Abstract
The collection of gene deletion mutants of Saccharomyces cerevisiae was used to screen for novel genes required for UV-induced mutagenesis. We found the SBF transcription factor (Swi4/Swi6 protein complex) to be required for wild-type levels of UV mutability in forward and reverse mutation assay. Expression of translesion polymerase ζ component Rev7 was identified as a target of SBF-dependent regulation.
Keywords: Yeast, UV Radiation, Mutagenesis, Translesion synthesis, Transcriptional regulation
Genetic instability resulting from enhanced mutagenesis is a severe consequence of exposing pro- and eukaryotic cells to DNA-damaging agents [1]. Especially for bulky adducts, such as UV-C induced pyrimidine dimers, point mutations opposite DNA lesions of reduced coding capacity mainly arise through active processes that ensure the completion of replication of a damaged template. In recent years, it became clear that high-fidelity replicative DNA polymerases are temporarily replaced at the lesion site by one of several translesion polymerases whose active sites can accommodate modified bases while lacking proofreading activity [2, 3]. A concerted action of one polymerase inserting a base opposite a lesion together with another polymerase extending from the imperfectly matched primer/template junction may be required [4]. The ensuing damage bypass may be error-free or error-prone, depending on type of lesion and polymerases involved. Most of the enhanced mutability following UV radiation is generated by such error-prone translesion synthesis. The majority of UV-induced and spontaneous mutations are dependent on polymerase ζ, a complex of Rev3 and Rev7 [5-7].
Our present understanding of the events leading to mutagenic translesion synthesis also assigns a major role to proliferating cell nuclear antigen (PCNA). PCNA is subject to ubiquitination by the Rad6/Rad18 complex [8]. Monoubiquitinated PCNA appears to facilitate the switch from replicative to bypass polymerase [9, 10] and thus, ubiquitin binding domains of bypass polymerases were found to be important for function [11-14]. Mutation of PCNA that prevents ubiquitination abolishes most of UV-induced mutability [15, 16].
In pro- and eukaryotic model organisms, the isolation of mutants that reduce DNA-damage induced mutability (anti-mutator mutants) was critical for defining the elements and mechanisms required for mutagenic bypass [17, 18]. Some of these mutants, such as rad6, were initially found among the collection of radiation-sensitive mutants. Additional screens were performed to specifically isolate mutants of the mutational process in budding yeast, irrespective of their radiation sensitivity [19-21]. The current list of genes required for UV mutagenesis in S. cerevisiae includes RAD6, RAD18, REV1, REV3 and REV7. Also, the inactivation of POL32 subunit of polymerase δ results in defective UV mutagenesis [22]. REV6 was recently shown to encode PCNA whose monoubiquitinated version appears to be important for the recruitment of translesion polymerases to sites of damage [15]. Additional proteins involved include the kinase Cdc7/Dbf4 [23, 24], thymidilate kinase Cdc8 [25], ribonucleotide reductase subunit Rnr4 [26], and defects were also found for genes required for checkpoint arrest [27].
The now commercially available collection of deletion mutants of non-essential yeast genes has previously been exploited to screen for mutants with enhanced spontaneous mutability and gross chromosomal rearrangements [28, 29]. Here, we used this collection to identify novel genes required for UV-induced mutagenesis that may have been missed previously. The identical approach has recently been reported in an independent study that lead to the isolation of many non-overlapping genes [30]. Here, we identify the SBF transcription factor as required for wild-type levels of UV mutability. We found the level of Rev7 to be one of the relevant targets of SBF-dependent regulation.
Material and methods
Yeast strains and plasmids
The wild type strains BY4741 (MATa his3Δ leu2Δ met15Δ ura3Δ) and Y300 (MATa ade2-1 ura3-1 trp1-1 his3-11,15 leu2-3,112 can1-100, originally from S. Ellledge) were used throughout the study. Various kanMX4-marked gene deletions were transferred into Y300 from existing deletion strains (purchased from OpenBiosystems), following propagation by PCR and transplacement by homologous recombination. Yeast transformation was performed as described [31]. Plasmid pELGH6yhRev7 (a gift from Dr. Zhigang Wang, University of Kentucky) was used for overexpression of Rev7 from a galactose-inducible promoter.
