Summary
Background
Cytoplasmic dynein mediates spindle positioning in budding yeast by powering sliding of microtubules along the cell cortex. Although previous studies have demonstrated cortical and plus-end targeting of dynein heavy chain (Dyn1/HC), the regulation of its recruitment to these sites remains elusive.
Results
Here we show that separate domains of Dyn1/HC confer differential localization to the dynein complex. The N-terminal tail domain targets Dyn1/HC to cortical Num1 receptor sites, whereas the C-terminal motor domain targets Dyn1/HC to microtubule plus-ends in a Bik1/CLIP-170 and Pac1/LIS1 dependent manner. Surprisingly, the isolated motor domain blocks plus-end targeting of Dyn1/HC, leading to a dominant negative effect on dynein function. Overexpression of Pac1/LIS1, but not Bik1/CLIP-170, rescues the dominant negativity by restoring Dyn1/HC to plus-ends. In contrast, the isolated tail domain has no inhibitory effect on Dyn1/HC targeting and function. However, cortical targeting of the tail construct is more robust than full-length Dyn1/HC, and occurs independently of Bik1/CLIP-170 or Pac1/LIS1.
Conclusions
Our results suggest that the cortical association domain is normally masked in the full-length dynein molecule. We propose that targeting of dynein to plus-ends unmasks the tail, priming the motor for off-loading to cortical Num1 sites.
Introduction
Cytoplasmic dynein is a minus-end-directed microtubule motor that participates in a variety of motile activities during mitosis, including centrosome separation [1], nuclear envelope breakdown [2], chromosome capture and congression [3], mitotic spindle assembly [4], spindle pole organization [5], spindle alignment [6, 7], and spindle checkpoint inactivation [8, 9]. The heavy chain of dynein consists of a ring of six AAA (ATPases associated with cellular activities) units [10, 11], which form the motor domain that powers movement along microtubules, and the tail domain, which binds accessory chains and adaptor molecules that attach the motor to diverse cellular cargoes [12-15]. To perform all of its tasks during mitosis, the cargo-binding tail domain must specify targeting of dynein to different sub-cellular sites in a spatially and temporally defined manner. Although various studies [2, 6-9, 16-20] have implicated the tail domain as playing a pivotal role, the mechanism by which this domain regulates dynein localization is not known. Additionally, whether the motor domain has any role in dynein targeting is not known.
In this study, we examined the roles of the tail and motor domains in dynein localization and function in the budding yeast S. cerevisiae. We found that these two domains play distinct but complementary roles in mediating the spatial targeting of dynein. We identified a novel role for the motor domain in dynein targeting, and revealed a new regulatory step mediated by the tail domain in the off-loading model for dynein function in spindle positioning.
