictive value independent of clinical factors associated with sensitivity to EGFR TKIs, such as sex, smoking history, and histology. The algorithm identified subgroups of smokers with favorable outcome after EGFR TKIs treatment, showing its benefit even in patients with clinical characteristics associated with low sensitivity to these drugs. For patients not treated with EGFR TKIs, the classification algorithm did not accurately classify patient outcomes.
Recently, an automated technology has been developed for the simultaneous measurement of serum peptides. Basically, peptides are captured and concentrated using reversed-phase batch processing in a magnetic particle-based format, automated on a liquid handling robot, and followed by MALDI-TOF MS. This technique is simple, scalable, and may provide better reproducibility, multidimensionality, and high throughput (95), but must be validated in larger populations and from several institutions.
Exhaled Breath Condensate and Sputum
EBC collection is a simple, safe, comfortable, and completely noninvasive method of sampling the lower respiratory tract. Because the condensate contains nonvolatile substances such as proteins, it is a potential diagnostic tool for lung diseases, and research has been done to apply this method as a screening tool for the early detection of lung cancer (96). By using two-dimensional gel electrophoresis, Griese and colleagues characterized proteins in EBC and saliva (97). Using immunoassay methods, endothelin-1 and interleukin-6 were both found to be increased in EBC of NSCLC patients when compared with control subjects (98, 99). Despite the ability to detect proteins in EBC, the use of proteomics to identify differential protein expression between EBC of patients with lung cancer and healthy control subjects has not been documented yet. Technical difficulties are related to the lack of normalization and standardization of the methodology, resulting in big variations between the results of different studies (8) and making difficult the translation of EBC analysis to clinical practice.
Sputum is even more complex to analyze by proteomics and, to date, there is no well-characterized MS-based proteomic alterations reported in the sputum of patients with lung cancer.
DATA ANALYSIS
Analysis and interpretation of the data derived from MS-based proteomic technologies represents unique challenges as well. From MALDI MS experiments, Spectra are generated in the mass-to-charge (m/z) 3,000 to 50,000. Internal calibration is performed using internal or external calibrants. The data processing consists of internal calibration, smoothing, baseline correction, normalization to the total ion current, feature selection with a signal-to-noise ratio, and binning of features. This processing results in 100 to 300 m/z peaks per spectrum on average, using conservative parameters. Statistical analyses of these data for biomarkers focus on the selection of MS features and differential expression levels between the study groups and on building class prediction models based on the selected features (68, 69, 100–102). The misclassification rate is typically estimated using the leave-one-out cross-validation.
From tandem MS analysis, raw data is extracted for individual spectra with filters applied to remove obvious background ions and low-quality spectra. These spectra yield a list of peptide sequences and the frequency that each peptide is detected. These sequences are searched against the NCBI protein database to generate candidate proteins from which they may have come. This later approach is less intuitive and is recently being facilitated by modern bioinformatics tools, enabling the analysis of proteomic digestion with different enzymes than trypsin and therefore increasing the likelihood of detecting increasing number of peptides mapping to the same protein; this, in turn, improves the confidence of identification (see Dr. Tabb's website from the Department of Biomedical Informatics of Vanderbilt University Medical Center with expertise in proteomics research: http://fenchurch.mc.vanderbilt.edu/lab/software.php). This list of candidate proteins is filtered in various ways to reduce the likelihood of false matches and the protein and hit count lists from different study groups are compared.
BIOMARKER VALIDATION AND IMPLICATIONS IN THE MANAGEMENT OF LUNG CANCER
The discovery of potential new biomarkers by MS techniques has to be followed by verification and validation strategies. Validation happens at multiple levels, from confirmation of detected changes in protein level by different techniques, to correlation with some biological outcomes of lung cancer such as early detection, chemosensitivity, or survival. Ultimately, population studies are required for the translation of biomarkers into clinical practice. This validation work is made easier with the access to repositories of well-annotated biological specimens. Tissue microarrays (TMAs) (103) allow high-throughput evaluation of candidate biomarkers on PPFE pathologic specimens by IHC, cytogenetic and molecular biology techniques (104). For each assay, one must assess accuracy, reproducibility, and variability within and across laboratories. The Early Detection Research Network (EDRN), an initiative of the National Cancer Institute (NCI), assists with the translation of biomarkers into clinical applications (105). It supports research to identify, develop, and validate biological markers for earlier cancer detection and risk assessment. By integrating basic and clinical science studies with computational, statistical, and epidemiologic approaches, it allows a comprehensive understanding of biomarkers.
