Abstract
Background: A genetic predisposition has been suggested to contribute to the risk for development of rotator cuff disease on the basis of observed family clusters of close relatives. We used a population-based resource combining genealogical data for Utah with clinical diagnosis data from a large Utah hospital to test the hypothesis of excess familial clustering for rotator cuff disease.
Methods: The Utah Population Database contains combined health and genealogical data on over two million Utah residents. Current Procedural Terminology, Fourth Revision, codes (29827, 23412, 23410, and 23420) and International Classification of Diseases, Ninth Revision, codes (726.1, 727.61, and 840.4) entered in patient records were used to identify patients with rotator cuff disease. We tested the hypothesis of excess familial clustering using two well-established methods (the Genealogical Index of Familiality test and the estimation of relative risks in relatives) in the overall study group (3091 patients) and a subgroup of the study group diagnosed before the age of forty years (652 patients).
Results: The Genealogical Index of Familiality test in patients diagnosed before the age of forty years showed significant excess relatedness for individuals with rotator cuff disease in close and distant relationships (as distant as third cousins) (p = 0.001). The relative risk of rotator cuff disease in the relatives of patients diagnosed before the age of forty years was significantly elevated for second degree (relative risk = 3.66, p = 0.0076) and third degree (relative risk = 1.81, p = 0.0479) relatives.
Conclusions: We analyzed a set of patients with diagnosed rotator cuff disease and a known genealogy to describe the familial clustering of affected individuals. The observations of significant excess relatedness of patients and the significantly elevated risks to both close and distant relatives of patients strongly support a heritable predisposition to rotator cuff disease.
Clinical Relevance: A better understanding of the familial risk of rotator cuff disease could lead to the identification of candidate genes predisposing individuals to rotator cuff disease. Gene identification will possibly allow the development of improved treatments, including biologic augmentations of rotator cuff repairs, which may improve tendon healing and repair outcomes.
Level of Evidence: Prognostic Level III. See Instructions to Authors for a complete description of levels of evidence.
The etiology of rotator cuff disease remains unclear. Possible theories regarding the pathogenesis of rotator cuff disease include decreased tendon vascularity, mechanical impingement on the undersurface of the acromion, or intrinsic degeneration1-4. Recently, some authors have suggested a genetic etiology of tendinopathies including rotator cuff disease5,6. Harvie et al. reported an increased risk for rotator cuff tearing in siblings compared with spouses of patients with rotator cuff tears6. To our knowledge, no study to date has evaluated the familial clustering of rotator cuff disease on a population-based, multigenerational level.
The Utah Population Database is a data resource combining genealogical information from more than two million Utah founders and their descendants with disease diagnosis data. This database includes birth, death, and family relationship information, with some family records extending back over ten generations. The University of Utah Health Sciences Center data warehouse, representing patient information from University of Utah Hospital and Clinics since 1994, has been linked to the Utah Population Database genealogical data. This combined database is a unique and invaluable resource for evaluating familial clustering for a variety of disease processes7-9.
The purpose of this study was to analyze the familial relationships among individuals identified in the Utah Population Database with a diagnosis indicating rotator cuff disease (rotator cuff tear or rotator cuff repair). We tested the hypothesis of excess familial clustering or heritable predisposition to rotator cuff disease using two well-established methods: the Genealogical Index of Familiality and the estimation of relative risks in relatives.
Materials and Methods
Data
The Utah Population Database includes genealogical data for over two million Utah founders and their descendants; some family records extend back for ten generations10. The University of Utah Health Sciences Center data warehouse is an electronic database including all patient-related information from inpatient hospital and outpatient clinic visits since 1994. The University of Utah Health Sciences Center database has been linked to the Utah Population Database genealogical data to allow study of the genetic relationships between affected individuals.
Methods for the analysis of the Utah Population Database genealogical and linked hospital data have been well reported and represent examination of a number of different phenotypes7-9. Analysis of the familial clustering includes comparing the average relatedness between all pairs of patients and the expected average relatedness with use of the Genealogical Index of Familiality. In addition, we estimated relative risks in first, second, and third-degree relatives of affected individuals from the Utah Population Database resource.