Results
The Tail Domain Targets Dynein to Cortical Num1 Sites
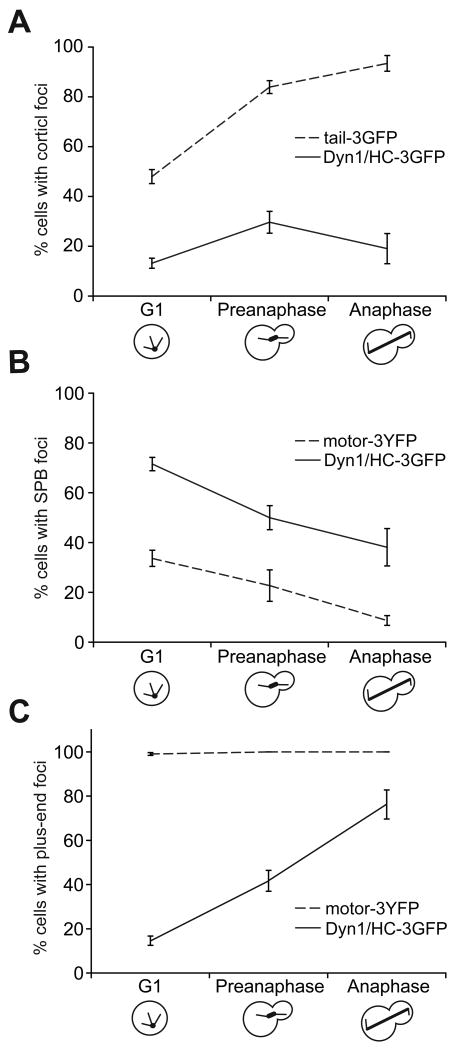
Dyn1/HC localizes as foci at the spindle pole body (SPB), microtubule plus-ends, and the cell cortex [17, 21-24]. We set out to test the hypothesis that the tail domain of Dyn1/HC is responsible for its targeting. We altered the DYN1 locus to express only the tail domain (amino acids 1-1363) fused with 3GFP under its native promoter. Haploid cells carrying TAIL-3GFP as their sole source of dynein exhibited cortical foci at all cell cycle stages (Figure 1B). Notably, these cells contained no SPB or plus-end associated foci. Cortical tail-3GFP foci were stationary (Movie S1) and were found at the mother and daughter cortex (Figure 1B), as previously described for Dyn1/HC-3GFP cortical foci [17]. In some cases (<10% of cells), we observed cytoplasmic tail-3GFP foci, which were not associated with microtubules and may represent tail-3GFP aggregates. TAIL-3GFP and DYN1/HC-3GFP strains exhibited similar levels of increase in the percentage of cortical-foci-containing cells as they entered pre-anaphase (Figure 2A; 2.2-fold for DYN1/HC-3GFP and 1.7-fold for TAIL-3GFP), suggesting that the temporal regulation of cortical association for tail-3GFP is similar to Dyn1/HC-3GFP. However, the likelihood of finding a cortical focus was significantly higher in the TAIL-3GFP strain than the DYN1/HC-3GFP strain. While cortical Dyn1/HC-3GFP foci were observed in 17.9% ± 1.9% (n = 431) of cells, cortical tail-3GFP foci were observed in 65.1% ± 2.0% (n = 581) of cells. Additionally, full 3-D stacks of confocal sections showed that there are 7.8 ± 3.3 (n = 87) cortical tail-3GFP foci per cell (Movie S2 and S3), compared to 3.8 cortical foci per cell for Dyn1/HC-3GFP [17]. This large discrepancy in cortical targeting between tail-3GFP and Dyn1/HC-3GFP could not be attributed to an affinity of Dyn1/HC-3GFP for microtubules (Figure S1F). Instead, the discrepancy indicates that the isolated tail domain is lacking a regulatory component for cortical association compared to Dyn1/HC. It suggests a role for the motor domain in masking the tail domain from cortical association in the context of the full-length molecule.
Figure 1. Isolated dynein tail and motor domains localize to distinct subcellular sites.
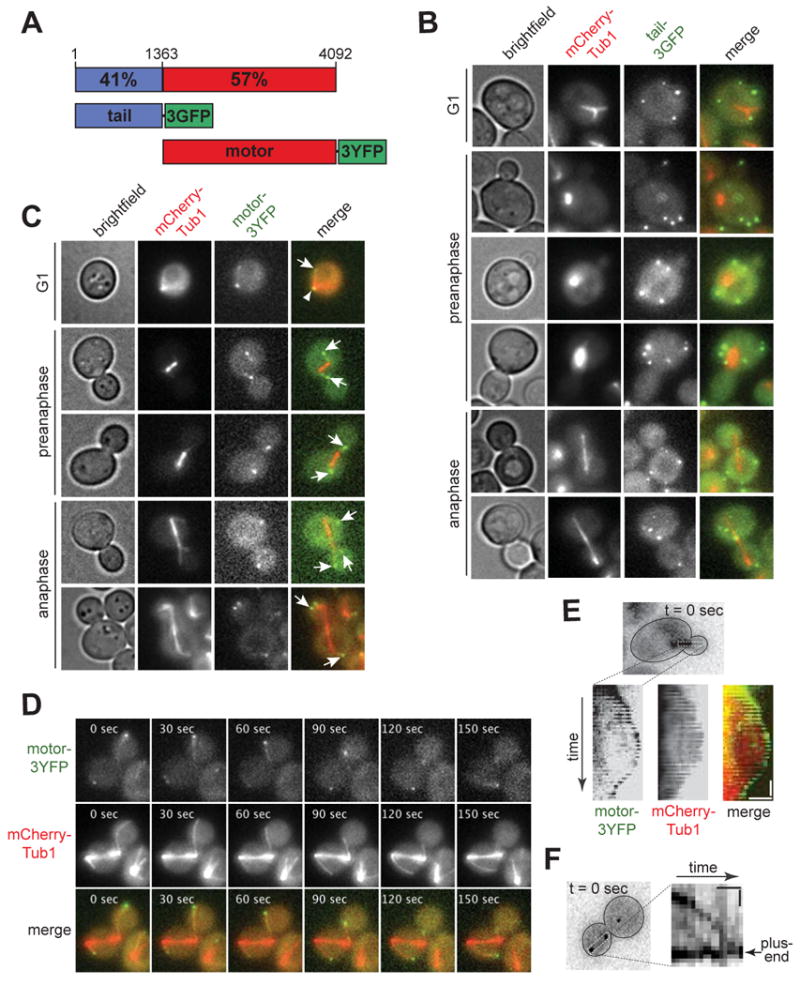
