Summary
It is important to understand the regulation of stem cell division because defects in this process can cause altered tissue homeostasis or cancer. The cyclin-dependent kinase inhibitor Dacapo (Dap), a p21/p27 homolog, acts downstream of the microRNA (miRNA) pathway to regulate the cell cycle in Drosophila melanogaster germline stem cells (GSCs). Tissue-extrinsic signals, including insulin, also regulate cell division of GSCs. We report that intrinsic and extrinsic regulators intersect in GSC division control; the Insulin receptor (InR) pathway regulates Dap levels through miRNAs, thereby controlling GSC division. Using GFP-dap 3′UTR sensors in vivo, we show that in GSCs the dap 3′UTR is responsive to Dicer-1, an RNA endonuclease III required for miRNA processing. Furthermore, the dap 3′UTR can be directly targeted by miR-7, miR-278 and miR-309 in luciferase assays. Consistent with this, miR-278 and miR-7 mutant GSCs are partially defective in GSC division and show abnormal cell cycle marker expression, respectively. These data suggest that the GSC cell cycle is regulated via the dap 3′UTR by multiple miRNAs. Furthermore, the GFP-dap 3′UTR sensors respond to InR but not to TGF-β signaling, suggesting that InR signaling utilizes Dap for GSC cell cycle regulation. We further demonstrate that the miRNA-based Dap regulation may act downstream of InR signaling; Dcr-1 and Dap are required for nutrition-dependent cell cycle regulation in GSCs and reduction of dap partially rescues the cell cycle defect of InR-deficient GSCs. These data suggest that miRNA- and Dap-based cell cycle regulation in GSCs can be controlled by InR signaling.
Keywords: Cell cycle, Dacapo, Stem cells, Drosophila
INTRODUCTION
Stem cells are a class of specialized cells that can divide for long periods of time to produce both new stem cells and daughter cells with the potential to differentiate into a specific cell lineage. They play important roles in maintaining homeostasis of tissues by providing new cells and replacing lost cells. The cell cycle is carefully controlled so that stem cells may respond properly to different physiological conditions including tissue injury, nutrition status and aging. Slow division or premature differentiation of stem cells may deplete the stem cell pool, whereas excessive proliferation may contribute to tumorigenesis. Therefore, it is important to understand cell cycle regulation in stem cells. Many adult stem cells reside in a niche, a restricted microenvironment with supporting cells (Fuller and Spradling, 2007; Morrison and Spradling, 2008; Yamashita et al., 2005). Extrinsic signaling between stem cells and niche cells is thought to play important roles in controlling intrinsic factors in stem cells, thereby affecting self-renewal, cell division and differentiation.
MicroRNAs (miRNAs) are endogenous, short (∼21 nucleotide) non-coding RNAs that regulate gene expression through incomplete sequence complementarity with miRNA response elements (MREs) in the 3′UTR of target mRNAs. Recent studies have shown that miRNAs serve important regulatory roles in a variety of tissues, including stem cells (Ambros, 2004; Bartel, 2004; Carrington and Ambros, 2003; Du and Zamore, 2005; Stadler and Ruohola-Baker, 2008; Yi et al., 2008). Mutations in, or misexpression of, miRNAs are found in several human cancers, indicating that miRNAs may also function as oncogenes or tumor suppressors (Croce and Calin, 2005; Esquela-Kerscher and Slack, 2006).
Analysis of essential miRNA biogenic factors [Dicer, Dgcr8 (Pasha - FlyBase)] has guided our understanding of the functions of miRNAs in vivo. Dicer1 knockout mice die as embryos owing to depletion of pluripotent stem cells (Bernstein et al., 2003) and the proliferation of mouse embryonic stem (ES) cells that are deficient for Dicer1 or Dgcr8 is reduced (Murchison et al., 2005; Wang et al., 2007). In Drosophila, disruption of Dcr-1 in germline stem cells (GSCs) also leads to a significant reduction of cell division and to the disruption of GSC maintenance (Hatfield et al., 2005; Jin and Xie, 2007; Park et al., 2007; Shcherbata et al., 2007). Expression of the Drosophila cyclin-dependent kinase inhibitor (CKI) Dacapo (Dap), a p21/p27 (Cdkn1a/Cdkn1b) homolog, is increased in Dcr-1 mutant GSCs, suggesting that miRNAs might regulate the cell cycle of GSCs by repressing Dap (Hatfield et al., 2005). Other components involved in miRNA biogenesis and function, such as loquacious (loqs) or Argonaute-1 (Ago1), are also required for GSC maintenance (Forstemann et al., 2005; Park et al., 2007; Yang et al., 2007). These results suggest that miRNAs play crucial roles in stem cell maintenance and division. Furthermore, conservation between miRNA function in stem cells is highlighted by miRNAs regulating p21cip1 in mouse ES cells and Dap in Drosophila GSCs (Sinkkonen et al., 2008; Wang et al., 2008; Hatfield et al., 2005).
Extrinsic signaling is crucial in regulating stem cells. For example, neural stem cell maintenance in mammals requires hedgehog signaling (Ahn and Joyner, 2005; Balordi and Fishell, 2007). Similarly, the chemokine Cxcl12 (Sdf1) is required to maintain bone marrow hematopoietic stem cells (Kollet et al., 2006; Sacchetti et al., 2007; Sugiyama et al., 2006). In Drosophila, TGF-β and insulin signaling regulate cell division of GSCs (Hsu et al., 2008; LaFever and Drummond-Barbosa, 2005; Xie and Spradling, 1998). Activation of the Insulin receptor (InR; Insulin-like receptor - FlyBase) in GSCs by Drosophila Insulin-like peptide (Dilp; Ilp1 - FlyBase) is required for the nutrient-dependent regulation of cell division (LaFever and Drummond-Barbosa, 2005). However, the molecular mechanisms downstream of InR signaling that directly regulate the GSC cell cycle remain unknown. Evidence from mammalian cells suggests that the CKIs p21cip1 and p27kip1 might regulate the cell cycle downstream of the InR signaling cascade through Foxo, a member of the forkhead transcription family (Medema et al., 2000; Nakae et al., 2003; Seoane et al., 2004; Alvarez et al., 2001; Burgering and Kops, 2002; Kops et al., 1999). Since InR signaling also regulates the cell cycle in Drosophila GSCs and miRNAs affect cell division by negatively regulating Dap levels in these cells, we explored the possibility that these pathways might interact to control the GSC cell cycle.
The Drosophila CKI Dap can inhibit the Cyclin E-Cdk2 (Cdc2c) complex that is required for the G1-S phase transition (de Nooij et al., 2000; Lane et al., 1996). Previous studies have shown increased Dap expression in Dcr-1 mutant GSCs. Furthermore, reduction of dap partially rescues the cell cycle defects in Dcr-1 mutant GSCs. This suggests that Dap acts downstream of miRNAs to regulate the cell cycle (Hatfield et al., 2005). We now show that the dap 3′UTR responds to miRNA activities in GSCs using heterologous reporters (sensors) consisting of a tubulin promoter driving the green fluorescent protein (GFP) gene fused to the dap 3′UTR. Using luciferase assays, we identified miRNAs that can directly target the dap 3′UTR, including miR-7, miR-278 and miR-309. GSCs deficient for miR-278 or miR-7 show mild division defects or abnormal expression of cell cycle markers, respectively. It is therefore possible that the control of cell division through Dap in GSCs requires the simultaneous function of multiple miRNAs. We further show that GFP-dap 3′UTR sensors respond to InR but not to TGF-β activity. Consistent with these findings, GSCs deficient for InR show a Dcr-1-like cell division defect: slow kinetics, increased frequency of staining for Cyclin E and Dap and decreased frequency of staining for Cyclin B. The genetic evidence places the miRNAs and Dap downstream of InR signaling in regulating cell division: cell division of Dcr-1 or dap mutant GSCs does not respond to nutrition, and reduction of dap partially rescues the cell cycle defects of InR mutant GSCs. Thus, our results suggest that InR can regulate the Drosophila GSC cell cycle through miRNAs and Dap.
MATERIALS AND METHODS
Plasmids and constructs
The following primers (5′ to 3′; sequence according to NM_057600) were used for cloning the dap 3′UTR:
dapL forward, GCTCTAGATTCGCTGGCCAACCC and reverse, GCTCTAGAATAGGCTCTGCCTATGT;
dapS forward, GCTCTAGACTCGTAACCAGTAATTAG and reverse, GCTCTAGAGCCCAGAGATCATAGCAA;
dapF forward, GCCTCTAGAACAACTAATGCTCCAGA and reverse, GCGACTAGTATTTATGTACTACCAAC.