By allowing high-throughput, sensitive, and specific analysis of proteins expressed in cells or tissues, proteomic profiling by MS is a valuable technique for the study of cancer proteomics. Unlike the study of a single protein, it enables a systematic overview, leading to a better understanding of the disease. Indeed, lung cancer is a heterogeneous disease at the cellular and molecular level, resulting in the expression of various proteins, so that looking for a combination of protein alterations (or profile) is likely to have greater utility than focusing on a specific tumor marker. This approach may improve the clinical management of lung cancer by identifying with higher sensitivity and specificity protein profiles that could serve as biomarkers to improve risk assessment, early diagnosis, diagnostic classification of lung tumors, and prediction of response to therapy and of survival. These protein profiles could also identify new molecular therapeutic targets. To prove the added value of the discovered biomarkers to the current standard of care, they should be evaluated in a clinical context. Whether they are independent from other clinical factors is a question to be addressed. If that is the case, a clinico-proteomic model incorporating clinical factors and protein biomarkers predictors of disease and/or prognosis can be developed.
LIMITATIONS OF THE MS-BASED APPROACH
Despite great advantages and insights provided to cancer biology, several pre-analytical, analytical, and post-analytical limitations are still associated with MS methodologies. Pre-analytical limitations are related to poor patient selection; nonstandardized sample collection, storage, and processing; and poor instrument calibration, all of which introduced important bias in the analyses. Analytical limitations are also numerous and depend on the methods used, but general considerations can be addressed. The first problem faced is the complex nature and large dynamic range of the proteome. Several depletion and fractionation methods improve but are not sufficient to reach the desired dynamic range. Second, sensitivity is limited to the most abundant proteins. Global MS techniques do not easily detect blood proteins at concentrations lower than 1 μg/ml (106), while known tumor markers in the serum are approximately 1,000 times less concentrated. Third, reproducibility between platforms and institutions is still a problem. Finally, some MS techniques such as MALDI MS applied to fresh tissue or blood samples do not readily allow direct identification of proteins. Some discriminatory peaks were revealed to be blood proteins or their proteolysis products produced ex vivo (after collection) and not related to cancer in vivo (107, 108). Post-analytical limitations are mainly due to bioinformatic/biostatistic artifacts. With a large number of spectra from a small number of samples, there is a risk of overfitting the data. Also, less than half of the proteins of a complex biological sample can be identified by current computational methods. Finally, only mandatory validation strategies including complementary methodologies would decide whether the discovered candidates could be proposed as new biomarkers. These limitations explain why these approaches have not been translated to clinical applications yet. Much effort is required to overcome those challenges and reach clinical utility.
STRATEGIES TO ADDRESS THE COMPLEXITY OF THE PROTEOME
The complexity of the proteome—with more than tens of thousands of different protein species (109), a wide range of concentrations, a large number of peptides generated from each protein, their post-translational modifications (109) and sequence variations among individuals (110)—presents current proteomic technologies with great challenges. To overcome these challenges, different complexity reduction strategies are used. In a first strategy, affinity depletion removes the most abundant serum proteins such as albumin (111–114) and detects low-abundance proteins, which are more informative as tumor-specific biomarkers but unfortunately are obscured by high-abundance proteins. Depletion procedures are limited by absence of standardization and problems of reproducibility. In a second strategy, proteins or peptides are fractionated using physicochemical properties such as size, residue charge, and hydropathy before analysis by MS (113, 115, 116). The fractionation has several limitations as it requires large sample amounts, is more expensive and more time-consuming, and increases the risk of variability within and between samples.
With shotgun proteomics combined with MudPIT (14, 117, 118) applied to the plasma of patients with lung cancer, 120 proteins have been shown to be exclusively expressed in the plasma of patients with lung ADC (119). In the last strategy, specific chemical probes are used to tag and facilitate isolation of a target peptide. After digestion of proteins with trypsin in the shotgun approach, analyses are complicated by the large number of redundant peptides from each protein. By targeting peptides containing unique or rare amino acids or post-translational modifications such as phosphorylation or glycosylation (120), we can reduce the complexity of the biological samples and analyze sub-proteomes. For example, Zhou and colleagues (121) developed a method for the high-throughput analysis of serum glycoproteins using solid-phase extraction of N-linked glycopeptides from glycoproteins (SPEG). Glycoproteins are conjugated to a solid support using hydrazide chemistry, nonglycosylated proteins are removed by trypsin digestion, and N-glycopeptides are specifically released via peptide-N-glycosidase F before finally being identified and quantified by tandem MS (122). Although quite appealing, these sub-proteomic strategies are early in development and require methodologic standardization.
Using a phosphoproteomic approach based on phosphopeptide immunoprecipitation and analysis by LC-MS/MS, Rikova and coworkers (123) characterized tyrosine kinase signaling across 41 NSCLC cell lines and 150 NSCLC tumors. They identified kinases already known as oncogenes (e.g., EGFR, c-MET) as well as kinases never previously implicated in NSCLC (PDGFRα, DDR1). The insights they provided are very important in the actual era of tyrosine kinase–based cancer therapeutics.