To identify appropriate controls for the Genealogical Index of Familiality and relative risk tests, we randomly selected matched control individuals who were hospital patients and who had linked genealogical data. All patients at the University of Utah Hospital and Clinics have been linked to the genealogical data in the Utah Population Database; however, only a subset of these records can be accessed at any time. A set of hospital patients representing 20% of all patients in the University of Utah Hospital and Clinics who had genealogical data were randomly selected. Controls for the analyses performed were selected randomly from this larger set of controls. To allow appropriate matching for characteristics that may influence the quality and quantity of genealogical data or record-linking success, we created cohorts for matching controls to patients. All 2.2 million individuals in the Utah Population Database with at least three generations of genealogical data were assigned to one of 264 cohorts. These cohorts are based on sex, birth year, birthplace (Utah or not Utah), and number of ancestors (more than six or not more than six). Sets of matched controls were chosen randomly from this set of all hospital controls for each group of patients.
Genealogical Index of Familiality
We used the Genealogical Index of Familiality statistic to test the hypothesis of no excess relatedness among affected individuals. This statistic was developed for the Utah Population Database11,12. The Genealogical Index of Familiality statistic measures the average relatedness of a set of individuals and compares the measure to the average relatedness expected in the Utah population. The Genealogical Index of Familiality analysis differs from the relative risk in that it includes all genetic relationships between individuals, both close and distant. The Genealogical Index of Familiality relatedness measure implements the Malécot coefficient of kinship, which is defined as the probability that randomly selected homologous genes from two individuals are identical by descent from a common ancestor13. The coefficient is 0.25 (1/22) for a sibling pair, 0.125 (1/23) for a grandparent-grandchild pair, 0.0625 (1/24) for a first cousin pair, and so forth. The contribution to the Genealogical Index of Familiality statistic is smaller for pairs of individuals with a greater genetic distance between them.
The case-Genealogical Index of Familiality is defined as the average of the coefficients of kinship between all possible pairs of affected individuals (×105). A control-Genealogical Index of Familiality can be estimated as the average of the coefficients of kinship between all possible pairs in a set of matched control individuals. The average pairwise relatedness of the patients is compared with the distribution of the average pairwise relatedness statistics estimated for 1000 independent sets of matched controls, providing an empirical test of significance. Controls were randomly selected from the set of hospital controls described previously.
Because both close and distant relationships are observed, the overall Genealogical Index of Familiality statistic shows excess relatedness in the presence of genetic effects or in the presence of familial, but nongenetic, effects (e.g., shared diet), or for a combination of both effects. The Genealogical Index of Familiality statistic can also be compared for patients and matched controls while ignoring all close relationships (relationships with a genetic path length of <3, or first and second-degree relatives). This allows a test of the hypothesis that the excess relatedness observed is significantly elevated for distant relatives, which further supports the existence of a heritable shared predisposition. We refer to this hypothesis test as the distance Genealogical Index of Familiality.
Relative Risks in Relatives
To estimate the relative risk of rotator cuff surgery or a tear among relatives of affected individuals, we compared the number of affected individuals observed among the relatives of the patients to the number of affected individuals expected. We estimated the number of affected individuals expected among the relatives of matched controls by counting affected relatives of five sets of matched hospital controls. We randomly selected five matched hospital controls for each patient, and counted the number of affected relatives of these matched controls (without duplication). We estimated the relative risk among relatives of patients as the classic case-control relative risk. The significance of the test of the null hypothesis relative risk = 1.0 was determined by a Fisher exact test for the 2 × 2 table. Confidence intervals for the relative risk were estimated as given by Agresti14.
No patient identifiers were used in this study; all analysis of genetic relationships between affected individuals was nonidentifiable. This study was approved by both the University of Utah institutional review board as well as the oversight body for the Utah Population Database.