Fragments of dap 3′UTR were amplified by PCR from Drosophila genomic DNA. Fragments of dapF (1185 bp), dapL (866 bp) and dapS (630 bp) were ligated into the tub-GFP plasmid. tub-firefly-luciferase (tub-fLuc) and tub-renilla-luciferase (tub-rLuc) plasmids were provided by the Cohen laboratory (Stark et al., 2003). dapF was ligated into the tub-fLuc (tub-fLuc-dapF) vector.
tub-miR-1, tub-miR-7 and tub-miR-309 cluster expression vectors were obtained from the Cohen laboratory (Stark et al., 2003); all other miRNA expression vectors were constructed from amplicons of 400-1000 bp containing pre-miRNA sequence and inserted into a tubulin-driven expression vector. The primers for primary miRNAs were:
miR-289 forward, CACGAAGGATCCAGTCCTGTGCCAG and reverse, CAGCAATCTAGAACCACTTCCAGCAC;
bantam forward, ATAGCGGCCGCGTTAACTGGCAGCATATAATTTC and reverse, ATTCTAGATTATAGGCAGATTTAACATGTGG;
miR-8 forward, ATAGCGGCCGCCGCGGTCACACG CACATTTCAATA and reverse, ATTCTAGAAATGGGAATTGGGAACGATCTCGC;
miR-303 forward, ATAGCGGCCGCTGCATTCGAAAGG CCAGGTGAA and reverse, ATTCTAGATTGTCCAGGATCTAACATGATTTCGT; let-7 (including pre-miR-125) forward, ATAGCGGCCGCGAAGATCAACAGCGATCCATTAAACA and reverse, ATTCTAGATTGCCGATACTTGTGCCTTGA;
miR-34 forward, ATAGCGGCCGCATTTGGCTTGCGCACACACT and reverse, ATTCTAGATTCGTTGTTCAGGCGTCTGGTT;
miR-278 forward, ATAGCGGCCGCTTGGCGCATTAACCGACGCTTT and reverse, ATTCTAGATCCTTGTGCACTCCCAGAAA.
Mutagenesis of the miRNA MREs
dapF was cut from tub-fLuc-dapF and inserted into the CS2p plasmid (Turner and Weintraub, 1994). Mutagenesis was by PCR using the following primers (introduced restriction sites in parentheses; mutated sequences underlined):
miR-7 mutation (NcoI) forward, GAATATTAATCGTTCCATGGCAACTACTCGTAACCAGTA and reverse, TTACGAGTAGTTGCCATGGAACGATTAATATTCGCAACT;
miR-8 mutation (NcoI) forward, CTGCGATTGTGTCCATGGTCCTAATTTTTTATTACGAACC and reverse, GTAATAAAAAATTAG GACCATGGACACAATCGCAGTGGCTT;
miR-309 mutation (BglII) forward, CTCATTTCTTAAAGATCTCTAAA AATGTCTTTTATGATTTG and reverse, CATAAAAGACATTTTTAGAGATCTTTAAGAAATGAGAGCG;
miR-278 mutation (BglII) forward, CGCTGGCCAAAGATCTGAATTGCAATTTGTAATTTTATTTTTTAC and reverse, TACAAATTGCAATTCAGATCTTTGGCCAGCGAATCTGGAGC.
Mutated dapF fragments were then cut from the CS2p plasmids and inserted into the tub-fLuc plasmid.
The bantam mutation was introduced by PCR using 5′-TAAAGATCTCTAAAAATGTCTTTTATGATTTGCTATCCATGGTGGGCAAATTATGAAAAC-3′ (with NcoI) and a T7 primer with the CS2p-dapF-miR-309 mutant as the template.
For UASp-miR-7, 432 nt containing the miR-7 precursor was amplified from pUAST-miR-7 with the following primers and ligated into pUASP: forward, CACGAAGGATCCGTCTAACCACCCATCCCCACAA and reverse, CAGCAATCTAGAATGGGAGGGTACTGGGGAGTTC.
S2 cell culture, transfection and luciferase assay
S2 cells were cultured in Schneider's Drosophila Medium (Gibco) with 10% heat-inactivated FBS and penicillin/streptomycin at room temperature (20°C). Transfection used Cellfectin (Invitrogen) according to the manufacturer's instruction. For the luciferase assays in Fig. 2, 1 μg of each miRNA expression plasmid, 100 ng of firefly luciferase reporter plasmid with or without dap 3′UTR, and 100 ng of renilla luciferase reporter plasmid were transfected into cells in a well of a 12-well plate. For the luciferase assays in Fig. S5 in the supplementary material, the amount of miRNA expression vector added in each group was: 0.6 μg miR-1; 0.2 μg miR-278 and 0.4 μg miR-1; 0.2 μg miR-278, 0.2 μg miR-309 and 0.2 μg miR-1; 0.2 μg miR-278, 0.2 μg miR-309 and 0.2 μg miR-7. Combined miRNA expression vectors and 50 ng of firefly luciferase reporter plasmid with or without dap 3′UTR, and 50 ng of renilla luciferase reporter plasmid were transfected into cells in a well of a 24-well plate. Luciferase assays (Dual Luciferase System, Promega) were performed 2 days after transfection. Renilla luciferase activity provided normalization for firefly luciferase activity. The relative luciferase activities of the cells transfected with tub-fLuc-dapF and the different tub-fLuc-mutant-dapF constructs were further normalized to the relative luciferase activities of tub-fLuc for each miRNA.
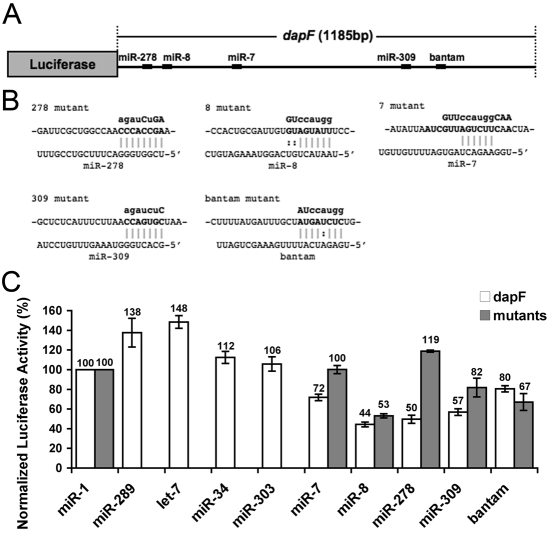
Fig. 2.
The dap 3′UTR is targeted directly by miR-7, miR-278 and miR-309. (A) The luciferase-dapF reporter consists of tubulin promoter-driven luciferase and the 3′UTR of dap. Predicted MREs are shown. (B) Mutations in the dap 3′UTR predicted MREs are shown in lowercase. miRNA/MRE complementarity is also shown. (C) Normalized luciferase activity upon co-expression of miRNAs. miR-1 served as a control. miR-7, -8, -278, -309 and bantam repress luciferase activity, and the predicted MREs for miR-7, -278 and -309 are required for repression. Mean±s.e. of at least three repeats.
Generation of GFP-dap 3′UTR sensors and UASp-miR-7 transgenic lines
Transgenic flies were generated by injection of purified plasmid DNA into w1118 Drosophila embryos (Rainbow Transgenic Flies, Newbury Park, CA, USA). These flies were crossed with w1118 and transformants were selected based on eye color. Twelve, nine, one and nine lines were generated for tub-GFP:dapF, tub-GFP:dapL, tub-EGFP:dapS and UASp-miR-7, respectively.
Recombination for FRT42DmiR-278KO, FRT42DmiR-278Gal4KI and FRT42DmiR-278Gal4KI,miR-7Δ1/CyO
Two miR-278 mutations were recombined into the FRT42D chromosome using standard meiotic recombination protocols (Xu and Rubin, 1993). FRT42DmiR-278Gal4KI was further recombined into the FRT42DmiR-7Δ1 chromosome.
Fly stocks
We used Drosophila stocks carrying the tub-GFP:2x(miR-7) reporter, control tub-GFP reporter (Stark et al., 2003), tub-GFP:dapL reporter, tub-GFP:dapF reporter, tub-GFP:dapS reporter, hsFLP;;UAST-GFPact>CD2> Gal4/TM3Sb, eyFlp;;FRT82BDcr-1Q1147X/TM3, FRT82BInRex52.1/TM6B, FRT82BInRex15/TM3, eyFlp;FRT82B, hsFlp;;FRT82BarmlacZ/TM3, hsFlp;;FRT82BUbi-GFP/TM3, FRT42Bdap4/CyO, FRT42B, eyFlp; FRT42D, hsFlp;FRT42BUbi-GFP/CyO, hsFlp;FRT42DUbi-GFP/CyO, FRT42DmiR-278KO/CyO, FRT42DmiR-278KI-gal4/CyO, FRT42DmiR-7Δ1/CyO, P{EP}blEP954, w-;FRT82Bput135/TM3, yw;Mad12FRT40A/CyO, dap[2x10]/Cyo, dap[g36]/Cyo, ftz-lacZ +;dap5gm.T:Hsap\MYC;+, w-;Sp/CyO; FRT82BInREX52.1/TM6B, yw hsFLP;Sp/CyO;FRT82BUbi-GFP/TM6B.
Generation of clones
Clones of GSCs were induced using the heat shock FLP-FRT system (Dang and Perrimon, 1992; Xu and Rubin, 1993). To generate GSCs clones during third instar larval or pupal stages, flies were heat shocked for 1 hour at 37°C for 2 consecutive days. To generate GSCs clones at the adult stage, newly eclosed flies (1-2 days) were collected and heat shocked twice per day for 30 minutes at 37°C for 2 consecutive days.