Screen for yeast deletion mutants with altered mutability
Clones of the systematic gene deletion collection (OpenBiosystems) were grown on YPD plates, transferred by replica-plating to synthetic medium plates containing 60 mg/l canavanine sulfate (US Biological) and exposed to UV-C radiation (254 nm, 40 J/m2) or 0.2 mg/l 4-nitroquinoline-N-oxide (4-NQO). Yeast media were as described elsewhere [32]. For the patches of each deletion clone, the number of arising canavanine-resistant mutants was compared between treated plates and untreated control plate. This assay was repeated for candidate deletion clones initially showing a low number of mutants on treated plates, or no mutants whatsoever.
Measurement of UV mutation frequencies
For determining UV-induced reversion frequencies, cells were grown to stationary phase on YPD plates. Appropriate numbers of cells were spread on synthetic tryptophane-free omission or canavanine-containing plates. Dilutions were plated on the identical medium supplemented with tryptophane or without canavanine, respectively, to determine survival of colony-forming cells. Cells were irradiated directly on solid media. In the case of Rev7 overexpression, cells were precultured and plated on media containing 1.5% galactose and 0.5% dextrose.
RNA detection
Total RNA was isolated using zirkonia beads and hot phenol, separated on a denaturing 1% agarose gel using formaldehyde/MOPS buffer and transferred to a nylon membrane as described [14]. Hybridization was performed in HybrisolR 1 (Serologicals Corp.) using DNA probes generated by PCR and labeled by random priming with HighPrime kit (Roche) and 32P-dCTP. Signal levels were quantified using ImageJ software. For RT-PCR, 50pg DNAse I-treated RNA samples served as templates for SuperScript™ one-step RT-PCR with Platinum Taq (Invitrogen), following manufacturer's instructions. Primer pairs used were TGTATACCCACCTCAGTCATTCGA and CACGTCAGAACCGACTAAAGA for REV7 and AGATATGGTCATCATCAGAAGA and TCTAGTTCTGTAGGTAGTACCG for PDA1, which served as a control. If UV treated-samples were to be analyzed, logarithmic-phase cells were exposed to 60 J/m2 UV-C in suspension under constant stirring in a petridish (15 ml, 2.5 × 107 cells/ml), resuspended in YPD and incubated at 30°C for various periods.
Protein detection
Chromosomal REV7 was tagged in strain BY4741 at the C-terminus with 3 copies of HA peptide by using microhomology-mediated recombination of a PCR product derived from plasmid-borne modules [33]. Protein samples were prepared using TCA and zirkonia beads [34]. Western blots were developed with a commercial HA antibody (Covance, 1:1,000 dilution), or a 3-phosphoglycerate kinase (Pgk1) antibody (Molecular Probes, 1:5,000) to detect the loading control.
Cell synchronization
Cells were arrested in G1 using α-factor (US Biological) as described [14]. After release, budding analysis revealed synchronous transversal of the cell cycle, with S-phase entry after about 40 min.
Results and discussion
Screen for deletion mutants with reduced UV mutability
In order to identify deletion mutants that show altered induced mutability, clones of the yeast gene deletion collection in the haploid strain BY4741 were surveyed as follows. Clones were expanded on YPD as an ordered array, replica-plated onto canavanine containing plates and UV-irradiated (Fig. 1A). Additional replica were placed on 4-NQO-containing plates and on untreated control plates. Low probability to form canavanine resistant mutants on UV-irradiated and 4-NQO-containing plates was semi-quantitatively confirmed for 108 deletion mutants (Supplemental Table 1). Only 2 deletion mutants showed this phenotype for 4-NQO alone.
Fig. 1.