We used the International Classification of Diseases, Ninth Revision (ICD-9) diagnosis codes and the Current Procedural Terminology, Fourth Revision (CPT-4) procedure codes to identify all patients at the University of Utah Hospital and Clinics with a diagnosis of rotator cuff tearing or rotator cuff surgery (Table I). We required that all patients also have at least three generations of genealogical data. The number of patients identified is shown in Table I. These counts include all patients with at least one medical record with each code, so that patients with more than one diagnosis or procedure code may be counted more than once in Table I; duplicate codes for the same individual were only counted once. Patients with a record of rotator cuff surgery also had a diagnosis record for rotator cuff tear. There was a total of 3091 patients with either rotator cuff surgery or tear diagnoses. Early onset of a disease has been determined to have an increased predisposition for a genetic contribution in several diseases including breast cancer and heart disease15-17. The rate of rotator cuff disease increases with age, usually starting around the age of forty18-21. Because age is a risk factor for rotator cuff disease, and early onset disease may indicate a stronger genetic contribution, we also separately analyzed patients who were diagnosed as having rotator cuff disease early (before the age of forty years). A total of 652 patients were less than forty years old with either rotator cuff surgery or tear diagnoses. These 652 patients were from the approximately two million patients in the combined database.
TABLE I.
Number of Individuals by International Classification of Diseases and Current Procedural Terminology Codes for Rotator Cuff Disease
| Code* | Definition | No. of Patients in Entire Cohort | No. of Patients <40 Yr Old |
|---|---|---|---|
| ICD-9 | |||
| 726.1 | Rotator cuff syndrome of shoulder and allied disorders | 1726 | 402 |
| 727.61 | Complete rupture of rotator cuff, nontraumatic | 45 | 7 |
| 840.4 | Rotator cuff traumatic strain | 1076 | 227 |
| CPT-4 | |||
| 29827 | Arthroscopy, shoulder, surgical; with rotator cuff repair | 123 | 7 |
| 23412 | Repair of ruptured musculotendinous cuff open; chronic | 34 | 2 |
| 23410 | Repair of ruptured musculotendinous cuff open; acute | 40 | 3 |
| 23420 | Reconstruction of complete shoulder (rotator) cuff avulsion, chronic (includes acromioplasty) | 47 | 4 |
| All individuals with any of the above codes | 3091 | 652 |
ICD-9 = International Classification of Diseases, Ninth Revision, and CPT-4 = Current Procedural Terminology, Fourth Revision.
Source of Funding
No substantial external funding was required for completion of this study.
Results
Genealogical Index of Familiality
We tested the hypothesis of no significant excess relatedness among affected individuals using the Genealogical Index of Familiality (Table II). Table II shows the number of affected individuals, the average relatedness of those individuals (case overall Genealogical Index of Familiality), the average relatedness from an analysis of 1000 sets of matched hospital controls (mean control Genealogical Index of Familiality), the empirical p value for the comparison for all patients and controls, and the empirical p value for the distance Genealogical Index of Familiality. Although the overall Genealogical Index of Familiality shows a significant excess relatedness for individuals with rotator cuff disease (p < 0.001), the distance Genealogical Index of Familiality test shows that the excess relatedness observed is not significant when close relationships are ignored (p = 0.848), suggesting common familial effects that might be genetic, but might be environmental.
TABLE II.
Results of Genealogical Index of Familiality Test of Excess Relatedness
| No. of Patients | Case Overall GIF* | Mean Control GIF* | Empirical P Values*
|
||
|---|---|---|---|---|---|
| Phenotype | Overall GIF | Distance GIF | |||
| Rotator cuff disease† | 3091 | 2.86 | 2.52 | <0.001 | 0.848 |
| Early onset of rotator cuff disease‡ | 652 | 2.53 | 1.84 | 0.001 | 0.004 |
GIF = Genealogical Index of Familiality.
All individuals who had been diagnosed with a rotator cuff tear or treated with rotator cuff surgery, including the early onset subset.
Only individuals who had early onset, which is defined as an age of less than forty years at the time of diagnosis.