Antibodies
The following were used: mouse anti-Adducin [clone 1B1, Developmental Studies Hybridoma Bank (DSHB)]; mouse anti-Cyclin B (DSHB); guinea pig anti-Cyclin E (T. Orr-Weaver, Whitehead Institute); mouse anti-Dap (I. Hariharan, University of California, Berkeley); Alexa 488-conjugated rabbit anti-GFP (Molecular Probes); Alexa 488, 555, 568 or 633-conjugated goat anti-mouse, anti-rabbit and anti-guinea pig antibodies (Molecular Probes).
Immunostaining and fluorescence microscopy
Ovaries were fixed as described previously: ovaries were dissected in PBS and fixed in 5% paraformaldehyde in PBS, then sequentially incubated in primary and secondary antibodies overnight at 4°C, followed by DAPI (1 μg/ml) for 15 minutes. Confocal microscopy and two-photon laser-scanning imaging were performed with a Leica TCS SP/MP or Leica SPE microscope.
Starvation procedure
The starvation condition was adapted from previous reports (Drummond-Barbosa and Spradling, 2001). For poor food conditions, adult flies were collected into plastic bottles containing molasses plates providing moisture and sugar, whereas for rich food conditions this was supplemented with wet yeast. Ovaries were dissected 10 or 14 days later.
Division frequency analysis
Only germaria containing both GFP (or β-gal)-positive and GFP (or β-gal)-negative GSCs were analyzed for cell division. The average sum of cystoblasts and cysts generated by individual GFP (or β-gal)-negative GSCs in region 1-2A of a germarium was normalized to that of individual GFP (or β-gal)-positive control heterozygous GSCs to obtain the division index.
Quantitation of GFP in GSCs
MetaMorph (Molecular Devices) software was used to quantify GFP fluorescence intensity in GSCs imaged on a Leica SP1 confocal microscope. Images of the brightest GFP optical slice were quantified by drawing an elliptical field of identical dimensions for each cell and reading the average intensity in the field.
Quantification of cell cycle markers in GSCs
Quantification of cell cycle marker levels in GSCs was determined with the histogram function in Adobe Photoshop. For a given GSC, intensity was determined by averaging intensities from three different regions within the cell. A background germline intensity value was determined by averaging intensities obtained from three different regions within region 1 of the germarium having low-level staining or background staining. For each antibody, the intensity fold above background was determined as the average GSC intensity divided by the average background intensity.
miRNA quantitative RT-PCR (qPCR)
RNA was isolated from 10-20 ovaries using Trizol (Invitrogen) and treated with RNase-free DNaseI (Fermentas). The extracted RNA (0.5 μg) was reversed transcribed with Omniscript reverse transcriptase (Qiagen). miRNA levels (dme-miR-8 and dme-miR-278) were quantified using TaqMan MicroRNA Assays (Applied Biosystems) as per manufacturer's instruction, using 10 ng of total RNA on an ABI 7300 real-time PCR system.
RESULTS
The dap 3′UTR is regulated in GSCs by miRNAs
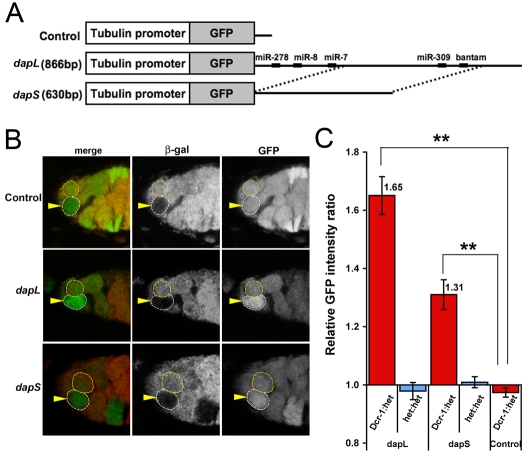
To test whether miRNAs regulate the dap 3′UTR in GSCs, we generated two transgenic lines containing different regions of the dap 3′UTR inserted downstream of GFP. dapL contains most of the dap 3′UTR, whereas some predicted MREs are deleted in the truncated dapS construct (Fig. 1A). A transgenic GFP line that lacks significant 3′UTR served as a control (control sensor; Fig. 1A). The GFP intensity was used to detect endogenous regulation through the dap 3′UTR. Reduction of miRNA production in Dcr-1 mutant (Dcr-1Q1147X) GSCs led to significant upregulation of GFP intensity of the dapL and dapS sensors, but not of the control sensor (Fig. 1B). Image quantification analysis showed that the GFP intensity of dapL and dapS sensors increased an average of 1.65±0.065-fold and 1.31±0.051-fold, respectively, in Dcr-1 null GSCs as compared with the neighboring control heterozygous GSCs. The control sensor showed no significant difference (Fig. 1C). Similarly, no significant difference was detected between two adjacent Dcr-1Q1147X/+ heterozygous GSCs for dapL or dapS (Fig. 1C). These results suggest that miRNAs can directly regulate Dap levels through the dap 3′UTR in Drosophila GSCs.
Fig. 1.
The Drosophila dap 3′UTR is repressed by miRNAs in the germline stem cells. (A) The control sensor lacks significant 3′UTR. The dapL and dapS sensors contain 866 bp and 630 bp of dap 3′UTR, respectively. Predicted miRNA response elements (MREs) are shown. (B) GFP expression was unchanged in Dcr-1 germline stem cells (GSCs) (arrowhead) in the control sensor background as compared with the neighboring control GSC, but was upregulated in the dapL and dapS sensor lines. (C) The dapL and dapS sensors were upregulated in Dcr-1 GSCs. Mean±s.e.; n≥23 pairs of GSCs. Student's t-test; **P<0.01.
miR-7, miR-278 and miR-309 can target the dap 3′UTR directly
To identify which miRNAs repress Dap directly through the dap 3′UTR, we used luciferase assays in S2 cells. Computational algorithms based on sequence complementarity, homology across species and RNA secondary structure predict that many miRNAs target the dap 3′UTR, including miR-7, -8, -34, -278, -289, -303, -309, let-7 and bantam (Fig. 2A,B) (Enright et al., 2003; Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2008; Grun et al., 2005; Long et al., 2007; Ruby et al., 2007). To test whether these predicted miRNAs are sufficient to regulate Dap, the full-length 3′UTR of dap, dapF, was fused downstream of a firefly luciferase gene. Partial precursor sequences of these miRNAs were cloned into an expression vector driven by a tubulin promoter. miR-1 was used as a control as there is no predicted miR-1 MRE in the dap 3′UTR. Whereas expression of miR-7, miR-8, miR-278, miR-309 and bantam inhibited luciferase activity significantly, miR-34, miR-289, miR-303 and let-7 did not repress the activity (Fig. 2C). It is notable that let-7 and miR-289 increased luciferase activity. One possible explanation for this result is that the negative control miRNA, miR-1, might still mildly repress the dap 3′UTR even though there is no predicted MRE for miR-1 and, therefore, let-7 and miR-289 increased luciferase activities after normalization to miR-1. It is also possible that let-7 and miR-289 increased the expression of luciferase through the dap 3′UTR (Vasudevan et al., 2007). Further experiments are required to test this hypothesis.
To test whether the predicted MREs are required for the repression by miRNAs, the sequences of putative MREs cognate to the miRNA seed regions were mutated individually to disrupt miRNA binding (Fig. 2B). The miRNA-dependent repression of luciferase activity was relieved significantly when the MRE site was disrupted for miR-7, miR-278 or miR-309, but not for bantam. When the MRE site for miR-8 was disrupted, the repression of luciferase activity was mildly relieved (Fig. 2C). These results suggest that miR-7, miR-278 and miR-309 can directly target the dap 3′UTR, whereas miR-8 and bantam may either target the dap 3′UTR indirectly or through cryptic binding sites that are not predicted by the current algorithms. To test the cooperative effect of multiple miRNAs on the dap 3′UTR, we co-transfected multiple miRNA expression constructs with the dap 3′UTR luciferase reporter (see Fig. S5 in the supplementary material). Expression of miR-7, miR-278 and miR-309 simultaneously repressed the luciferase activity significantly more than expression of miR-278 alone, suggesting that multiple miRNAs might act on Dap expression.
miR-278 and miR-7 affect the cell cycle of GSCs
We further examined whether miR-278 and miR-7 might regulate GSC division. Two miR-278 null lines, miR-278KO and miR-278Gal4KI, were examined (Teleman et al., 2006). Quantitative RT-PCR (qPCR) was used to detect mature miRNAs in FRT42D control or FRT42DmiR-278KO mutant ovaries. miR-8 was used as a control miRNA as it is expressed in the GSCs (Shcherbata et al., 2007). miR-278 expression was more than 300-fold higher in FRT42D control ovaries than in miR-278KO ovaries. By contrast, the control miRNA miR-8 was expressed at similar levels in the ovaries in both animals (Fig. 3A; see Table S1 in the supplementary material). These results show that miR-278 is expressed in the ovary.