Activity-based MS proteomic analysis of lung cancer microenvironment investigates interactions and functions of proteins in normal and diseased organs. For example, the nature and post-translational modifications of proteases, which are involved in many aspects of cancer development, have been assayed using an activity-based proteomic profiling with molecular tags specific for enzymatic activity (124). This approach has been associated with multidimensional proteomic LC-MS to discover the roles of active proteases in tumor cells and their microenvironment (125). Measuring changes in many enzymes' activity with active site-directed probes, investigators identified human carcinoma enzyme activities selectively expressed in culture or xenograft tumors, as well as mouse stromal enzyme activities infiltrating or excluded from xenograft tumors, showing the importance of specific host components for breast cancer development.
Targeted proteomics using multiple reaction monitoring (LC-MRM MS) allows the verification of candidate biomarkers by accurate quantitation of proteins/peptides. In this strategy, which requires a triple quadrupole-class tandem MS (126), there are two stages of mass selection: the first stage (MS1) selects a limited number of precursor ions with pre-specified m/z values that will undergo fragmentation, while the second stage (MS2) gives spectra only for a specific pre-identified fragment ion associated with a given precursor. The association of two mass filters leads to a very specific and sensitive identification. Several “precursor ion/fragment ion” pairs can be specified in a single LC-MS/MS run, allowing parallel quantification of several proteins/peptides. The quantification can be achieved by iTRAQ (isobaric Tags for Relative and Absolute Quantification; covalent linkage to lysines, -NH2 termini) (127) at the MS2 level or by label-free methodology at the MS1 level. C-reactive protein (59), apolipoprotein A-I (128), human growth hormone (129), and prostate-specific antigen (130) have been measured in plasma or serum using the MRM approach.
CONCLUSIONS
Major advances have been brought to the biology of lung cancer by MS-based proteomic analysis. We summarized some of the progress made in the field in the recent years. Although proteomic profiling and high-throughput sensitive MS-based technologies have not been proven of clinical utility yet, they are rapidly developing and promise to advance the areas of early detection of lung cancer, prediction of response to therapy, and prognostic evaluation.
Supported by CA102353, lung SPORE CA90949, the Damon Runyon Cancer Research Foundation (Ci # 19–03 to P.P.M.), and Veterans Affairs Merit Review Grant. S.O. was supported by a grant from the Universite Catholique de Louvain, Belgium. P.C. acknowledges financial support from the NIH/NIGMS (2R01 GM58008–09) and the Department of Defense (W81XWH-05–1-0179).
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet 2000;355:479–485. [DOI] [PubMed] [Google Scholar]
- 3.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA 2004;101:10143–10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schreiber G, McCrory DC. Performance characteristics of different modalities for diagnosis of suspected lung cancer: summary of published evidence. Chest 2003;123:115S–128S. [DOI] [PubMed] [Google Scholar]
- 5.Giaccone G. Clinical impact of novel treatment strategies. Oncogene 2002;21:6970–6981. [DOI] [PubMed] [Google Scholar]
- 6.Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, Cormier Y, Goss G, Inculet R, Vallieres E, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589–2597. [DOI] [PubMed] [Google Scholar]
- 7.Pisters KM, Le Chevalier T. Adjuvant chemotherapy in completely resected non-small-cell lung cancer. J Clin Oncol 2005;23:3270–3278. [DOI] [PubMed] [Google Scholar]
- 8.Chanin TD, Merrick DT, Franklin WA, Hirsch FR. Recent developments in biomarkers for the early detection of lung cancer: perspectives based on publications 2003 to present. Curr Opin Pulm Med 2004;10:242–247. [DOI] [PubMed] [Google Scholar]
- 9.Chen G, Gharib TG, Huang CC, Thomas DG, Shedden KA, Taylor JM, Kardia SL, Misek DE, Giordano TJ, Iannettoni MD, et al. Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin Cancer Res 2002;8:2298–2305. [PubMed] [Google Scholar]
- 10.Beavis RC, Chait BT. Rapid, sensitive analysis of protein mixtures by mass spectrometry. Proc Natl Acad Sci USA 1990;87:6873–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormack AL, Schieltz DM, Goode B, Yang S, Barnes G, Drubin D, Yates JR III. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal Chem 1997;69:767–776. [DOI] [PubMed] [Google Scholar]
- 12.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR III. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 1999;17:676–682. [DOI] [PubMed] [Google Scholar]
- 13.Wolters DA, Washburn MP, Yates JR III. An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem 2001;73:5683–5690. [DOI] [PubMed] [Google Scholar]
- 14.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 2001;19:242–247. [DOI] [PubMed] [Google Scholar]