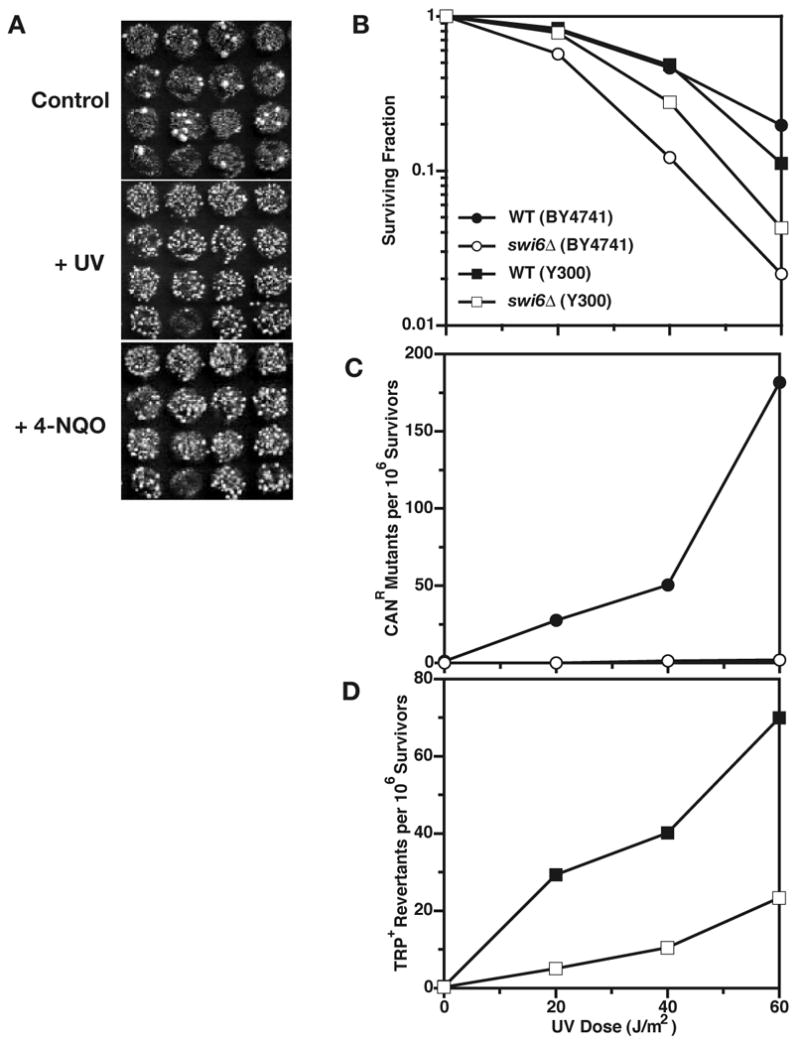
Identification and characterization of yeast deletion mutants with altered mutability. (A) Replica of clones of the systematic gene deletion collection were transferred to plates containing canavanine and exposed to UV-C radiation or 4-NQO. Compared to the untreated control, many canavanine-resistant mutants emerged in most replica. Those with an unexpectedly low number represent candidate genes that may be required for induced mutagenesis. (B) Survival of colony-forming cells, (C) frequency of canavanine resistant mutants and (D) of revertants of trp1-1 among surviving cells following treatment of stationary-phase cells with UV. Responses of wild-type (WT) (filled symbols) and swi6Δ∷KanMX4 cells (open symbols) were compared in two different genetic backgrounds, BY4741 (◯●) and Y300 (□■). Representative single-experiment data are shown.
Certain genes were eliminated because of their known role in established DNA repair pathways and, most likely, induced mutants were undetectable in our screening because of their high UV sensitivity. Additionally, many of the identified genes had no obvious direct connection with the known molecular mechanisms of UV mutagenesis and were more likely to affect the efficiency of the forward mutation-based screening system. We reasoned that many mutations that enhance toxicity towards canavanine could limit the yield of resistant mutants since cells may loose viability on the selection plate before a mutationally inactivated arginine permease takes effect. Increased uptake of canavanine or defective detoxification may be underlying causes. For example, a number of VPS gene deletions (for vacular protein sorting) were isolated (Supplemental Table 1). In an older study, 7 genes required for normal frequency of UV-induced canavanine resistance mutants had been isolated by conventional mutant screening (umr1-umr7) [21]. Enhanced canavanine toxicity was detectable in some but not all umr mutants.
After evaluating canavanine sensitivity in a gradient assay and quantitatively characterizing UV-induced forward mutation frequencies (data not shown), the number of potential deletion mutants reducing UV mutagenesis was reduced to 32 (Supplemental Table 1, underlined gene names). Obviously, this screening system was subject to substantial inherent variability, e.g. the amount of cells treated was initially not controlled. Also, a high spontaneous background may have mitigated the detection of UV-induced mutants. We successfully re-isolated deletions of REV3, REV7 and RAD6, known to be critically involved in induced mutagenesis, however, we failed to isolate deletions of other known genes with similar role, such as REV1 or RAD18. We conclude that the applied screen was appropriate but non-exhaustive.
