Abstract
We describe a novel germline mutation in BMPR1A in a family with Juvenile Polyposis and colon cancer. This mutation is that of two consecutive substitutions (735-6 TG>AT) which cause two nonsense mutations (Y245X, G246X), inherited in an autosomal dominant fashion, on one parental chromosome. This mutation caused protein truncation, and represents a unique case of consecutive nonsense mutations in human disease.
Keywords: Juvenile Polyposis, nonsense mutations, BMPR1A
Introduction
Juvenile Polyposis (JP; OMIM 174900) is an autosomal dominant condition where affected individuals develop hamartomatous polyps of the colon, rectum, and less frequently, the stomach[1]. JP patients are at significant risk for developing GI cancer[2, 3], and some may also have hereditary hemorrhagic telangiectasia[4, 5]. Two predisposing genes have been identified in JP, SMAD4 from 18q21[6, 7], and BMPR1A from 10q22-23[8]. Both of these genes are members of the transforming growth factor beta superfamily, and mutations of the coding sequence of each gene have been found in approximately 20% of JP cases[9]. About half of the germline mutations thus far described have been microdeletions, and the other half substitutions. In this report, we describe the novel finding of two consecutive substitutions leading to two consecutive nonsense codons in a family with JP.
Clinical Report
The proband (II-3, Figure 1) developed lower GI bleeding at age 5, and on sigmoidoscopy, 6 hamartomatous polyps were removed. Over the following 18 years of surveillance, 107 polyps were removed, and all were hamartomatous without adenomatous features. He was found to have gastric polyps at age 21. The proband's mother had a negative colonoscopy at age 42 and was still healthy at age 59 and without symptoms. His father was deemed to be healthy, but died at age 66, reportedly without polyps. The proband's paternal grandmother died of colorectal cancer at age 66. His brother had colonoscopy and an upper GI series at age 18, which were negative.
Figure 1.
Hungarian JP kindred. Solid shading=affected; ?=at risk.
The family was lost to follow-up from 1989-1997. The proband was next seen at age 31, when he presented with jaundice, weight loss, and a bowel obstruction. At surgery, he was found to have colon cancer with liver metastases, and he died 3 months later. His older brother (II-2) resurfaced at about this time for surveillance, and was found to have numerous polyps in the colon, 3 of which had foci of adenocarcinoma. He underwent a left hemicolectomy, and has been polyp free since then. His two children (III-1, III-2) had no polyps found at colonoscopy at ages 20 and 21, respectively. The proband's daughter was 9 years of age at last follow-up, and had not had colonoscopy.
Results and Discussion
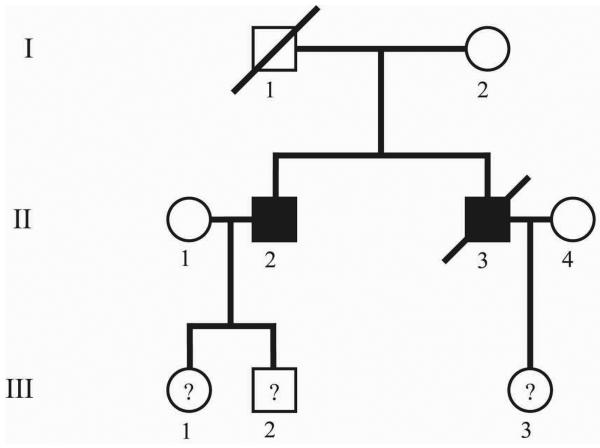
Blood samples were obtained from individuals II-2, III-1, III-2, and III-3. DNA was extracted, and individual II-2 was sequenced for all exons and intron-exon boundaries of SMAD4 and BMPR1A. Two substitutions were found in consecutive nucleotides of exon 7 of BMPR1A (Figure 2a), 735-6 TG>AT. Interestingly, each of these substitutions would change the corresponding amino acid into a stop codon (Y245X, G246X). Individual III-1 also had this mutation (and had no known polyps at age 20), as did III-3 (who was 9 years old at last follow-up, and had not had colonoscopy). Individual III-2 did not have the mutation and had negative colonoscopic screening at age 21 years.
Figure 2.
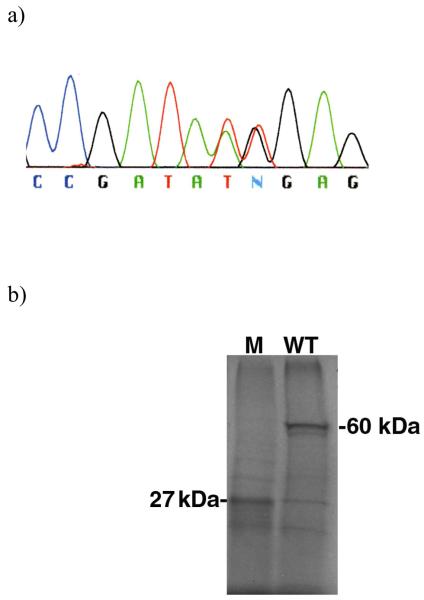
(a) BMPR1A exon7 sequence of affected family member. (b) Autoradiogram of protein truncation test results, labeled using S35-methionine, with electrophoresis through 10% SDS-PAGE. M= mutant truncated BMPR1A protein (27 kDa). WT= wild type full length BMPR1A protein (60 kDa).