- 15.Liebler DC. Shotgun mass spec goes independent. Nat Methods 2004;1:16–17. [DOI] [PubMed] [Google Scholar]
- 16.March RE. An introduction to quadrupole ion trap mass spectrometry. J Mass Spectrom 1997;32:351–369. [Google Scholar]
- 17.Schwartz JC, Senko MW, Syka JE. A two-dimensional quadrupole ion trap mass spectrometer. J Am Soc Mass Spectrom 2002;13:659–669. [DOI] [PubMed] [Google Scholar]
- 18.Amster IJ. Fourier transform mass spectrometry. J Mass Spectrom 1996;31:1325–1337. [Google Scholar]
- 19.Cooper HJ, Hakansson K, Marshall AG. The role of electron capture dissociation in biomolecular analysis. Mass Spectrom Rev 2005;24:201–222. [DOI] [PubMed] [Google Scholar]
- 20.Mayya V, Rezaul K, Cong YS, Han D. Systematic comparison of a two-dimensional ion trap and a three-dimensional ion trap mass spectrometer in proteomics. Mol Cell Proteomics 2005;4:214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comisaro MB, Marshall AG. Fourier-transform ion-cyclotron resonance spectroscopy. Chem Phys Lett 1974;25:282–283. [Google Scholar]
- 22.Marshall AG. Milestones in fourier transform ion cyclotron resonance mass spectrometry technique development. Int J Mass Spectrom 2000;200:331–356. [Google Scholar]
- 23.Syka JEP, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, et al. Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J Proteome Res 2004;3:621–626. [DOI] [PubMed] [Google Scholar]
- 24.Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. The orbitrap: a new mass spectrometer. J Mass Spectrom 2005;40:430–443. [DOI] [PubMed] [Google Scholar]
- 25.Macek B, Waanders LF, Olsen JV, Mann M. Top-down protein sequencing and ms3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol Cell Proteomics 2006;5:949–958. [DOI] [PubMed] [Google Scholar]
- 26.Olsen JV, de Godoy LMF, Li GQ, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a c-trap. Mol Cell Proteomics 2005;4:2010–2021. [DOI] [PubMed] [Google Scholar]
- 27.Yates JR, Cociorva D, Liao LJ, Zabrouskov V. Performance of a linear ion trap-orbitrap hybrid for peptide analysis. Anal Chem 2006;78:493–500. [DOI] [PubMed] [Google Scholar]
- 28.Chaurand P, Schwartz SA, Caprioli RM. Profiling and imaging proteins in tissue sections by MS. Anal Chem 2004;76:87A–93A. [PubMed] [Google Scholar]
- 29.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med 2001;7:493–496. [DOI] [PubMed] [Google Scholar]
- 30.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res 2006;5:2889–2900. [DOI] [PubMed] [Google Scholar]
- 31.Caprioli RM. Deciphering protein molecular signatures in cancer tissues to aid in diagnosis, prognosis, and therapy. Cancer Res 2005;65:10642–10645. [DOI] [PubMed] [Google Scholar]
- 32.Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal Chem 2004;76:1145–1155. [DOI] [PubMed] [Google Scholar]
- 33.Chaurand P, Sanders ME, Jensen RA, Caprioli RM. Proteomics in diagnostic pathology: profiling and imaging proteins directly in tissue sections. Am J Pathol 2004;165:1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaurand P, Schwartz SA, Reyzer ML, Caprioli RM. Imaging mass spectrometry: principles and potentials. Toxicol Pathol 2005;33:92–101. [DOI] [PubMed] [Google Scholar]
- 35.Reyzer ML, Hsieh Y, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom 2003;38:1081–1092. [DOI] [PubMed] [Google Scholar]
- 36.Reyzer ML, Caldwell RL, Dugger TC, Forbes JT, Ritter CA, Guix M, Arteaga CL, Caprioli RM. Early changes in protein expression detected by mass spectrometry predict tumor response to molecular therapeutics. Cancer Res 2004;64:9093–9100. [DOI] [PubMed] [Google Scholar]
- 37.Goodlett DR, Keller A, Watts JD, Newitt R, Yi EC, Purvine S, Eng JK, von Haller P, Aebersold R, Kolker E. Differential stable isotope labeling of peptides for quantitation and de novo sequence derivation. Rapid Commun Mass Spectrom 2001;15:1214–1221. [DOI] [PubMed] [Google Scholar]
- 38.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 1999;17:994–999. [DOI] [PubMed] [Google Scholar]
- 39.Ji JY, Chakraborty A, Geng M, Zhang X, Amini A, Bina M, Regnier F. Strategy for qualitative and quantitative analysis in proteomics based on signature peptides. J Chromatogr B Analyt Technol Biomed Life Sci 2000;745:197–210. [DOI] [PubMed] [Google Scholar]
- 40.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 2005;35:265–273. [DOI] [PubMed] [Google Scholar]
- 41.Mason DE, Liebler DC. Quantitative analysis of modified proteins by LC-MS-MS of peptides labeled with phenyl isocyanate. J Proteome Res 2003;2:265–272. [DOI] [PubMed] [Google Scholar]
- 42.Munchbach M, Quadroni M, Miotto G, James P. Quantitation and facilitated de novo sequencing of proteins by isotopic N-terminal labeling of peptides with a fragmentation directing moiety. Anal Chem 2000;72:4047–4057. [DOI] [PubMed] [Google Scholar]
- 43.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods 2003;29:124–130. [DOI] [PubMed] [Google Scholar]
- 44.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 2004;3:1154–1169. [DOI] [PubMed] [Google Scholar]