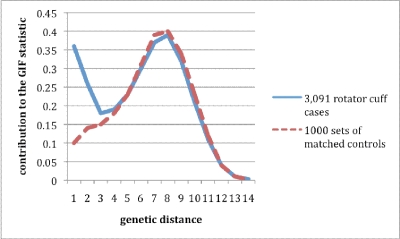
Figure 1 shows the distribution of the Genealogical Index of Familiality statistic for the different pairs of genetic relationships observed for affected individuals compared with controls. The X axis represents the genetic distance between the pairs of individuals (1 = parent-offspring pairs, 2 = sibling or grandparent-grandchild pairs, 3 = avuncular pairs, and so forth). The Y axis shows the value of the Genealogical Index of Familiality statistic that was contributed for the different genetic distances. Figure 1 shows that most of the excess relatedness observed in affected individuals compared with controls was observed in parent-offspring pairs (genetic distance = 1) and sibling pairs (genetic distance = 2). The relatedness observed for more distant relationships was similar for affected individuals and controls. This evidence for significant excess relatedness suggests familial effects that might be either environmental or genetic, but we cannot determine which.
Fig. 1.
Comparison of the distributions of relatedness measure from pairwise relationships for 3091 patients with rotator cuff disease and the average distribution of relatedness measure from pairwise relationships for 1000 sets of matched controls. The lines plotted represent the contribution to the overall relatedness measure of the Genealogical Index of Familiality (GIF) statistic (shown on the Y axis) for each pairwise genetic distance (X axis) for pairs of patients and for pairs of controls. Genetic distance (X axis) refers to different types of pairwise relationships, increasing from close relationships (1 = parent-offspring and 2 = siblings or grandparent-grandchild) to more distant relationships (6 = second cousins and 8 = third cousins).
Because age is a risk factor for rotator cuff disease, and early onset disease may indicate a stronger genetic contribution, we also separately analyzed patients who were diagnosed with rotator cuff disease early (before the age of forty years) (Table II). When only those 652 individuals whose rotator cuff disease exhibited itself before the age of forty years are considered, both the overall Genealogical Index of Familiality (p = 0.001) and the distance Genealogical Index of Familiality (p = 0.004) tests show that significant excess relatedness is observed. These results strongly suggest a heritable contribution to predisposition to early rotator cuff disease.
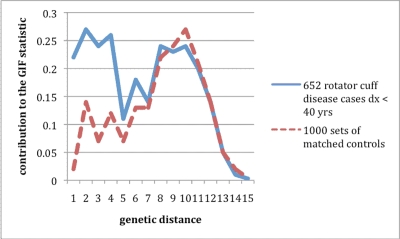
Figure 2 shows the contribution of the Genealogical Index of Familiality statistic by genetic relationship for the 652 individuals diagnosed with rotator cuff disease before the age of forty years. In contrast to Figure 1, the excess relatedness observed for patients compared with controls includes relationships out to a genetic distance of 8 (third cousins). This figure shows excess relatedness for both close and distant relationships and strongly supports a genetic predisposition to rotator cuff disease.
Fig. 2.
Comparison of distributions of pairwise relationships for 652 patients with rotator cuff disease diagnosed before the age of forty years. The lines plotted represent the contribution to the overall relatedness measure of the Genealogical Index of Familiality (GIF) statistic (shown on the Y axis) for each pairwise genetic distance (X axis) for pairs of patients and for pairs of controls.
Relative Risks
Table III shows the estimated relative risk for rotator cuff disease in first, second, and third-degree relatives of individuals with rotator cuff disease. Significantly elevated relative risks were observed in first and second-degree relatives. Although the estimated relative risk for third-degree relatives was elevated, it was not significantly greater than 1.0. This corresponds with the Genealogical Index of Familiality results in Figure 1, in which we observed an excess for first degree (genetic distance 1 and 2) and second-degree (genetic distance 3) relationships, but not for third-degree relationships (genetic distance 4).
TABLE III.