Fig. 3.
miR-278-deficient GSCs proliferate more slowly than control GSCs. (A) qPCR analysis of the mature miRNA expression level of miR-8 and miR-278 in the whole ovary. RNA from miR-278KO ovaries served as negative control for the expression of miR-278. The miRNA expression level is calculated from the cycle threshold (CT) (see Table S1 in the supplementary material) by 2CT(FRT42D control)/2CT(miR-278KO). Mean±s.e. for at least two repeats. (B) miR-278-deficient GSCs produce fewer progeny than control GSCs. A mutant GSC (arrowhead) and its progeny are marked by the lack of GFP. Add, Adducin. (C) The division kinetics of miR-278 GSCs was reduced to 75-79% as compared with the neighboring heterozygous control GSCs or the FRT42D control GSC clones. Division kinetics of miR-7 GSCs were unaffected by comparison with control. The division kinetics of GSCs mutant for both miR-7 and miR-278 were reduced to 82% as compared with the control. Mean±s.e. of three repeats. Student's t-test; *P<0.05. The number of homozygous GSCs counted: hsFLP;FRT42D, n=32; hsFLP;FRT42DmiR-278KO, n=42; hsFLP;FRT42DmiR-278Gal4KI, n=35; hsFLP;FRT42DmiR-7Δ1, n=55; hsFLP;FRT42DmiR-278Gal4KI,miR-7Δ1, n=41.
We generated GSC clones deficient for miR-278 using the heat shock FLP-FRT system (Fig. 3B). GSCs can be identified by their position adjacent to the cap cells, and by the shape and position of the fusome stained by anti-Adducin (Add; Hu li tai shao - FlyBase) antibody (Fig. 3B). After generation of clones, the sum of cystoblasts and cysts generated by each mutant GSC in regions 1-2A of the germarium was counted and compared with that generated by the neighboring control GSCs (see Materials and methods). The miR-278 mutant GSCs showed a 21-25% reduction in division index, whereas the FRT42D control clones divided normally (Fig. 3C; see Table S2 in the supplementary material). Similarly, GSCs in transheterozygous (miR-278KO/miR-278Gal4KI) flies showed a 28% reduction in the number of cystoblasts and cysts at region 1-2A in germaria as compared with GSCs in heterozygous (miR-278KO/+) animals 13-14 days after eclosion. These results suggest that miR-278 plays a role in regulating the cell division of GSCs.
Since the miR-7 mutant is homozygous lethal (Li and Carthew, 2005), qPCR analysis of homozygous miR-7 mutant ovaries (comparable to the miR-278 analysis shown in Fig. 3A) could not be performed. Therefore, in order to determine whether miR-7 is expressed in the GSCs, we analyzed a sensor line with a constitutively driven GFP transgene bearing two perfectly matched target sites for miR-7 (Li and Carthew, 2005; Stark et al., 2003). Compared with a control sensor driven by the same promoter but lacking miRNA complementarity, the GFP-miR-7 sensor was strongly downregulated in the germline cells in the anterior of germaria, from the GSCs to region 3, as well as in the somatic tissue of the germarium (Fig. 4A). Furthermore, the GFP-miR-7 sensor was upregulated in Dcr-1 mutant GSCs in comparison with the heterozygous neighboring control GSCs (Fig. 4B). Image quantification analysis showed that the GFP level was 1.528±0.065-fold higher (mean±s.e. of three repeats; n=71 germaria) in the Dcr-1 mutant GSCs than in neighboring control (heterozygous) GSCs. These results showed that the miR-7 sensor in the GSCs is responsive to Dcr-1 activity, suggesting that miR-7 is expressed in the GSCs.
Fig. 4.
miR-7 expression in GSCs. (A) The GFP-miR-7 sensor is repressed in the anterior-most germline (bracket), as compared with control GFP sensor lacking significant 3′UTR content. Adducin (Add), red; DAPI, blue; GFP, green. (B) Dcr-1 GSC clones lacking β-gal (arrowhead) exhibit elevated GFP-miR-7 sensor expression relative to neighboring heterozygous cells. (C) miR-7Δ1 GSC clones exhibit an elevated frequency of CycE-positive staining compared with heterozygous GSCs. Ectopic expression of miR-7 in miR-7Δ1 mutant GSCs returns the frequency of CycE staining to near control levels. Mean±s.e. of three repeats. Student's t-test; *P<0.05, **P<0.01. The number of GSCs counted: FRT42DmiR-7Δ1/CyO;nosGal4/TM6, n=523; FRT42DmiR-7Δ1;nosGal4/TM6, n=87; FRT42DmiR-7Δ1;nosGal4/UAS-miR-7, n=110. (D,E) Ectopic expression of miR-7 in the GFP-positive follicle cell clones (y w hsFLP;EP{954};pUAST-GFP-act>CD2>Gal4) decreases the frequency of Dap staining compared with the neighboring wild-type cells. Mean±s.e. of three repeats. Student's t-test; **P<0.01.
To characterize the effect of miR-7 on the GSC cell cycle, we examined the expression of the cell cycle marker Cyclin E (CycE) in miR-7 mutant (FRT42DmiR-7Δ1) GSCs (Li and Carthew, 2005). It has been shown that Dap can trap the CycE-Cdk2 complex in a stable but inactive form (de Nooij et al., 1996). Increased levels of Dap result in cell cycle arrest at the G1-S transition and prolonged expression of CycE protein (Shcherbata et al., 2004). We detected an increase in the frequency of CycE staining in miR-7Δ1 GSCs relative to the heterozygous neighboring control GSCs (Fig. 4C). However, the defects observed in the miR-7 mutant GSCs were insufficient to cause an obvious reduction in division index (Fig. 3C; see Fig. S1 and Table S2 in the supplementary material). Since miR-7 resides in an intron of a host gene, bancal (Charroux et al., 1999), we asked whether the elevated frequency of GSCs positive for CycE was due to the loss of bancal or miR-7. Expressing miR-7 with UASp-miR-7 driven by a germline-specific nanos-Gal4 driver returned the frequency of GSCs that stained positive for CycE to wild-type levels, thereby showing that it is the loss of miR-7 and not bancal that is responsible for the increased frequency of CycE staining (Fig. 4C). We found further evidence that miR-7 is sufficient to regulate Dap expression: follicle cell clones in stage 2-4 egg chambers overexpressing miR-7 with an enhancer trap driver, P{EP}blEP954 (Li and Carthew, 2005), exhibited a decreased frequency of Dap staining compared with wild-type control cells (Fig. 4D,E).
Although disruption of miR-278 or miR-7 showed mild cell division defects or abnormal cell cycle marker expression, respectively, neither alone nor in combination displayed as dramatic a perturbation of the cell cycle as Dcr-1-deficient GSCs (Fig. 3C). These results suggest that regulation of the GSC cell cycle might require miRNAs in addition to miR-7 and miR-278, and possibly a combination of multiple miRNAs.
The dap 3′UTR is regulated by InR but not TGF-β signaling
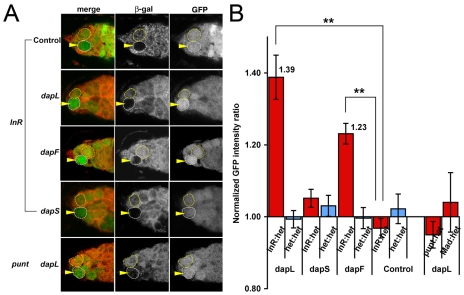
GSCs, like many other stem cells, change their division rate in response to extrinsic factors such as nutrition-dependent InR and TGF-β signaling from the niche cells (LaFever and Drummond-Barbosa, 2005; Xie and Spradling, 1998). To analyze whether these signaling pathways affect cell division through miRNA-based Dap regulation, we tested the responsiveness of the GFP-dap 3′UTR sensor lines to both TGF-β and InR activity. Interestingly, the GFP intensity of the dap 3′UTR sensors dapL and dapF was upregulated in InR mutant GSCs (Fig. 5A). By contrast, the GFP intensity in GSCs deficient for Mad or punt, two key components in the TGF-β pathway, was not affected in the GFP-dap 3′UTR sensor dapL (Fig. 5A,B). This result demonstrates that InR signaling, but not TGF-β signaling, regulates the dap 3′UTR, suggesting that InR signaling might affect Dap expression in GSCs through miRNAs. Quantitation showed that the GFP intensity of the dapL and dapF sensor in InR mutant GSCs increased 1.39-fold and 1.23-fold, respectively, in comparison with the neighboring control heterozygous GSCs (Fig. 5B). Interestingly, the GFP intensity of the dapS sensor was not significantly affected in InR mutant GSCs (Fig. 5A,B), suggesting that the InR-responsive dap 3′UTR region is absent from the dapS construct.
Fig. 5.
The dap 3′UTR responds to InR but not TGF-β signaling in GSCs. (A) In InR mutant GSCs (first four rows, arrowhead), the GFP expression is upregulated in dapL, dapF and dapS, but not in the control sensor. In punt mutant GSCs (bottom row, arrowhead), the GFP expression in dapL is not upregulated. (B) Quantification of GFP intensity in InR, punt or Mad mutant GSCs compared with the neighboring heterozygous GSCs. The GFP intensity is upregulated in InR mutant GSCs with dapL and dapF by 1.39-fold and 1.23-fold, respectively. Mean±s.e. for all pairs of GSCs (at least 12 pairs for InRex52.1 clones, five pairs for punt135 clones, and 24 pairs for Mad12 clones). Student's t-test; **P<0.01.