- 45.Ono M, Shitashige M, Honda K, Isobe T, Kuwabara H, Matsuzuki H, Hirohashi S, Yamada T. Label-free quantitative proteomics using large peptide data sets generated by nano-flow liquid chromatography and mass spectrometry. Mol Cell Proteomics 2006;5:1338–1347. [DOI] [PubMed] [Google Scholar]
- 46.Ru QHC, Zhu LWA, Silberman J, Shriver CD. Label-free semiquantitative peptide feature profiling of human breast cancer and breast disease sera via two-dimensional liquid chromatography-mass spectrometry. Mol Cell Proteomics 2006;5:1095–1104. [DOI] [PubMed] [Google Scholar]
- 47.Silva JC, Gorenstein MV, Li GZ, Vissers JPC, Geromanos SJ. Absolute quantification of proteins by LCMSE - a virtue of parallel MS acquisition. Mol Cell Proteomics 2006;5:144–156. [DOI] [PubMed] [Google Scholar]
- 48.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 2005;4:1487–1502. [DOI] [PubMed] [Google Scholar]
- 49.Gade D, Thiermann J, Markowsky D, Rabus R. Evaluation of two-dimensional difference gel electrophoresis for protein profiling. J Mol Microbiol Biotechnol 2003;5:240–251. [DOI] [PubMed] [Google Scholar]
- 50.Lilley KS, Friedman DB. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev Proteomics 2004;1:401–409. [DOI] [PubMed] [Google Scholar]
- 51.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 2001;1:377–396. [DOI] [PubMed] [Google Scholar]
- 52.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 1997;18:2071–2077. [DOI] [PubMed] [Google Scholar]
- 53.Von Eggeling F, Gawriljuk A, Fiedler W, Ernst G, Claussen U, Klose J, Romer I. Fluorescent dual colour 2D-protein gel electrophoresis for rapid detection of differences in protein pattern with standard image analysis software. Int J Mol Med 2001;8:373–377. [DOI] [PubMed] [Google Scholar]
- 54.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 2003;3:36–44. [DOI] [PubMed] [Google Scholar]
- 55.Friedman DB, Hill S, Keller JW, Merchant NB, Levy SE, Coffey RJ, Caprioli RM. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 2004;4:793–811. [DOI] [PubMed] [Google Scholar]
- 56.Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog 2006;2:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedman DB, Wang SE, Whitwell CW, Caprioli RM, Arteaga CL. Multivariable difference gel electrophoresis and mass spectrometry: a case study on transforming growth factor-beta and erbb2 signaling. Mol Cell Proteomics 2007;6:150–169. [DOI] [PubMed] [Google Scholar]
- 58.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 2006;5:573–588. [DOI] [PubMed] [Google Scholar]
- 59.Kuhn E, Wu J, Karl J, Liao H, Zolg W, Guild B. Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and C-13-labeled peptide standards. Proteomics 2004;4:1175–1186. [DOI] [PubMed] [Google Scholar]
- 60.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA 2003;100:6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh JM, Brichory F, Puravs E, Kuick R, Wood C, Rouillard JM, Tra J, Kardia S, Beer D, Hanash S. A database of protein expression in lung cancer. Proteomics 2001;1:1303–1319. [DOI] [PubMed] [Google Scholar]
- 62.Bergman AC, Benjamin T, Alaiya A, Waltham M, Sakaguchi K, Franzen B, Linder S, Bergman T, Auer G, Appella E, et al. Identification of gel-separated tumor marker proteins by mass spectrometry. Electrophoresis 2000;21:679–686. [DOI] [PubMed] [Google Scholar]
- 63.Gharib TG, Chen G, Wang H, Huang CC, Prescott MS, Shedden K, Misek DE, Thomas DG, Giordano TJ, Taylor JM, et al. Proteomic analysis of cytokeratin isoforms uncovers association with survival in lung adenocarcinoma. Neoplasia 2002;4:440–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen G, Gharib TG, Wang H, Huang CC, Kuick R, Thomas DG, Shedden KA, Misek DE, Taylor JM, Giordano TJ, et al. Protein profiles associated with survival in lung adenocarcinoma. Proc Natl Acad Sci USA 2003;100:13537–13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alfonso P, Catala M, Rico-Morales ML, Durante-Rodriguez G, Moro-Rodriguez E, Fernandez-Garcia H, Escribano JM, Alvarez-Fernandez E, Garcia-Poblete E. Proteomic analysis of lung biopsies: differential protein expression profile between peritumoral and tumoral tissue. Proteomics 2004;4:442–447. [DOI] [PubMed] [Google Scholar]
- 66.Campa MJ, Wang MZ, Howard B, Fitzgerald MC, Patz EF Jr. Protein expression profiling identifies macrophage migration inhibitory factor and cyclophilin a as potential molecular targets in non-small cell lung cancer. Cancer Res 2003;63:1652–1656. [PubMed] [Google Scholar]
- 67.Howard BA, Wang MZ, Campa MJ, Corro C, Fitzgerald MC, Patz EF Jr. Identification and validation of a potential lung cancer serum biomarker detected by matrix-assisted laser desorption/ionization-time of flight spectra analysis. Proteomics 2003;3:1720–1724. [DOI] [PubMed] [Google Scholar]
- 68.Yanagisawa K, Shyr Y, Xu BJ, Massion PP, Larsen PH, White BC, Roberts JR, Edgerton M, Gonzalez A, Nadaf S, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 2003;362:433–439. [DOI] [PubMed] [Google Scholar]
- 69.Shyr Y, Kim K. Weighted flexible compound covariate method for classifying microarray data. In: Berrar D, editor. A practical approach to microarray data analysis. New York: Kluwer Academic; 2003. pp. 186–200.