Relative Risk in Relatives of Individuals with Rotator Cuff Disease
| Relationship | No. of Relatives (Patients/Controls) | No. of Affected (Patients/Controls) | P Value | Relative Risk | 95% Confidence Interval |
|---|---|---|---|---|---|
| First degree | 23,700/117,063 | 202/409 | <0.0001 | 2.44 | (2.06, 2.89) |
| Second degree | 68,600/317,179 | 155/577 | 0.0177 | 1.24 | (1.04, 1.48) |
| Third degree | 157,292/631,409 | 246/915 | 0.2866 | 1.08 | (0.94, 1.24) |
Table IV shows the estimated relative risk for early onset rotator cuff disease in the relatives of the 652 individuals diagnosed with rotator cuff disease before the age of forty years. There is a substantial decrease in the sample sizes when only early onset disease is considered. First, second, and third-degree relative risks were all elevated, but only the relative risk in second and third-degree relatives was significantly elevated. These results correspond to the Genealogical Index of Familiality results in Figure 2, in which an excess of second and third-degree relationships (and more distant relationships) was observed, and they strongly support a heritable predisposition for early onset rotator cuff disease.
TABLE IV.
Relative Risk for Early Rotator Cuff Disease in Relatives of 652 Individuals Diagnosed with Rotator Cuff Disease Before the Age of Forty Years
| Relationship | No. of Relatives (Patients/Controls) | No. of Affected (Patients/Controls) | P Value | Relative Risk | 95% Confidence Interval |
|---|---|---|---|---|---|
| First degree | 3714/19,299 | 6/18 | 0.2614 | 1.73 | (0.69, 4.37) |
| Second degree | 8268/41,624 | 8/11 | 0.0076 | 3.66 | (1.47, 9.11) |
| Third degree | 21,642/104,306 | 18/48 | 0.0479 | 1.81 | (1.05, 3.11) |
Discussion
The preliminary natural history data on rotator cuff disease support a familial component5,6,22. Our data support previous suggestions for a hereditary pattern of rotator cuff disease. When all patients are considered, significantly elevated risks were observed for first and second-degree relatives. Because age is a recognized risk factor for rotator cuff disease, we also analyzed only those patients in whom the disease was exhibited at an early age. When only those individuals with a diagnosis before the age of forty years were considered, we observed significantly elevated risks. The analysis of relatedness in the younger cohort confirmed these findings, with excess relatedness observed for patients over matched controls out to a genetic distance of third cousins. Both analyses are strongly suggestive of a genetic predisposition to rotator cuff disease.
Rotator cuff tearing usually presents in the fifth and sixth decades. The natural history of rotator cuff disease and shoulder magnetic resonance imaging, ultrasound, and arthrography data from asymptomatic individuals suggest that full-thickness rotator cuff tears are extremely uncommon in patients under the age of forty years18-21. Because tearing is so uncommon in this young age group, a genetic predisposition might be more likely to be exhibited in this group, as we observed.
Overall, there is only very preliminary published evidence suggesting a genetic etiology of tendinopathies. The largest body of data evaluating the genetic link between tendinopathies, as far as we know, has been reported for patients with Achilles tendinopathy or tearing. Several studies have found a correlation between blood group O or the A/O ratio and Achilles tendon ruptures23-25. The ABO gene is located on chromosome 9 and encodes for enzymes that produce the major antigens in the ABO blood group system. Two genes have been localized to the same region on chromosome 9 as the ABO gene, tenascin C (TNC) and collagen type-V α 1 (COL5A1), and they have been shown to have polymorphisms associated with the presence of Achilles tendon injuries26,27. Variants in either of these genes are ideal candidate genes for Achilles tendon injuries and potentially other tendinopathies including rotator cuff disease. No study has evaluated potential candidate genes for rotator cuff disease. The results of the current study define a potential population of patients, those with tears before the age of forty years, in which to study candidate genes for rotator cuff disease.