InR-deficient GSCs show abnormal cell cycle marker expression
Since InR signaling regulates the dap 3′UTR in GSCs, we further investigated how InR signaling regulates the GSC cell cycle. Previous results have shown that disruption of InR signaling reduces the cell division of GSCs (LaFever and Drummond-Barbosa, 2005). We analyzed the cell cycle characteristics of GSC clones deficient for InR with several cell cycle markers (Fig. 6B; see Figs S2 and S3 in the supplementary material). In the InR mutant, there was a 2-fold increase in the frequency of CycE- and Dap-positive GSCs and 1.5-fold decrease in the frequency of CycB-positive GSCs (Fig. 6A). FRT82B control GSC clones showed a similar frequency of CycE, CycB and Dap staining as neighboring heterozygous GSCs (Fig. 6A). Based on the expression of these cell cycle markers, our results suggest that the cell cycle characteristics of InR-deficient GSCs are similar to those of Dcr-1-deficient GSCs and are consistent with our hypothesis that InR signaling may regulate the cell cycle through miRNAs and Dap in GSCs (Hatfield et al., 2005).
Fig. 6.
InR-deficient GSCs show abnormal frequencies of cell cycle marker expression. (A) The percentage of CycE-positive GSCs in two different InR alleles (hsFLP;;FRT82BInRex52.1/FRT82BGFP and hsFLP;;FRT82BInRex15/FRT82BGFP) increased 1.7-fold as compared with the control neighboring GSCs. The percentage of Dap-positive GSCs increased 2-fold, whereas the percentage of CycB-positive GSCs decreased 1.5-fold in InR mutant GSCs. The percentage of dap-5gm-positive GSCs remained the same in InR mutant GSCs as in the control heterozygous neighboring GSCs. Flies were dissected 8 or 12 days after larval/pupal heat shock. Mean±s.e. of two to three repeats. Student's t-test; *P<0.05, **P<0.01. The number of homozygous GSCs counted was as follows. For CycE staining: FRT82B, n=14; InRex52.1, n=58; InRex15, n=47. For Dap staining: FRT82B, n=58; InRex52.1, n=57. For CycB staining: FRT82B, n=71; InRex52.1, n=71. For dap-5gm: InRex52.1, n=43. (B) Dap expression in an InR mutant GSC (yellow arrowhead, expression 2.4-fold higher than background).
The dap-5gm construct does not respond to InR activity
As shown above, Dap expression, and specifically the dap 3′UTR, are responsive to InR activity in GSCs. To test whether other regions of the dap gene show responsiveness, we used dap-5gm, a genomic construct that contains the dap promoter region responsible for GSC expression followed by the dap coding sequence fused to six Myc-tag coding sequences and lacking most of the dap 3′UTR (Meyer et al., 2002; Hatfield et al., 2005). Expression of dap-5gm can be determined with an antibody against Myc. Expression of the Myc tag was the same in InR-deficient and neighboring control heterozygous GSCs (Fig. 6A; see Fig. S4 in the supplementary material). These data suggest that the regions of the dap gene contained in dap-5gm, including the GSC-specific promoter region, are not responsive to InR signaling, supporting our notion that InR regulation of Dap in GSCs occurs through the dap 3′UTR. Furthermore, in accordance with the dap 3′UTR sensor data (Fig. 6), the full-length dap 3′UTR is responsive to InR activity, whereas the dap 3′UTR region remaining in dap-5gm is not.
miRNAs and dap act downstream of InR in regulating cell division
To determine whether miRNAs are required for InR-dependent regulation of cell division, we used a protein-restricted diet to reduce InR signaling (Drummond-Barbosa and Spradling, 2001) and tested whether Dcr-1 affected the phenotype. We generated GSC clones homozygous for the Dcr-1 null allele (Dcr-1Q1147X), as well as parental control FRT82B, during larval/pupal stages. Two days after eclosion, the adult flies were kept under two different diet conditions: rich food with wet yeast and poor food (starvation) without wet yeast. Whereas starvation reduced cell division of the control GSCs 1.7-fold, the low cell division index of the Dcr-1-deficient GSCs observed under rich food conditions did not change under poor food conditions (Fig. 7A). This result is consistent with our hypothesis that miRNAs act downstream of InR signaling in regulating cell division.
Fig. 7.
miRNAs and dap act downstream of nutritional/InR signaling to regulate cell division. The average number of progeny in germarial region 1-2A counted under conditions of differing nutrition. (A) The cell division of FRT82B control GSCs was reduced dramatically under poor food as compared with rich food conditions, whereas the cell division of Dcr-1 GSCs was not significantly reduced. (B) The cell division of control GSCs was dramatically reduced under poor food as compared with rich food conditions, whereas cell division of dap GSCs was not significantly reduced. The cell division of dap4 GSCs was not significantly increased compared with FRT42B control GSCs in poor food conditions (P=0.14075, Student's t-test). (C) Reduction of dap4 partially rescues cell cycle defects in InR-deficient GSCs. Other dap alleles gave similar results (CyO/+;InRex52.1, 50% division index; dap2x10/+;InRex52.1, 68.75% division index). Mean±s.e. of three repeats. Student's t-test; *P<0.05, **P<0.01. n=∼40 per condition.
To test whether InR signaling acts through Dap in GSCs, we analyzed cell division kinetics under differing nutritional conditions for GSC clones homozygous for the dap null allele dap4 (Lane et al., 1996), with FRT42B as a control. The GSC clones were generated in young adults and flies were subsequently housed under different dietary conditions. Whereas starvation reduced cell division of the control GSCs more than 2-fold, the cell division of the GSCs deficient for dap was not significantly reduced (Fig. 7B), suggesting that dap is required for the starvation-dependent reduction of the cell cycle.
To further examine the interaction between dap and InR signaling, we generated GSC clones deficient for InR in a dap4 or dap2x10 heterozygous background. InR mutant GSCs show a strong cell division defect (Fig. 7C) (LaFever and Drummond-Barbosa, 2005). This defect can be partially rescued by reducing dap: the cell division index of InR-deficient GSCs increased from 40 to 60% when dap was reduced (dap4; a 1.5-fold increase) (Fig. 7C). Other strong dap mutant alleles provided similar results (dap2x10; a 1.4-fold increase) (Fig. 7C, legend). These results suggest that dap acts downstream of InR in regulating the cell cycle. Together, these data support our hypothesis that the InR pathway regulates the GSC cell cycle by reducing the levels of Dap (Fig. 8).
Fig. 8.
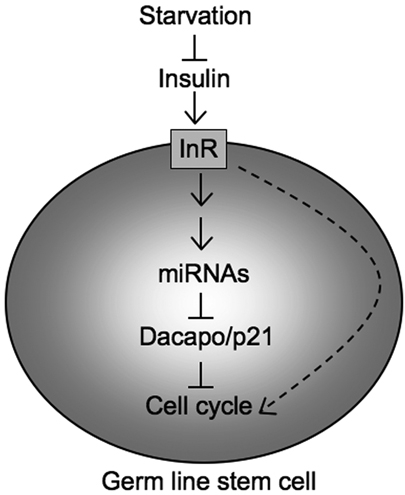
A model for activation of miRNAs by InR signaling, which inhibits Dap expression and accelerates Drosophila GSC division. Reduced InR signaling reduces the levels of miRNAs that repress Dap. Therefore, Dap is upregulated and the cell cycle slows in GSCs. It is also possible that InR signaling regulates GSC division by additional mechanisms (dashed arrow).