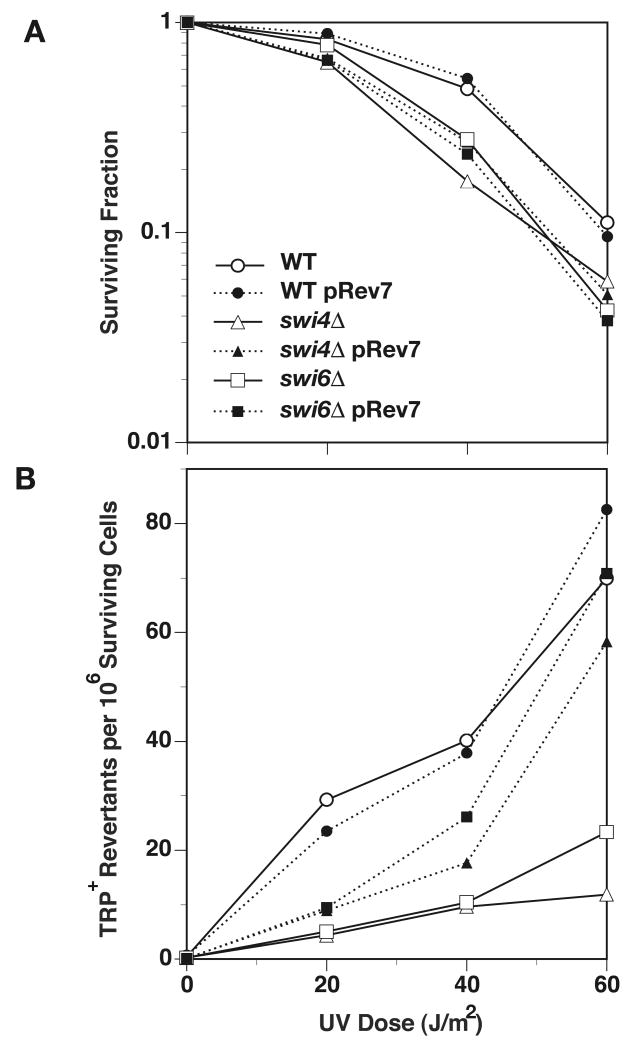
To demonstrate that the mutation phenotype was independent of strain and mutational system used, we transferred selected gene deletions into an unrelated genetic background (Y300) and measured UV mutability in the trp1-1 reversion system. According to molecular processes affected by these genes, we chose the following 9 genes out of 32 as most likely to be involved in the general process of mutagenesis: HNT3, RSC1, ARP5, INO2, SDS3, LDB19, LDB7, SWI6, DOA4. Of these, only two genes were found to be reproducibly required for normal levels of UV mutagenesis both in forward and reverse mutation systems, irrespective of genetic background: SWI6 (Fig. 1B-D) and DOA4 (data not shown). The remaining candidate genes await further characterization and it is possible that the use of the more specific reversion assay has eliminated genes that do not have an effect on all types of mutational events.
SBF transcription factor is required for normal UV mutagenesis by stimulating REV7 expression
Here, we present a more detailed characterization of the influence of the Swi6 transcription factor component on induced mutagenesis. (A study of the ubiquitin-recycling protein Doa4 will be presented elsewhere.) Swi6 forms a complex with the DNA-binding protein Swi4 termed SBF (for Swi4-Swi6 cell cycle box binding factor) or, independently, with Mbp1 forming MSB (for MluI binding factor) [35-39]. Both transcription factors are primarily involved in the regulation of genes preferentially expressed at the G1/S transition [40-44]. No evidence for S-phase specific expression of the respective transcripts have been reported for most genes essential for the mutagenic process, such as RAD6, RAD18, REV1, REV3, REV7, CDC7, or CDC8, making them unlikely targets of SBF or MSB regulation. However, when transcript levels of REV1, REV3, REV7, and RAD6 were compared to wild type, we unexpectedly found a reduced level of REV7 but not of other transcripts in swi6 deletion mutants; the same was true for swi4 but not mbp1 deletion strains (Fig. 2A, quantified in B, and data not shown). The results of Northern blot analysis were verified by RT-PCR (Fig. 2C, quantified in 2E). At the same time, we showed that REV7 transcript has no detectable UV inducibility (Fig. 2D,E). We conclude that SBF transcription factor is required for constitutive REV7 expression.