Very little research has been performed regarding the influence of genetics on the development of rotator cuff tendon injuries. Yamaguchi et al. used a questionnaire to evaluate 586 patients who had a diagnostic shoulder ultrasound for shoulder pain and found a strong trend between a reported family history of rotator cuff tear (10.9% compared with 6.9%; p = 0.06) and the risk of rotator cuff tear22. Harvie et al. retrospectively evaluated the 129 siblings of a cohort of 205 patients diagnosed as having full-thickness rotator cuff tears by ultrasound to identify the prevalence of rotator cuff tearing in this sibling population6. With use of the 150 spouses of the patients as a control group, the relative risk of full-thickness tears in siblings compared with controls was 2.42 (95% confidence interval, 1.77 to 3.31), while the relative risk of symptomatic full-thickness tears in siblings compared with controls was 4.65 (95% confidence interval, 2.42 to 8.63). This increased risk for tears in siblings implies that genetic factors play a major role in the development of full-thickness rotator cuff tears. Drawbacks of the study by Harvie et al. included limiting the review to close relatives only (first-degree relatives), bias associated with a very high percentage of patients who were lost to follow-up, and a relatively small sample size for a familial analysis. Nevertheless, both of these studies set the stage for further investigations regarding familial or genetic influences on the development of rotator cuff tears, including our population-based analysis.
In an attempt to improve on the limitations recognized in previous studies, we analyzed the potential for a genetic influence on rotator cuff disease in a similar fashion as has been previously performed to identify a genetic predisposition for other chronic diseases and cancers. The Utah Population Database is a combination of a computerized genealogy of the Utah pioneers and their descendants with various data including the Utah Cancer Registry, Utah death certificates, and the University of Utah Hospital and Clinics10. This resource has been used to define the genetic contribution to many other phenotypes and was the basis of the identification in Utah pedigrees of BRCA1, BRCA2, and p1628-30. Using this resource and similar methodology, we explored the hypothesis of a genetic contribution to rotator cuff disease and have shown a strong genetic predisposition for disease development.
A limitation of this study is the use of ICD-9 code information. Because diagnosis codes are dependent on the clinician, a diagnosis of rotator cuff tearing may be less accurate from a nonorthopaedist compared with that from a shoulder specialist. We attempted to limit this bias by limiting identification of patients to diagnoses of a rotator cuff tear, excluding all other codes associated with shoulder pain or dysfunction. The use of CPT-4 codes for rotator cuff surgery is very accurate since all of these patients presumably had a tear that was repaired and therefore improves the accuracy of the analysis. We included both the rotator cuff tear ICD-9 and CPT-4 codes to increase the overall number of patients analyzed, improving the ability to perform a population-based analysis. In this analysis, we were able to identify only the patients at the University of Utah Hospital and Clinics who were coded with a diagnosis or procedure code indicating the presence of rotator cuff disease and who also had at least three generations of genealogical data. This censoring of diagnosis occurs uniformly across the data resource, to relatives of both patients and controls, and has been shown to have no effect on the overall results. Finally, there is a very high concentration of patients of the Mormon faith in the region of the country from which the study population is derived. Consequently, there may be cultural or behavioral factors that may influence the prevalence of rotator cuff disease in these patients. Despite these potential differences, our results still suggest that there is a genetic component to the disease, taking into consideration these potential environmental effects.
Rotator cuff disease likely has a multifactorial etiology, which includes mechanical and environmental influences. We have additionally shown a strong genetic predisposition for the disease. Future applications of these data include high-risk pedigree studies to identify the predisposition gene(s) responsible for these observations, and the analysis of potential candidate genes associated with rotator cuff disease. A better understanding of abnormal genetics for rotator cuff disease may allow for early identification of at-risk individuals as well as the development of potential biologic augmentations of rotator cuff repairs, which may have implications on prevention, repair healing, and outcomes. 