DISCUSSION
Previous studies have shown that miRNAs may regulate the Drosophila CKI Dap, thereby controlling the cell division of GSCs (Hatfield et al., 2005). Here, we show that the dap 3′UTR directly responds to miRNA activities in GSCs. Using luciferase assays, we identify miR-7, miR-278 and miR-309 as miRNAs that can directly repress Dap through the dap 3′UTR in vitro. Although miR-278 and miR-7 play a role in regulating GSC division and cell cycle marker expression, respectively, neither of these mutants showed as dramatic a defect in the GSC cell cycle as Dcr-1-deficient GSCs (Hatfield et al., 2005). Thus, the dap 3′UTR may serve to integrate the effect of multiple miRNAs during cell cycle regulation. It remains possible that some miRNAs involved in this process remain to be identified. We further show that InR signaling controls the dap 3′UTR in GSCs. This led us to explore the interaction between InR signaling and miRNA/Dap cell cycle regulation. GSCs deficient for InR or Dcr-1 show similar cell cycle defects. Using starvation to control InR signaling, we show that both Dcr-1 and dap are required for proper InR signaling-dependent regulation of GSC division. Further, reduction of dap partially rescues the cell division defect of the InR mutant GSCs, suggesting that InR signaling regulates the cell cycle via Dap. Our results suggest that miRNAs and Dap act downstream of InR signaling to regulate GSC division (Fig. 8).
miRNAs target the dap 3′UTR
Our data suggest that multiple miRNAs can regulate the 3′UTR of dap: miR-7, miR-278 and miR-309 can regulate the dap 3′UTR directly, whereas bantam and miR-8 may regulate it indirectly, or through cryptic MREs in the dap 3′UTR. Using GFP sensor assays, we also show that the dap 3′UTR may be directly regulated by miRNAs in the GSCs in vivo. However, which specific miRNAs control endogenous Dap levels in Drosophila GSCs remains unknown. Mammalian p21cip1 has also been shown to be a direct target for specific miRNAs of the miR-106 family, including miR-290s and miR-372 (Ivanovska et al., 2008; Sinkkonen et al., 2008). Further, the mouse miR-290 family has recently been identified as regulating the G1-S transition (Wang et al., 2008). In addition, miR-221 and miR-222 have been shown to regulate p27kip1, thereby promoting cell division in different mammalian cancer cell lines (Galardi et al., 2007; le Sage et al., 2007; Visone et al., 2007). Neither the miR-290 nor miR-220 family is conserved in Drosophila. Together, these results indicate that the CKIs (Dap) might be a common target for miRNAs in regulating the cell cycle in stem cells. However, the specific miRNAs that regulate the CKIs might vary between organisms.
miR-7 and miR-278
Our study reveals novel regulatory roles for miR-7 and miR-278 in the GSC cell cycle. We have shown by luciferase assays that miR-7 and miR-278 can directly target Dap. GSCs deficient for miR-278 show a mild but significant reduction in cell proliferation. Ectopic expression of miR-7 in follicle cells reduces the proportion of cells that stain positive for Dap. Furthermore, ablation of miR-7 in GSCs results in a perturbation of the frequency of CycE-positive GSCs. However, the cell division kinetics of miR-7 mutant GSCs is not reduced, by contrast with the dramatic reduction of cell division in Dcr-1-deficient GSCs. It is plausible that miR-7 and miR-278 act in concert with other miRNAs to regulate the level of Dap in GSCs and thereby contribute to cell cycle control in GSCs. Recently, the 3′UTR of nerfin-1, a Drosophila zinc-finger transcription factor gene required for axon pathfinding, has been shown to be regulated by multiple miRNAs in the developing nervous system (Kuzin et al., 2007). Although we have shown that the dap 3′UTR lies downstream of the miRNA pathway, it is still possible that some miRNAs control Dap expression indirectly.
The interaction of multiple miRNAs with the dap 3′UTR might integrate information from multiple pathways. Further studies will reveal what regulates miR-7 and miR-278 expression in GSCs and which other miRNAs might act together in Dap regulation. It is known that miR-7 and the transcriptional repressor Yan (Anterior open - FlyBase) mutually repress one another in the eye imaginal disk (Li and Carthew, 2005). In this model, Yan prevents transcription of miR-7 until Erk in the Egfr pathway downregulates Yan activity by phosphorylation, thereby permitting expression of miR-7. Conversely, miR-7 can repress the translation of Yan. Thus, a single pulse of Egfr signaling results in stable expression of miR-7 and repression of Yan. Whether similar regulation will be observed between miR-7 and the signaling pathways that regulate GSC division remains to be seen. It has been suggested that miR-7 might regulate downstream targets of Notch, such as Enhancer of split and Bearded (Stark et al., 2003). Thus, miR-7 may have a mild repressive effect on multiple targets in GSCs. Further experiments might illuminate this possibility.
miR-278, on the other hand, has been implicated in tissue growth and InR signaling (Teleman et al., 2006). Overexpression of miR-278 promotes tissue growth in eye and wing imaginal discs. Deficiency of miR-278 leads to a reduced fat body, which is similar to the effect of impaired InR signaling in adipose tissue. Interestingly, miR-278 mutants have elevated insulin/Dilp production and a reduction of insulin sensitivity. Furthermore, miR-278 regulates expanded, which may modulate growth factor signaling including InR. Since InR signaling plays important roles in tissue growth and cell cycle control (Edgar, 2006; Taguchi and White, 2008; Wu and Brown, 2006), it will be interesting to further test how miR-278 may regulate InR signaling, and whether InR signaling might regulate miR-278 in a feedback loop in GSCs.
Other miRNAs or miRNA-dependent mechanisms might also play roles in Drosophila GSCs. For example, the miRNA bantam is required for GSC maintenance (Shcherbata et al., 2007). A recent study has shown that the Trim-NHL-containing protein Mei-P26, which belongs to the same family as Brain tumor (Brat), affects bantam levels and restricts cell growth and proliferation in the GSC lineage (Neumuller et al., 2008). Interestingly, most miRNAs are upregulated in mei-P26 mutant flies. By contrast, overexpression mei-P26 in bag of marbles (bam) mutants broadly reduces miRNA levels. This suggests that Mei-P26 regulates proliferation and maintenance of GSC lineages via miRNA levels. Since InR signaling cell-autonomously regulates GSC division but not maintenance, the possible interaction between Mei-P26 and InR signaling might be complex.
InR signaling regulates Dap and the cell cycle cell-autonomously
The systemic compensatory effect of insulin secretion in mammals with defective InR signaling is well documented. Insulin levels in mice with liver-specific InR (Insr - Mouse Genome Informatics) knockout are ∼20-fold higher than those of control animals owing to the compensatory response of the pancreatic β-cells and impairment of insulin clearance by the liver (Michael et al., 2000). Knockout of the neuronal InR also leads to a mild hyperinsulinemia, indicating whole-body insulin resistance (Bruning et al., 2000). Furthermore, the knockout of components in the InR signaling pathway, such as Akt2 and the regulatory and catalytic subunits of PI3 kinase, also leads to hyperinsulinemia and glucose intolerance (Brachmann et al., 2005; Cho et al., 2001; MacDonald et al., 2004; Ueki et al., 2002). Therefore, a systemic decrease in InR signaling may lead to compensatory responses.
To understand the roles of InR signaling in the GSCs while avoiding any systemic compensatory effect we analyzed the phenotypes of GSC clones. Using a panel of cell cycle markers, we find that InR mutant GSCs show cell cycle defects similar to those of Dcr-1 mutant GSCs: a reduction of cell division rate, an increased frequency of cells staining positive for Dap and CycE, and a decreased frequency of cells staining positive for CycB. Using GFP-dap 3′UTR sensors, we show that the dap 3′UTR responds to InR signaling in GSCs, suggesting that InR signaling can regulate Dap expression through the dap 3′UTR. This, together with our genetic data indicating that InR/starvation-dependent cell cycle regulation requires Dcr-1 and dap, led us to propose the hypothesis that InR signaling regulates the cell cycle through miRNAs that further regulate Dap levels (Fig. 8). Since a reduction in dap only partially rescues the cell cycle defects of InR mutant GSCs, it is possible that InR signaling might also regulate GSC division by additional mechanisms (Fig. 8, dashed arrow).
Starvation, InR signaling and cell cycle control
InR signaling regulates the cell cycle through multiple mechanisms, mainly through the G1-S, but also partly through the G2-M, transition. Recent work has shown a delay in the G2-M transition in GSCs during C. elegans dauer formation (Narbonne and Roy, 2006). Starvation and InR deficiency may also affect the G2-M checkpoint in Drosophila GSCs (Hsu et al., 2008). Here we dissect one possible molecular pathway that InR signaling utilizes to regulate the Drosophila GSC G1-S transition and show that InR signaling can control the cell cycle through miRNA-based regulation of Dap.
Many studies have connected InR and CKIs to Tor (Target of rapamycin) or Foxo pathways downstream of InR signaling. In S. cerevisiae, the yeast homolog of p21/p27 is upregulated when Tor signaling is inhibited (Zinzalla et al., 2007). Foxo, a transcription factor that can be repressed by InR signaling, is known to play important roles in nutrition-dependent cell cycle regulation by upregulating p21 and p27 (Medema et al., 2000; Nakae et al., 2003; Seoane et al., 2004) and by repressing cyclin D1/D2 (Park et al., 2005; Schmidt et al., 2002). In C. elegans, starvation causes L1 cell cycle arrest mediated by InR (daf-2) and Foxo (daf-16): InR represses the function of Foxo, thereby downregulating the CKI (cki-1) and upregulating the miRNA lin-4 (Baugh and Sternberg, 2006). We have now shown that a miRNA-based regulation of Dap can be coordinated by InR in Drosophila GSCs.