mRNA was extracted from patient II-2, converted to cDNA by reverse-transcriptase PCR (using primers GGATCCTAATACGACTCACTATAGGGAGCCACCATGCCTCAGCTATACATTTAC and ATGGGTTAACCATCAGATTTTTACATCTTGGGATTCAACCATCTTGGC), and subcloned into a plasmid vector. After transformation, different recombinant clones were sequenced in order to identify both the wild-type and mutated sequences. Both substitutions were confirmed to be on the same mutant allele. Wild-type clones were 10 fold more abundant than those with mutations, suggesting a possible issue with RNA stability of the mutant sequence. Protein was synthesized using a coupled reticulocyte lysate system kit (Promega TNT). The results from SDS-PAGE revealed a truncated protein of 27 kDa corresponding to the mutant clone and the expected size of 60 kDa from the wild-type clone (Figure 2b).
There have been several reports of two consecutive substitutions and missense mutations in human disease genes. Van Belzen et al. described a Dutch family with ataxia telangiectasia (autosomal recessive) where an affected brother and sister were both homozygous for 2 consecutive substitutions in exon 55 of the ATM gene, leading to 2 consecutive missense mutations (D2625E/A2626P)[10]. Mullan et al. found substitutions in 2 families with early onset Alzheimer's disease that created missense mutations at 2 contiguous codons of the β-amyloid precursor protein (K670N, M671L)[11]. Another example of 2 missense mutations in a human gene was reported by Hassan et al. in the hemoglobin C gene in three affected family members, but these were not contiguous (E6V and P58R)[12].
There have also been several reports of nonsense mutations followed by missense mutations. Longy et al. described consecutive substitutions of PTEN in a Bannaya-Riley-Ruvalcaba syndrome patient (1735-6TA>AT), which led to a stop codon followed by missense change (Y178X, S179C)[13]. Kawabe et al. found a homozygous mutation in the dysferlin gene (3817-8TG>AA) in a patient with limb-girdle muscular dystrophy, which resulted in a nonsense mutation followed by a missense mutation (Y1148X, G1149R)[14]. De Rosa et al. described consecutive substitutions in APC (3225-6TC>AA) in a patient with classic familial adenomatous polyposis and congenital hypertrophy of the retinal pigment epithelium. This results in a nonsense followed by a missense mutation (Y1075X, P1076T)[15].
To our knowledge, there has not been a description of two consecutive substitutions leading to consecutive nonsense mutations in human disease genes. Smit et al. described a patient with cystic fibrosis and a single nucleotide insertion (2307insA) in CFTR that led to a frameshift in codon 726, which in turn created two consecutive stop codons at amino acids 729 and 730. The patient appeared to be homozygous for this mutation[16]. Becker et al. reported a patient with congenital ichthyosiform erythroderma, which is autosomal recessive, with 2 nonsense mutations in the transglutaminase 1 gene. Each of the child's unaffected parents was heterozygous for one of the mutations (7780C>G, Y503X and 8533C>G, S669X), and therefore the nonsense mutations in this case were on separate chromosomes, nor were they at consecutive codons[17]. In our patient, subcloning of each parental copy of BMPR1A revealed that both nonsense mutations were inherited on the same chromosome. Our case also contrasted with these others in that JP is an autosomal dominant disorder, and inheritance of a single deleterious allele causes the phenotype. It would therefore be difficult to establish that functionally both mutations would lead to further impairment of gene function over one or the other by itself. If the disease is related to protein truncation, then there should be no difference. However, whether two premature stop codons could result in an increased level of nonsense mediated decay is less clear. The severity of disease in this family seemed more pronounced than in many others with germline BMPR1A mutations, as two affected individuals developed colon cancer, and one had gastric juvenile polyps, both of which are more common in JP patients with SMAD4 rather than BMPR1A mutations[18, 19].