Disclosure: In support of their research for or preparation of this work, one or more of the authors received, in any one year, outside funding or grants in excess of $10,000 from the National Institutes of Health-National Library of Medicine (NLM R01 LM009331). Partial support (less than $10,000) for all datasets within the Utah Population Database was provided by the University of Utah Huntsman Cancer Institute. Neither they nor a member of their immediate families received payments or other benefits or a commitment or agreement to provide such benefits from a commercial entity. No commercial entity paid or directed, or agreed to pay or direct, any benefits to any research fund, foundation, division, center, clinical practice, or other charitable or nonprofit organization with which the authors, or a member of their immediate families, are affiliated or associated.
Investigation performed at the University of Utah School of Medicine, Salt Lake City, Utah
References
- 1.Hashimoto T, Nobuhara K, Hamada T. Pathologic evidence of degeneration as a primary cause of rotator cuff tear. Clin Orthop Relat Res. 2003;415:111-20. [DOI] [PubMed] [Google Scholar]
- 2.Neer CS 2nd. Anterior acromioplasty for the chronic impingement syndrome in the shoulder: a preliminary report. J Bone Joint Surg Am. 1972;54:41-50. [PubMed] [Google Scholar]
- 3.Rathbun JB, Macnab I. The microvascular pattern of the rotator cuff. J Bone Joint Surg Br. 1970;52:540-53. [PubMed] [Google Scholar]
- 4.Rudzki JR, Adler RS, Warren RF, Kadrmas WR, Verma N, Pearle AD, Lyman S, Fealy S. Contrast-enhanced ultrasound characterization of the vascularity of the rotator cuff tendon: age- and activity-related changes in the intact asymptomatic rotator cuff. J Shoulder Elbow Surg. 2008;17(1 Suppl):96S-100S. [DOI] [PubMed] [Google Scholar]
- 5.September AV, Schwellnus MP, Collins M. Tendon and ligament injuries: the genetic component. Br J Sports Med. 2007;41:241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvie P, Ostlere SJ, Teh J, McNally EG, Clipsham K, Burston BJ, Pollard TC, Carr AJ. Genetic influences in the aetiology of tears of the rotator cuff. Sibling risk of a full-thickness tear. J Bone Joint Surg Br. 2004;86:696-700. [DOI] [PubMed] [Google Scholar]
- 7.Albright FS, Orlando P, Pavia AT, Jackson GG, Cannon Albright LA. Evidence for a heritable predisposition to death due to influenza. J Infect Dis. 2008;197:18-24. [DOI] [PubMed] [Google Scholar]
- 8.Teerlink CC, Hegewald MJ, Cannon-Albright LA. A genealogical assessment of heritable predisposition to asthma mortality. Am J Respir Crit Care Med. 2007;176:865-70. [DOI] [PubMed] [Google Scholar]
- 9.Weires MB, Tausch B, Haug PJ, Edwards CQ, Wetter T, Cannon-Albright LA. Familiality of diabetes mellitus. Exp Clin Endocrinol Diabetes. 2007;115:634-40. [DOI] [PubMed] [Google Scholar]
- 10.Skolnick M. The Utah genealogical database: a resource for genetic epidemiology. In: Cairns J, Lyon JL, Skolnick M, editors. Cancer incidence in defined populations. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories; 1980. p 285-97. Banbury Report 4.
- 11.Hill JR. A kinship survey of cancer in the Utah Mormon population. [PhD thesis.] Salt Lake City, UT: University of Utah; 1980a.
- 12.Hill JR. A survey of cancer sites by kinship in the Utah Mormon population. In: Cairns J, Lyon JL, Skolnick M, editors. Cancer incidence in defined populations. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories; 1980. p 299-318. Banbury Report 4.
- 13.Malécot G. Les Mathematiques de L'Heredite. Paris: Masson; 1948.
- 14.Agresti A. Categorical data analysis. New York: Wiley; 1990.