- 70.Xu BJ, Gonzalez AL, Kikuchi T, Yanagisawa K, Massion PP, Wu H, Mason SE, Olson SJ, Shyr Y, Carbone DP, et al. MALDI-MS derived prognostic protein markers for resected non-small cell lung cancer. Proteomics Clin Appl 2008;2:1508–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rahman SM, Shyr Y, Yildiz PB, Gonzalez AL, Li H, Zhang X, Chaurand P, Yanagisawa K, Slovis BS, Miller RF, et al. Proteomic patterns of preinvasive bronchial lesions. Am J Respir Crit Care Med 2005;172:1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yanagisawa K, Tomida S, Shimada Y, Yatabe Y, Mitsudomi T, Takahashi T. A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J Natl Cancer Inst 2007;99:858–867. [DOI] [PubMed] [Google Scholar]
- 73.Amann JM, Chaurand P, Gonzalez A, Mobley JA, Massion PP, Carbone DP, Caprioli RM. Selective profiling of proteins in lung cancer cells from fine-needle aspirates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Cancer Res 2006;12:5142–5150. [DOI] [PubMed] [Google Scholar]
- 74.Groseclose MR, Massion PP, Chaurand P, Caprioli RM. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using maldi imaging mass spectrometry. Proteomics 2008;8:3715–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seemann MD, Beinert T, Furst H, Fink U. An evaluation of the tumour markers, carcinoembryonic antigen (CEA), cytokeratin marker (CYFRA 21–1) and neuron-specific enolase (NSE) in the differentiation of malignant from benign solitary pulmonary lesions. Lung Cancer 1999;26:149–155. [DOI] [PubMed] [Google Scholar]
- 76.Pastor A, Menendez R, Cremades MJ, Pastor V, Llopis R, Aznar J. Diagnostic value of SCC, CEA and CYFRA 21.1 in lung cancer: a Bayesian analysis. Eur Respir J 1997;10:603–609. [PubMed] [Google Scholar]
- 77.Kulpa J, Wojcik E, Reinfuss M, Kolodziejski L. Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21–1, and neuron-specific enolase in squamous cell lung cancer patients. Clin Chem 2002;48:1931–1937. [PubMed] [Google Scholar]
- 78.Sidransky D, Irizarry R, Califano JA, Li X, Ren H, Benoit N, Mao L. Serum protein maldi profiling to distinguish upper aerodigestive tract cancer patients from control subjects. J Natl Cancer Inst 2003;95:1711–1717. [DOI] [PubMed] [Google Scholar]
- 79.Banez LL, Prasanna P, Sun L, Ali A, Zou Z, Adam BL, McLeod DG, Moul JW, Srivastava S. Diagnostic potential of serum proteomic patterns in prostate cancer. J Urol 2003;170:442–446. [DOI] [PubMed] [Google Scholar]
- 80.Mobley JA, Lam YW, Lau KM, Pais VM, L'Esperance JO, Steadman B, Fuster LM, Blute RD, Taplin ME, Ho SM. Monitoring the serological proteome: the latest modality in prostate cancer detection. J Urol 2004;172:331–337. [DOI] [PubMed] [Google Scholar]
- 81.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002;359:572–577. [DOI] [PubMed] [Google Scholar]
- 82.Petricoin EF III, Ornstein DK, Paweletz CP, Ardekani A, Hackett PS, Hitt BA, Velassco A, Trucco C, Wiegand L, Wood K, et al. Serum proteomic patterns for detection of prostate cancer. J Natl Cancer Inst 2002;94:1576–1578. [DOI] [PubMed] [Google Scholar]
- 83.Winter SF, Sekido Y, Minna JD, McIntire D, Johnson BE, Gazdar AF, Carbone DP. Antibodies against autologous tumor cell proteins in patients with small-cell lung cancer: association with improved survival. J Natl Cancer Inst 1993;85:2012–2018. [DOI] [PubMed] [Google Scholar]
- 84.Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z, et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res 2002;62:3609–3614. [PubMed] [Google Scholar]
- 85.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem 2002;48:1296–1304. [PubMed] [Google Scholar]
- 86.Patz EF Jr, Campa MJ, Gottlin EB, Kusmartseva I, Guan XR, Herndon JE II. Panel of serum biomarkers for the diagnosis of lung cancer. J Clin Oncol 2007;25:5578–5583. [DOI] [PubMed] [Google Scholar]
- 87.Yildiz PB, Shyr Y, Rahman JS, Wardwell NR, Zimmerman LJ, Shakhtour B, Gray WH, Chen S, Li M, Roder H, et al. Diagnostic accuracy of maldi mass spectrometric analysis of unfractionated serum in lung cancer. J Thorac Oncol 2007;2:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cho WC, Yip TT, Yip C, Yip V, Thulasiraman V, Ngan RK, Lau WH, Au JS, Law SC, Cheng WW, et al. Identification of serum amyloid a protein as a potentially useful biomarker to monitor relapse of nasopharyngeal cancer by serum proteomic profiling. Clin Cancer Res 2004;10:43–52. [DOI] [PubMed] [Google Scholar]