Insulin and insulin-like growth factors (Igf1 and Igf2) are known to play important roles in regulating metabolic and developmental processes in many stem cells (Mourkioti and Rosenthal, 2005; Saltiel and Kahn, 2001; Ye and D'Ercole, 2006). In mammals, Igf signaling is required by different stem cell types, including human and mouse ES cells for survival and self-renewal (Bendall et al., 2007; Hallmann et al., 2003; Rubin et al., 2007; Wang et al., 2007), neural stem cells for expediting the G1-S transition and cell cycle re-entry (Hodge et al., 2004), and skeletal muscle satellite cells for promoting the G1-S transition via p27kip1 downregulation (Chakravarthy et al., 2000). Here we have dissected the molecular mechanism of the InR pathway in another adult stem cell type, Drosophila GSCs, showing that InR signaling can regulate stem cell division through miRNA-based downregulation of the G1-S inhibitor Dap. Further studies will reveal whether miRNAs also mediate InR signaling in other stem cell types.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/9/1497/DC1
Supplementary Material
Acknowledgments
We thank Drs A. Spradling, S. Cohen, T. Orr-Weaver, D. Drummond-Barbosa, T. Xie, E. Hafen, B. Edgar, L. Pick, I. Hariharan, C. Lehner, J. Secombe, R. Eisenman, R. Carthew and members of Ruohola-Baker laboratory for suggestions, flies and reagents. We thank Volodya Shchherbatyy, Sarah Mahoney, David Chang, Kathrine Park and Emily Kerr for plasmids preparation and fly injection.
This work was supported by AHA fellowships for J.Y.Y., H.R.S. and S.H.R., Schultz fellowship for S.D.H. and MOD and NIH grants and the Tietze fellowship for H.R.B. Deposited in PMC for release after 12 months.
References
- Ahn, S. and Joyner, A. L. (2005). In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 437, 894-897. [DOI] [PubMed] [Google Scholar]
- Alvarez, B., Martinez, A. C., Burgering, B. M. and Carrera, A. C. (2001). Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413, 744-747. [DOI] [PubMed] [Google Scholar]
- Ambros, V. (2004). The functions of animal microRNAs. Nature 431, 350-355. [DOI] [PubMed] [Google Scholar]
- Balordi, F. and Fishell, G. (2007). Hedgehog signaling in the subventricular zone is required for both the maintenance of stem cells and the migration of newborn neurons. J. Neurosci. 27, 5936-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281-297. [DOI] [PubMed] [Google Scholar]
- Baugh, L. R. and Sternberg, P. W. (2006). DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 16, 780-785. [DOI] [PubMed] [Google Scholar]
- Bendall, S. C., Stewart, M. H., Menendez, P., George, D., Vijayaragavan, K., Werbowetski-Ogilvie, T., Ramos-Mejia, V., Rouleau, A., Yang, J., Bosse, M. et al. (2007). IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature 448, 1015-1021. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Kim, S. Y., Carmell, M. A., Murchison, E. P., Alcorn, H., Li, M. Z., Mills, A. A., Elledge, S. J., Anderson, K. V. and Hannon, G. J. (2003). Dicer is essential for mouse development. Nat. Genet. 35, 215-217. [DOI] [PubMed] [Google Scholar]
- Brachmann, S. M., Ueki, K., Engelman, J. A., Kahn, R. C. and Cantley, L. C. (2005). Phosphoinositide 3-kinase catalytic subunit deletion and regulatory subunit deletion have opposite effects on insulin sensitivity in mice. Mol. Cell. Biol. 25, 1596-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning, J. C., Gautam, D., Burks, D. J., Gillette, J., Schubert, M., Orban, P. C., Klein, R., Krone, W., Muller-Wieland, D. and Kahn, C. R. (2000). Role of brain insulin receptor in control of body weight and reproduction. Science 289, 2122-2125. [DOI] [PubMed] [Google Scholar]
- Burgering, B. M. and Kops, G. J. (2002). Cell cycle and death control: long live Forkheads. Trends Biochem. Sci. 27, 352-360. [DOI] [PubMed] [Google Scholar]
- Carrington, J. C. and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336-338. [DOI] [PubMed] [Google Scholar]
- Chakravarthy, M. V., Abraha, T. W., Schwartz, R. J., Fiorotto, M. L. and Booth, F. W. (2000). Insulin-like growth factor-I extends in vitro replicative life span of skeletal muscle satellite cells by enhancing G1/S cell cycle progression via the activation of phosphatidylinositol 3′-kinase/Akt signaling pathway. J. Biol. Chem. 275, 35942-35952. [DOI] [PubMed] [Google Scholar]
- Charroux, B., Angelats, C., Fasano, L., Kerridge, S. and Vola, C. (1999). The levels of the bancal product, a Drosophila homologue of vertebrate hnRNP K protein, affect cell proliferation and apoptosis in imaginal disc cells. Mol. Cell. Biol. 19, 7846-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H., Mu, J., Kim, J. K., Thorvaldsen, J. L., Chu, Q., Crenshaw, E. B., 3rd, Kaestner, K. H., Bartolomei, M. S., Shulman, G. I. and Birnbaum, M. J. (2001). Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science 292, 1728-1731. [DOI] [PubMed] [Google Scholar]
- Croce, C. M. and Calin, G. A. (2005). miRNAs, cancer, and stem cell division. Cell 122, 6-7. [DOI] [PubMed] [Google Scholar]
- Dang, D. T. and Perrimon, N. (1992). Use of a yeast site-specific recombinase to generate embryonic mosaics in Drosophila. Dev. Genet. 13, 367-375. [DOI] [PubMed] [Google Scholar]
- de Nooij, J. C., Letendre, M. A. and Hariharan, I. K. (1996). A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87, 1237-1247. [DOI] [PubMed] [Google Scholar]
- de Nooij, J. C., Graber, K. H. and Hariharan, I. K. (2000). Expression of the cyclin-dependent kinase inhibitor Dacapo is regulated by cyclin E. Mech. Dev. 97, 73-83. [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa, D. and Spradling, A. C. (2001). Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev. Biol. 231, 265-278. [DOI] [PubMed] [Google Scholar]
- Du, T. and Zamore, P. D. (2005). microPrimer: the biogenesis and function of microRNA. Development 132, 4645-4652. [DOI] [PubMed] [Google Scholar]
- Edgar, B. A. (2006). How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7, 907-916. [DOI] [PubMed] [Google Scholar]
- Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C. and Marks, D. S. (2003). MicroRNA targets in Drosophila. Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher, A. and Slack, F. J. (2006). Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259-269. [DOI] [PubMed] [Google Scholar]
- Forstemann, K., Tomari, Y., Du, T., Vagin, V. V., Denli, A. M., Bratu, D. P., Klattenhoff, C., Theurkauf, W. E. and Zamore, P. D. (2005). Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3, e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, M. T. and Spradling, A. C. (2007). Male and female Drosophila germline stem cells: two versions of immortality. Science 316, 402-404. [DOI] [PubMed] [Google Scholar]
- Galardi, S., Mercatelli, N., Giorda, E., Massalini, S., Frajese, G. V., Ciafre, S. A. and Farace, M. G. (2007). miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 282, 23716-23724. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Grocock, R. J., van Dongen, S., Bateman, A. and Enright, A. J. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Saini, H. K., van Dongen, S. and Enright, A. J. (2008). miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154-D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun, D., Wang, Y. L., Langenberger, D., Gunsalus, K. C. and Rajewsky, N. (2005). microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann, D., Trumper, K., Trusheim, H., Ueki, K., Kahn, C. R., Cantley, L. C., Fruman, D. A. and Horsch, D. (2003). Altered signaling and cell cycle regulation in embryonal stem cells with a disruption of the gene for phosphoinositide 3-kinase regulatory subunit p85alpha. J. Biol. Chem. 278, 5099-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield, S. D., Shcherbata, H. R., Fischer, K. A., Nakahara, K., Carthew, R. W. and Ruohola-Baker, H. (2005). Stem cell division is regulated by the microRNA pathway. Nature 435, 974-978. [DOI] [PubMed] [Google Scholar]
- Hodge, R. D., D'Ercole, A. J. and O'Kusky, J. R. (2004). Insulin-like growth factor-I accelerates the cell cycle by decreasing G1 phase length and increases cell cycle reentry in the embryonic cerebral cortex. J. Neurosci. 24, 10201-10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, H. J., LaFever, L. and Drummond-Barbosa, D. (2008). Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev. Biol. 313, 700-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska, I., Ball, A. S., Diaz, R. L., Magnus, J. F., Kibukawa, M., Schelter, J. M., Kobayashi, S. V., Lim, L., Burchard, J., Jackson, A. L. et al. (2008). MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 28, 2167-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z. and Xie, T. (2007). Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 17, 539-544. [DOI] [PubMed] [Google Scholar]
- Jordan, K. C., Schaeffer, V., Fischer, K. A., Gray, E. E. and Ruohola-Baker, H. (2006). Notch signaling through tramtrack bypasses the mitosis promoting activity of the JNK pathway in the mitotic-to-endocycle transition of Drosophila follicle cells. BMC Dev. Biol. 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollet, O., Dar, A., Shivtiel, S., Kalinkovich, A., Lapid, K., Sztainberg, Y., Tesio, M., Samstein, R. M., Goichberg, P., Spiegel, A. et al. (2006). Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat. Med. 12, 657-664. [DOI] [PubMed] [Google Scholar]
- Kops, G. J., de Ruiter, N. D., De Vries-Smits, A. M., Powell, D. R., Bos, J. L. and Burgering, B. M. (1999). Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398, 630-634. [DOI] [PubMed] [Google Scholar]