- 15.Rissanen AM. Familial occurrence of coronary heart disease: effect of age at diagnosis. Am J Cardiol. 1979;44:60-6. [DOI] [PubMed] [Google Scholar]
- 16.Meiner V, Friedlander Y, Milo H, Sharon N, Bel-Avi L, Shpitzen S, Leitersdorf E, Siscovick DS, Schwartz SM. Cholesteryl ester transfer protein (CETP) variation and early onset of non-fatal myocardial infarction. Ann Hum Genet. 2008;72(Pt 6):732-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King ML. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684-9. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi K, Ditsios K, Middleton WD, Hildebolt CF, Galatz LM, Teefey SA. The demographic and morphological features of rotator cuff disease. A comparison of asymptomatic and symptomatic shoulders. J Bone Joint Surg Am. 2006;88:1699-704. [DOI] [PubMed] [Google Scholar]
- 19.Milgrom C, Schaffler M, Gilbert S, van Holsbeeck M. Rotator-cuff changes in asymptomatic adults. The effect of age, hand dominance and gender. J Bone Joint Surg Br. 1995;77:296-8. [PubMed] [Google Scholar]
- 20.Sher JS, Uribe JW, Posada A, Murphy BJ, Zlatkin MB. Abnormal findings on magnetic resonance images of asymptomatic shoulders. J Bone Joint Surg Am. 1995;77:10-5. [DOI] [PubMed] [Google Scholar]
- 21.Tempelhof S, Rupp S, Seil R. Age-related prevalence of rotator cuff tears in asymptomatic shoulders. J Shoulder Elbow Surg. 1999;8:296-9. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi K, Baumgarten K, Gerlach DJ, Ditsios K, Teefey SA, Middleton WD. The demographics and morphology of rotator cuff disease: a comparison of asymptomatic and symptomatic shoulders. Presented at the Ninth International Congress on Surgery of the Shoulder meeting; 2004 May 2-5; Washington, DC.
- 23.Kannus P, Natri A. Etiology and pathophysiology of tendon ruptures in sports. Scand J Med Sci Sports. 1997;7:107-12. [DOI] [PubMed] [Google Scholar]
- 24.Jozsa L, Balint JB, Kannus P, Reffy A, Barzo M. Distribution of blood groups in patients with tendon rupture. An analysis of 832 cases. J Bone Joint Surg Br. 1989;71:272-4. [DOI] [PubMed] [Google Scholar]
- 25.Kujala UM, Järvinen M, Natri A, Lehto M, Nelimarkka O, Hurme M, Virta L, Finne J. ABO blood groups and musculoskeletal injuries. Injury. 1992;23:131-3. [DOI] [PubMed] [Google Scholar]
- 26.Mokone GG, Schwellnus MP, Noakes TD, Collins M. The COL5A1 gene and Achilles tendon pathology. Scand J Med Sci Sports. 2006;16:19-26. [DOI] [PubMed] [Google Scholar]
- 27.Mokone GG, Gajjar M, September AV, Schwellnus MP, Greenberg J, Noakes TD, Collins M. The guanine-thymine dinucleotide repeat polymorphism within the tenascin-C gene is associated with Achilles tendon injuries. Am J Sports Med. 2005;33:1016-21. [DOI] [PubMed] [Google Scholar]
- 28.Cannon-Albright LA, Goldgar DE, Meyer LJ, Lewis CM, Anderson DE, Fountain JW, Hegi ME, Wiseman RW, Petty EM, Bale AE, et al. Assignment of a locus for familial melanoma, MLM, to chromosome 9p13-p22. Science. 1992;258:1148-52. [DOI] [PubMed] [Google Scholar]
- 29.Kamb A, Shattuck-Eidens D, Eeles R, Liu Q, Gruis NA, Ding W, Hussey C, Tran T, Miki Y, Weaver-Feldhaus J, McClure M, Aitken JF, Anderson DE, Bergman W, Frants R, Goldgar DE, Green A, MacLennan R, Martin NG, Meyer LJ, Youl P, Zone JJ, Skolnick MH, Cannon-Albright LA. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:23-6. [DOI] [PubMed] [Google Scholar]
- 30.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugen-Strano A, Katchre H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow P, Norris F, Helvering L, Morrison P, Rosteck P, Lai M, Barrett J, Lewis C, Neuhasen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick M. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66-71. [DOI] [PubMed] [Google Scholar]