Fig. 2.
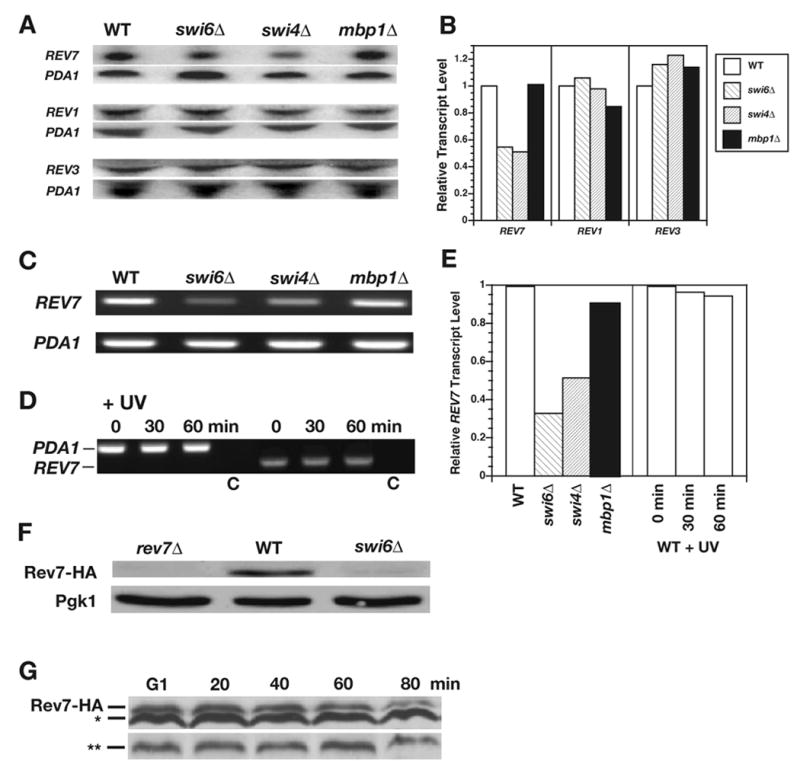
Level of REV7 transcript and protein in wild-type, swi6, swi4 and mbp1 deletion mutants. (A) Transcript levels of REV7 and PDA1 (control) as determined by Northern blotting. (B) Quantified REV7 transcript levels of A, normalized for PDA1 loading controls. (C) Transcript levels of REV7 and PDA1 determined by RT-PCR. (D) Transcript levels of REV7 after UV treatment. Control lanes (marked C) correspond to mock reactions that included all components except reverse transcriptase. (E) Quantified REV7 transcript levels of C, D, normalized for PDA1. (F) Analysis of Rev7-HA protein levels in wild type and swi6Δ. An isogenic rev7Δ mutant was included to verify band identity. Pgk1 was used as loading control. (G) Absence of cell-cycle stage-dependent regulation of Rev7. Cells were released from α-factor arrest in G1. Unspecific bands (*, **) can serve as loading controls.
In order to confirm this result at the level of protein expression, chromosomal REV7 was C-terminally tagged with 3xHA [33]. As predicted by previous RNA analysis, deletion of SWI6 resulted in reduced Rev7-HA levels that were frequently difficult to detect at all (Fig. 2F). Additionally, in synchronized cells released from α-factor arrest in G1, we explored possible cell-cycle stage-dependent expression levels. The protein levels of Rev7 were found to be relatively constant throughout the cell cycle and no evidence for higher levels in G1/S were obtained (Fig. 2G).