- 89.Tolson J, Bogumil R, Brunst E, Beck H, Elsner R, Humeny A, Kratzin H, Deeg M, Kuczyk M, Mueller GA, et al. Serum protein profiling by SELDI mass spectrometry: detection of multiple variants of serum amyloid alpha in renal cancer patients. Lab Invest 2004;84:845–856. [DOI] [PubMed] [Google Scholar]
- 90.Khan N, Cromer CJ, Campa M, Patz EF Jr. Clinical utility of serum amyloid a and macrophage migration inhibitory factor as serum biomarkers for the detection of nonsmall cell lung carcinoma. Cancer 2004;101:379–384. [DOI] [PubMed] [Google Scholar]
- 91.Benson MD, Eyanson S, Fineberg NS. Serum amyloid a in carcinoma of the lung. Cancer 1986;57:1783–1787. [DOI] [PubMed] [Google Scholar]
- 92.Han KQ, Huang G, Gao CF, Wang XL, Ma B, Sun LQ, Wei ZJ. Identification of lung cancer patients by serum protein profiling using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Am J Clin Oncol 2008;31:133–139. [DOI] [PubMed] [Google Scholar]
- 93.Yang SY, Xiao XY, Zhang WG, Zhang LJ, Zhang W, Zhou B, Chen G, He DC. Application of serum seldi proteomic patterns in diagnosis of lung cancer. BMC Cancer 2005;5:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taguchi F, Solomon B, Gregorc V, Roder H, Gray R, Kasahara K, Nishio M, Brahmer J, Spreafico A, Ludovini V, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst 2007;99:838–846. [DOI] [PubMed] [Google Scholar]
- 95.Villanueva J, Philip J, Entenberg D, Chaparro CA, Tanwar MK, Holland EC, Tempst P. Serum peptide profiling by magnetic particle-assisted, automated sample processing and MALDI-TOF mass spectrometry. Anal Chem 2004;76:1560–1570. [DOI] [PubMed] [Google Scholar]
- 96.Kharitonov SA, Barnes PJ. Exhaled markers of pulmonary disease. Am J Respir Crit Care Med 2001;163:1693–1722. [DOI] [PubMed] [Google Scholar]
- 97.Griese M, Noss J, Bredow Cv C. Protein pattern of exhaled breath condensate and saliva. Proteomics 2002;2:690–696. [DOI] [PubMed] [Google Scholar]
- 98.Carpagnano GE, Foschino-Barbaro MP, Resta O, Gramiccioni E, Carpagnano F. Endothelin-1 is increased in the breath condensate of patients with non-small-cell lung cancer. Oncology 2004;66:180–184. [DOI] [PubMed] [Google Scholar]
- 99.Carpagnano GE, Resta O, Foschino-Barbaro MP, Gramiccioni E, Carpagnano F. Interleukin-6 is increased in breath condensate of patients with non-small cell lung cancer. Int J Biol Markers 2002;17:141–145. [DOI] [PubMed] [Google Scholar]
- 100.Yamagata N, Shyr Y, Yanagisawa K, Edgerton M, Dang TP, Gonzalez A, Nadaf S, Larsen P, Roberts JR, Nesbitt JC, et al. A training-testing approach to the molecular classification of resected non-small cell lung cancer. Clin Cancer Res 2003;9:4695–4704. [PubMed] [Google Scholar]
- 101.Tukey JW. Tightening the clinical trial. Control Clin Trials 1993;14:266–285. [DOI] [PubMed] [Google Scholar]
- 102.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med 2001;344:539–548. [DOI] [PubMed] [Google Scholar]
- 103.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–847. (see comments). [DOI] [PubMed] [Google Scholar]
- 104.Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, Westfall MD, Roberts JR, Pietenpol JA, Carbone DP, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res 2003;63:7113–7121. [PubMed] [Google Scholar]
- 105.Sullivan Pepe M, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, Winget M, Yasui Y. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 2001;93:1054–1061. [DOI] [PubMed] [Google Scholar]
- 106.Diamandis EP. Point: Proteomic patterns in biological fluids: do they represent the future of cancer diagnostics? Clin Chem 2003;49:1272–1275. [DOI] [PubMed] [Google Scholar]
- 107.Koomen JM, Li D, Xiao LC, Liu TC, Coombes KR, Abbruzzese J, Kobayashi R. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J Proteome Res 2005;4:972–981. [DOI] [PubMed] [Google Scholar]