- Kuzin, A., Kundu, M., Brody, T. and Odenwald, W. F. (2007). The Drosophila nerfin-1 mRNA requires multiple microRNAs to regulate its spatial and temporal translation dynamics in the developing nervous system. Dev. Biol. 310, 35-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever, L. and Drummond-Barbosa, D. (2005). Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science 309, 1071-1073. [DOI] [PubMed] [Google Scholar]
- Lane, M. E., Sauer, K., Wallace, K., Jan, Y. N., Lehner, C. F. and Vaessin, H. (1996). Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell 87, 1225-1235. [DOI] [PubMed] [Google Scholar]
- le Sage, C., Nagel, R., Egan, D. A., Schrier, M., Mesman, E., Mangiola, A., Anile, C., Maira, G., Mercatelli, N., Ciafre, S. A. et al. (2007). Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 26, 3699-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. and Carthew, R. W. (2005). A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123, 1267-1277. [DOI] [PubMed] [Google Scholar]
- Long, D., Lee, R., Williams, P., Chan, C. Y., Ambros, V. and Ding, Y. (2007). Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 14, 287-294. [DOI] [PubMed] [Google Scholar]
- MacDonald, P. E., Joseph, J. W., Yau, D., Diao, J., Asghar, Z., Dai, F., Oudit, G. Y., Patel, M. M., Backx, P. H. and Wheeler, M. B. (2004). Impaired glucose-stimulated insulin secretion, enhanced intraperitoneal insulin tolerance, and increased beta-cell mass in mice lacking the p110gamma isoform of phosphoinositide 3-kinase. Endocrinology 145, 4078-4083. [DOI] [PubMed] [Google Scholar]
- Medema, R. H., Kops, G. J., Bos, J. L. and Burgering, B. M. (2000). AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404, 782-787. [DOI] [PubMed] [Google Scholar]
- Meyer, C. A., Kramer, I., Dittrich, R., Marzodko, S., Emmerich, J. and Lehner, C. F. (2002). Drosophila p27Dacapo expression during embryogenesis is controlled by a complex regulatory region independent of cell cycle progression. Development 129, 319-328. [DOI] [PubMed] [Google Scholar]
- Michael, M. D., Kulkarni, R. N., Postic, C., Previs, S. F., Shulman, G. I., Magnuson, M. A. and Kahn, C. R. (2000). Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87-97. [PubMed] [Google Scholar]
- Morrison, S. J. and Spradling, A. C. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourkioti, F. and Rosenthal, N. (2005). IGF-1, inflammation and stem cells: interactions during muscle regeneration. Trends Immunol. 26, 535-542. [DOI] [PubMed] [Google Scholar]
- Murchison, E. P., Partridge, J. F., Tam, O. H., Cheloufi, S. and Hannon, G. J. (2005). Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 102, 12135-12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae, J., Kitamura, T., Kitamura, Y., Biggs, W. H., 3rd, Arden, K. C. and Accili, D. (2003). The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell 4, 119-129. [DOI] [PubMed] [Google Scholar]
- Narbonne, P. and Roy, R. (2006). Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development 133, 611-619. [DOI] [PubMed] [Google Scholar]
- Neumuller, R. A., Betschinger, J., Fischer, A., Bushati, N., Poernbacher, I., Mechtler, K., Cohen, S. M. and Knoblich, J. A. (2008). Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454, 241-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. K., Liu, X., Strauss, T. J., McKearin, D. M. and Liu, Q. (2007). The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 17, 533-538. [DOI] [PubMed] [Google Scholar]
- Park, Y., Maizels, E. T., Feiger, Z. J., Alam, H., Peters, C. A., Woodruff, T. K., Unterman, T. G., Lee, E. J., Jameson, J. L. and Hunzicker-Dunn, M. (2005). Induction of cyclin D2 in rat granulosa cells requires FSH-dependent relief from FOXO1 repression coupled with positive signals from Smad. J. Biol. Chem. 280, 9135-9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, R., Arzumanyan, A., Soliera, A. R., Ross, B., Peruzzi, F. and Prisco, M. (2007). Insulin receptor substrate (IRS)-1 regulates murine embryonic stem (mES) cells self-renewal. J. Cell. Physiol. 213, 445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby, J. G., Stark, A., Johnston, W. K., Kellis, M., Bartel, D. P. and Lai, E. C. (2007). Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17, 1850-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti, B., Funari, A., Michienzi, S., Di Cesare, S., Piersanti, S., Saggio, I., Tagliafico, E., Ferrari, S., Robey, P. G., Riminucci, M. et al. (2007). Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 131, 324-336. [DOI] [PubMed] [Google Scholar]
- Saltiel, A. R. and Kahn, C. R. (2001). Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799-806. [DOI] [PubMed] [Google Scholar]
- Schmidt, M., Fernandez de Mattos, S., van der Horst, A., Klompmaker, R., Kops, G. J., Lam, E. W., Burgering, B. M. and Medema, R. H. (2002). Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol. Cell. Biol. 22, 7842-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane, J., Le, H. V., Shen, L., Anderson, S. A. and Massague, J. (2004). Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211-223. [DOI] [PubMed] [Google Scholar]
- Shcherbata, H. R., Althauser, C., Findley, S. D. and Ruohola-Baker, H. (2004). The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development 131, 3169-3181. [DOI] [PubMed] [Google Scholar]
- Shcherbata, H. R., Ward, E. J., Fischer, K. A., Yu, J. Y., Reynolds, S. H., Chen, C. H., Xu, P., Hay, B. A. and Ruohola-Baker, H. (2007). Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell 1, 698-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen, L., Hugenschmidt, T., Berninger, P., Gaidatzis, D., Mohn, F., Artus-Revel, C. G., Zavolan, M., Svoboda, P. and Filipowicz, W. (2008). MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 15, 259-267. [DOI] [PubMed] [Google Scholar]
- Stadler, B. M. and Ruohola-Baker, H. (2008). Small RNAs: keeping stem cells in line. Cell 132, 563-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russell, R. B. and Cohen, S. M. (2003). Identification of Drosophila MicroRNA targets. PLoS Biol. 1, E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, T., Kohara, H., Noda, M. and Nagasawa, T. (2006). Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25, 977-988. [DOI] [PubMed] [Google Scholar]
- Taguchi, A. and White, M. F. (2008). Insulin-like signaling, nutrient homeostasis, and life span. Annu. Rev. Physiol. 70, 191-212. [DOI] [PubMed] [Google Scholar]
- Teleman, A. A., Maitra, S. and Cohen, S. M. (2006). Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 20, 417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, D. L. and Weintraub, H. (1994). Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434-1447. [DOI] [PubMed] [Google Scholar]
- Ueki, K., Yballe, C. M., Brachmann, S. M., Vicent, D., Watt, J. M., Kahn, C. R. and Cantley, L. C. (2002). Increased insulin sensitivity in mice lacking p85beta subunit of phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA 99, 419-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, S., Tong, Y. and Steitz, J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931-1934. [DOI] [PubMed] [Google Scholar]
- Visone, R., Russo, L., Pallante, P., De Martino, I., Ferraro, A., Leone, V., Borbone, E., Petrocca, F., Alder, H., Croce, C. M. et al. (2007). MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr. Relat. Cancer 14, 791-798. [DOI] [PubMed] [Google Scholar]
- Wang, L., Schulz, T. C., Sherrer, E. S., Dauphin, D. S., Shin, S., Nelson, A. M., Ware, C. B., Zhan, M., Song, C. Z., Chen, X. et al. (2007). Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood 110, 4111-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Medvid, R., Melton, C., Jaenisch, R. and Blelloch, R. (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Baskerville, S., Shenoy, A., Babiarz, J. E., Baehner, L. and Blelloch, R. (2008). Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 40, 1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. and Brown, M. R. (2006). Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 51, 1-24. [DOI] [PubMed] [Google Scholar]
- Xie, T. and Spradling, A. C. (1998). decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94, 251-260. [DOI] [PubMed] [Google Scholar]
- Xu, T. and Rubin, G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223-1237. [DOI] [PubMed] [Google Scholar]
- Yamashita, Y. M., Fuller, M. T. and Jones, D. L. (2005). Signaling in stem cell niches: lessons from the Drosophila germline. J. Cell Sci. 118, 665-672. [DOI] [PubMed] [Google Scholar]
- Yang, L., Chen, D., Duan, R., Xia, L., Wang, J., Qurashi, A., Jin, P. and Chen, D. (2007). Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development 134, 4265-4272. [DOI] [PubMed] [Google Scholar]
- Ye, P. and D'Ercole, A. J. (2006). Insulin-like growth factor actions during development of neural stem cells and progenitors in the central nervous system. J. Neurosci. Res. 83, 1-6. [DOI] [PubMed] [Google Scholar]
- Yi, R., Poy, M. N., Stoffel, M. and Fuchs, E. (2008). A skin microRNA promotes differentiation by repressing `stemness'. Nature 452, 225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla, V., Graziola, M., Mastriani, A., Vanoni, M. and Alberghina, L. (2007). Rapamycin-mediated G1 arrest involves regulation of the Cdk inhibitor Sic1 in Saccharomyces cerevisiae. Mol. Microbiol. 63, 1482-1494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.