In order to prove that reduced Rev7 levels are responsible for the reduced UV-mutability of a swi6 mutant, we overexpressed plasmid-borne REV7 from a galactose-inducible promoter. Since RNA analysis suggested that the SBF complex, consisting of Swi4 and Swi6, controls REV7 expression, we also analyzed UV survival and mutation frequencies in swi4 with and without overexpression of REV7 (Fig. 3A,B). REV7 overexpression did not change the mutability of the wild type and transformation with the empty vector plasmid had no detectable effect in any strain (data not shown). In contrast, REV7 overexpression alleviated the defects in UV reverse mutagenesis that were evident in both swi4 and swi6 mutants (Fig. 3B). Even when Rev7 was overexpressed, the strains did not quite reach wild-type mutation frequencies. One has to assume that additional mechanisms are responsible for the reduced mutability of swi4 or swi6 mutants that are independent of Rev7 levels. Also, the somewhat enhanced UV sensitivity of the mutant strains is not corrected (Fig. 3A).
Fig. 3.
(A) Survival of colony-forming cells and (B) frequency of trp1-1 revertants among surviving cells following treatment of stationary-phase WT (Y300, ◯●), swi6Δ (Δ) and swi4Δ cells (□■) with UV. Cells were transformed with Rev7 overexpression plasmid and induced with galactose (closed symbols). These data were compared to untransformed cells (open symbols).
In summary, we have identified the SBF transcription factor as required for UV mutagenesis. The underlying mechanism involves regulation of expression of REV7, required for processivity of polymerase ζ. This is a somewhat unexpected finding since SBF regulates predominantly, though not exclusively, genes with increased expression at the G1/S boundary but there is no evidence for significant fluctuations of REV7 transcript or protein during cell cycle progression. However, cell-cycle dependent regulation may still be revealed under suboptimal growth conditions or during meiosis. It is interesting to note that the mammalian Rev7 homolog appears to have additional roles that are cell-cycle stage-specific [45].
Other yeast genes involved in mutagenesis express transcripts that fluctuate during cell cycle, e.g. POL30, POL32, DBF4, but they lack an obvious SBF binding site in their promoters. This sequence is found in the promoter of the UV-inducible RNR4 gene whose inactivation leads to depressed UV mutation frequencies [26, 30]. However, no difference in Rnr4-GFP-fusion protein levels was noted between wild-type and SWI6-deleted cells (data not shown).
As indicated by the absence of SBF binding sequence in its promoter region, REV7 does not appear to be a direct target of SBF [41]. A number of transcription factors are themselves regulated by SBF but REV7 promoter has not yet been identified as one of their targets [42]. It remains to be seen if on yet another level of regulation a transcription factor cascade regulated by SBF can provide the direct link to REV7.
For the first time, this study has identified a transcription factor as critical for DNA-damage induced mutagenesis in eukaryotes. If conserved, these novel aspects of regulation of error-prone bypass may open up important avenues for advancing human health by modifying the efficiency of translesion synthesis.
Supplementary Material
Acknowledgments
We thank Nimrat Kaur and Aniruth Sethi for experimental help. These studies were supported by National Institutes of Health grant ES011163.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. second. American Society of Microbiology; Washington D.C.: 2005. [Google Scholar]
- 2.Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nature Rev Mol Cell Biol. 2005;6:943–953. doi: 10.1038/nrm1781. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence CW. Mutagenesis in Saccharomyces cerevisiae. Adv Genet. 1982;21:173–254. doi: 10.1016/s0065-2660(08)60299-0. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence CW. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair. 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 7.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 8.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 9.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 11.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 12.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol Cell Biol. 2006;26:8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of polymerase η in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–889. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pabla R, Rozario D, Siede W. Regulation of Saccharomyces cerevisiae DNA polymerase η transcript and protein. Radiat Environ Biophys. 2008;47:157–168. doi: 10.1007/s00411-007-0132-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Gibbs PE, Lawrence CW. The Saccharomyces cerevisiae rev6-1 mutation, which inhibits both the lesion bypass and the recombination mode of DNA damage tolerance, is an allele of POL30, encoding proliferating cell nuclear antigen. Genetics. 2006;173:1983–1989. doi: 10.1534/genetics.106.058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence CW. Mechanisms of induced mutagenesis in yeast. In: Sugimura T, Kondo S, Takebe H, editors. Environmental Mutagens and Carcinogens. Univ. Tokyo/Alan R Liss; Tokyo/New York: 1982. pp. 128–136. [Google Scholar]