- 108.Marshall J, Kupchak P, Zhu W, Yantha J, Vrees T, Furesz S, Jacks K, Smith C, Kireeva I, Zhang R, et al. Processing of serum proteins underlies the mass spectral fingerprinting of myocardial infarction. J Proteome Res 2003;2:361–372. [DOI] [PubMed] [Google Scholar]
- 109.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 2002;1:845–867. [DOI] [PubMed] [Google Scholar]
- 110.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Investigating diversity in human plasma proteins. Proc Natl Acad Sci USA 2005;102:10852–10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, et al. The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics 2003;3:1345–1364. [DOI] [PubMed] [Google Scholar]
- 112.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics 2003;3:422–432. [DOI] [PubMed] [Google Scholar]
- 113.Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics 2002;1:947–955. [DOI] [PubMed] [Google Scholar]
- 114.Liu T, Qian WJ, Chen WN, Jacobs JM, Moore RJ, Anderson DJ, Gritsenko MA, Monroe ME, Thrall BD, Camp DG II, et al. Improved proteome coverage by using high efficiency cysteinyl peptide enrichment: the human mammary epithelial cell proteome. Proteomics 2005;5:1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shen Y, Jacobs JM, Camp DG II, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. Ultra-high-efficiency strong cation exchange LC/RPLC/MS/MS for high dynamic range characterization of the human plasma proteome. Anal Chem 2004;76:1134–1144. [DOI] [PubMed] [Google Scholar]
- 116.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics 2003;2:1096–1103. [DOI] [PubMed] [Google Scholar]
- 117.Chen EI, Hewel J, Felding-Habermann B, Yates JR III. Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MUDPIT). Mol Cell Proteomics 2006;5:53–56. [DOI] [PubMed] [Google Scholar]
- 118.Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR III, Jeffrey SS, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods 2005;2:691–697. [DOI] [PubMed] [Google Scholar]
- 119.Fujii K, Nakano T, Kanazawa M, Akimoto S, Hirano T, Kato H, Nishimura T. Clinical-scale high-throughput human plasma proteome analysis: lung adenocarcinoma. Proteomics 2005;5:1150–1159. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Z, Bast RC Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res 2004;64:5882–5890. [DOI] [PubMed] [Google Scholar]
- 121.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem 2007;79:5826–5837. [DOI] [PubMed] [Google Scholar]
- 122.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol 2003;21:660–666. [DOI] [PubMed] [Google Scholar]
- 123.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190–1203. [DOI] [PubMed] [Google Scholar]
- 124.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci USA 2004;101:10000–10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR III, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci USA 2004;101:13756–13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science 2006;312:212–217. [DOI] [PubMed] [Google Scholar]
- 127.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci USA 2007;104:5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barr JR, Maggio VL, Patterson DG Jr, Cooper GR, Henderson LO, Turner WE, Smith SJ, Hannon WH, Needham LL, Sampson EJ. Isotope dilution–mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem 1996;42:1676–1682. [PubMed] [Google Scholar]
- 129.Wu CC, MacCoss MJ. Shotgun proteomics: tools for the analysis of complex biological systems. Curr Opin Mol Ther 2002;4:242–250. [PubMed] [Google Scholar]
- 130.Barnidge DR, Goodmanson MK, Klee GG, Muddiman DC. Absolute quantification of the model biomarker prostate-specific antigen in serum by LC-MS/MS using protein cleavage and isotope dilution mass spectrometry. J Proteome Res 2004;3:644–652. [DOI] [PubMed] [Google Scholar]
- 131.Chaurand P, Friedman DB, Caprioli RM. Mass spectrometry in cancer biology. In: Mendelsohn J, Howley P, Israel M, Gray J, Thompson C, editors. The molecular basis of cancer, 3rd ed. New York: Elsevier; 2001. pp. 293–308.
- 132.Yildiz PB, Shyr Y, Rahman JS, Wardwell NR, Zimmerman LJ, Shakhtour B, Gray WH, Chen S, Li M, Roder H, et al. Diagnostic accuracy of MALDI mass spectrometric analysis of unfractionated serum in lung cancer. J Thorac Oncol 2007;2:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]