Acknowledgements
Thanks to Victor McKusick at Johns Hopkins University and Peter Stenson at the Human Genome Mutation Database for their help in searching for similar cases of mutation. This study was funded by NIH Grant 5RO1-09813-03 and the Roy J. Carver Charitable Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.JR Howe. Juvenile Polyposis Syndrome. In: Scriver CR, Beaudet AL, Sly WS, et al., editors. The Metabolic and Molecular Bases of Inherited Disease. 8th Edition, Eighth ed. Vol. 1. McGraw-Hill; New York: 2001. pp. 805–818. [Google Scholar]
- 2.Jarvinen HJ, Franssila KO. Familial juvenile polyposis coli: Increased risk of colorectal cancer. Gut. 1984;25:792–800. doi: 10.1136/gut.25.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howe JR, Mitros FA, Summers RW. The risk of gastrointestinal carcinoma in familial juvenile polyposis. Ann Surg Oncol. 1998;5:751–756. doi: 10.1007/BF02303487. [DOI] [PubMed] [Google Scholar]
- 4.Gallione CJ, Repetto GM, Legius E, Ruastgi AK, Schelley SL, Dunlop M, Mitchell G, Drouin E, Westermann CJ, Marchuk DA. Mutations in SMAD4 cause a combined hereditary hemorrhagic telangiectasia-Juvenile Polyposis syndrome. Am J Hum Genet. 2003;73:A248. [Google Scholar]
- 5.Gallione CJ, Repetto GM, Legius E, Rustgi AK, Schelley SL, Tejpar S, Mitchell G, Drouin E, Westermann CJ, Marchuk DA. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4) Lancet. 2004;363(9412):852–9. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- 6.Howe JR, Ringold JC, Summers RW, Mitros FA, Nishimura DY, Stone EM. A gene for familial juvenile polyposis maps to chromosome 18q21.1. Am J Hum Genet. 1998;62:1129–1136. doi: 10.1086/301840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IPM, Houlston R, Bevan S, Mitros FA, Stone EM, Aaltonen LA. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 8.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nature Genetics. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 9.Howe JR, Sayed MG, Ahmed AF, Ringold J, Larsen-Haidle J, Merg A, Mitros FA, Vaccaro CA, Petersen GM, Giardiello FM, Tinley ST, Aaltonen LA, Lynch HT. The prevalence of MADH4 and BMPR1A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations. J Med Genet. 2004;41(7):484–91. doi: 10.1136/jmg.2004.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Belzen MJ, Hiel JA, Weemaes CM, Gabreels FJ, van Engelen BG, Smeets DF, van den Heuvel LP. A double missense mutation in the ATM gene of a Dutch family with ataxia telangiectasia. Hum Genet. 1998;102(2):187–91. doi: 10.1007/s004390050675. [DOI] [PubMed] [Google Scholar]
- 11.Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, Lannfelt L. A pathogenic mutation for probable Alzheimer's disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1(5):345–7. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 12.Hassan W, Basset P, Oudart JL, Goossens M, Rosa J. Properties of the double substituted hemoglobin C Ziguinchor alpha2A beta 2 6 Glu replaced by Val 58 Pro replaced by Arg. Hemoglobin. 1977;1(5):487–501. doi: 10.3109/03630267709027866. [DOI] [PubMed] [Google Scholar]
- 13.Longy M, Coulon V, Duboue B, David A, Larregue M, Eng C, Amati P, Kraimps JL, Bottani A, Lacombe D, Bonneau D. Mutations of PTEN in patients with Bannayan-Riley-Ruvalcaba phenotype. J Med Genet. 1998;35(11):886–9. doi: 10.1136/jmg.35.11.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawabe K, Goto K, Nishino I, Angelini C, Hayashi YK. Dysferlin mutation analysis in a group of Italian patients with limb-girdle muscular dystrophy and Miyoshi myopathy. Eur J Neurol. 2004;11(10):657–61. doi: 10.1111/j.1468-1331.2004.00755.x. [DOI] [PubMed] [Google Scholar]
- 15.De Rosa M, Scarano MI, Panariello L, Morelli G, Riegler G, Rossi GB, Tempesta A, Romano G, Renda A, Pettinato G, Izzo P. The mutation spectrum of the APC gene in FAP patients from southern Italy: detection of known and four novel mutations. Hum Mutat. 2003;21(6):655–6. doi: 10.1002/humu.9151. [DOI] [PubMed] [Google Scholar]
- 16.Smit LS, Nasr SZ, Iannuzzi MC, Collins FS. An African-American cystic fibrosis patient homozygous for a novel frameshift mutation associated with reduced CFTR mRNA levels. Hum Mutat. 1993;2(2):148–51. doi: 10.1002/humu.1380020217. [DOI] [PubMed] [Google Scholar]
- 17.Becker K, Csikos M, Sardy M, Szalai ZS, Horvath A, Karpati S. Identification of two novel nonsense mutations in the transglutaminase 1 gene in a Hungarian patient with congenital ichthyosiform erythroderma. Exp Dermatol. 2003;12(3):324–9. doi: 10.1034/j.1600-0625.2003.120313.x. [DOI] [PubMed] [Google Scholar]
- 18.Sayed MG, Ahmed AF, Ringold JC, M.E. A, Bair JL, Mitros FA, Lynch HT, Petersen GM, Giardiello FM, Vogelstein B, Howe JR. Germline SMAD4 or BMPR1A mutations and phenotype of Juvenile Polyposis. Ann Surg Oncol. 2002;9(9):901–906. doi: 10.1007/BF02557528. [DOI] [PubMed] [Google Scholar]
- 19.Friedl W, Uhlhaas S, Schulman K, Stolte M, Loff S, Back W, Mangold E, Stern M, Knaebel H-P, Sutter C, Weber R, Pistorius S, Burger B, Propping P. Juvenile Polyposis: massive gastric polyposis is more common in MADH4 mutation carriers than in BMPR1A mutation carriers. Hum Genet. 2002;111:108–111. doi: 10.1007/s00439-002-0748-9. [DOI] [PubMed] [Google Scholar]