- 18.Lawrence CW. Historical reflections. Following the RAD6 pathway. DNA Repair. 2007;6:676–686. doi: 10.1016/j.dnarep.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence CW, Krauss BR, Christensen RB. New mutations affecting induced mutagenesis in yeast. Mutat Res. 1985;150:211–216. doi: 10.1016/0027-5107(85)90117-4. [DOI] [PubMed] [Google Scholar]
- 20.Lemontt JF. Mutants of yeast defective in mutation induction by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemontt JF. Pathways of ultraviolet mutability in Saccharomyces cerevisiae. III. Genetic analysis and properties of mutants resistant to ultraviolet-induced forward mutation. Mutat Res. 1977;43:179–204. doi: 10.1016/0027-5107(77)90003-3. [DOI] [PubMed] [Google Scholar]
- 22.Huang ME, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic repair bypass pathway. Curr Genet. 2000;38:78–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 23.Njagi GDE, Kilbey BJ. Mutagenesis in cdc7 strains of yeast. The fate of premutational lesions induced by ultraviolet light. Mutat Res. 1982;105:313–318. doi: 10.1016/0165-7992(82)90099-9. [DOI] [PubMed] [Google Scholar]
- 24.Pessoa-Brandão L, Sclafani R. CDC7/DBF4 functions in the translesion synthesis branch of the RAD6 epistasis group in Saccharomyces cerevisiae. Genetics. 2004;167:1597–1610. doi: 10.1534/genetics.103.021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prakash L, Hinkle D, Prakash S. Decreased UV mutagenesis in cdc8, a DNA replication mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1979;172:249–258. doi: 10.1007/BF00271724. [DOI] [PubMed] [Google Scholar]
- 26.Strauss M, Grey M, Henriques JA, Brendel M. RNR4 mutant alleles pso3-1 and rnr4Δ block induced mutation in Saccharomyces cerevisiae. Curr Genet. 2007;51:221–231. doi: 10.1007/s00294-007-0120-7. [DOI] [PubMed] [Google Scholar]
- 27.Paulovich AG, Armour CD, Hartwell LH. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics. 1998;150:75–93. doi: 10.1093/genetics/150.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myung K, Chen C, Kolodner R. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 30.Lis ET, O'Neill BM, Gil-Lamaignere C, Chin JK, Romesberg FE. Identification of pathways controlling DNA damage induced mutation in Saccharomyces cerevisiae. DNA Repair. 2008;7:801–810. doi: 10.1016/j.dnarep.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gietz RD, Schiestl RH. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 32.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor: 2005. [Google Scholar]
- 33.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 34.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol. 1994;14:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Primig M, Sockanathan S, Auer H, Nasmyth K. Anatomy of a transcription factor important for the start of the cell cycle in Saccharomyces cerevisiae. Nature. 1992;358:593–597. doi: 10.1038/358593a0. [DOI] [PubMed] [Google Scholar]
- 36.Moll T, Dirick L, Auer H, Bonkovsky J, Nasmyth K. SWI6 is a regulatory subunit of two different cell cycle START-dependent transcription factors in Saccharomyces cerevisiae. J Cell Sci Suppl. 1992;16:87–96. doi: 10.1242/jcs.1992.supplement_16.11. [DOI] [PubMed] [Google Scholar]
- 37.Andrews BJ, Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342:830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- 38.Andrews BJ, Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989;57:21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 39.Sidorova J, Breeden L. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1069–1077. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 41.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 42.Horak CE, Luscombe NM, Qian J, Bertone P, Piccirrillo S, Gerstein M, Snyder M. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002;16:3017–3033. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dirick L, Moll T, Auer H, Nasmyth K. A central role for SWI6 in modulating cell cycle start-specific transcription in yeast. Nature. 1992;357:508–512. doi: 10.1038/357508a0. [DOI] [PubMed] [Google Scholar]
- 44.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Shi Y, Wu H, Wan B, Li P, Zhou L, Shi H, Huo K. Hepatocellular carcinoma-associated gene 2 interacts with MAD2L2. Mol Cell Biochem. 2007;304:297–304. doi: 10.1007/s11010-007-9512-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.