(A) Schematic representation of Dyn1/HC constructs used in this study. Percent similarity to corresponding rat DYNC1H1 domain is indicated (see Figure S1A). Numbering indicates amino acid residues. (B) Tail-3GFP localizes as stationary foci at the cell cortex throughout the cell cycle (see Movie S1). (C) Motor-3YFP localizes to SPB and microtubule plus-ends throughout the cell cycle. Arrows indicate plus-end foci; arrowhead indicates SPB focus. Brightfield and maximum intensity projection of wide-field fluorescence images of cells expressing either (B) tail-3GFP or (C) motor-3YFP under the control of the endogenous DYN1 promoter, and mCherry-Tub1 under the control of the MET3 promoter. Merged image on the right depicts tail-3GFP or motor-3YFP in green and mCherry-Tub1 in red. (D) Representative movie frames of motor-3YFP and mCherry-Tub1 depicting motor-3YFP at shrinking and growing microtubules in the same cell. Each image is a maximum intensity projection of a 2 μm Z-stack of wide-field images. The time elapsed in seconds is indicated (see Movie S4). (E) Kymograph depicting motor-3YFP tip-tracking on a polymerizing and depolymerizing cytoplasmic microtubule (see Movie S5). Vertical bar represents 20 s; horizontal bar represents 2 μm. (F) Kymograph depicting directional movement of motor-3YFP along a cytoplasmic microtubule towards the plus-end (see Movie S6). Vertical bar represents 10 s; horizontal bar represents 1 μm. Plus-end associated motor-3YFP was used as a reference point to crop images, allowing for the plus-end to appear stationary with respect to the focus migrating towards the plus-end.
Figure 2. Cell cycle dependent targeting of Dyn1/HC, tail-3GFP, and motor-3YFP.
The percentage of cells in strains carrying DYN1/HC-3GFP (n = 431 cells), TAIL-3GFP (n = 581 cells) or MOTOR-3YFP (n = 301 cells) that display fluorescent foci at (A) the cell cortex, (B) the SPB or (C) the plus-end is plotted for the different cell cycle stages. Each strain also expressed fluorescently labeled microtubules. A 2 μm Z-stack of images was collected at 5 s intervals for Dyn1/HC-3GFP, tail-3GFP, or motor-3YFP with mCherry-Tub1. Stationary cortical foci, SPB foci and plus-end foci were identified in two-color movies and scored accordingly. Cells in G1, pre-anaphase and anaphase were identified by brightfield and spindle morphology. Error bars represent standard error of proportion.
We previously proposed a dynein off-loading model in which Dyn1/HC is delivered by microtubule plus-ends to the cortex, where Dyn1/HC becomes anchored at cortical Num1 sites in order to generate forces for pulling the spindle [22, 24]. We asked whether tail-3GFP localizes to Num1 sites, as would be expected if the tail domain is responsible for anchoring dynein to Num1. We observed that the majority of tail-3GFP foci colocalized with a cortical Num1-mCherry focus (Figure 3A; 80.3% ± 3.4%, n = 137). Furthermore, we detected fluorescence resonance energy transfer (FRET) from tail-3GFP to Num1-mCherry (Figure 3B, left). The mean calculated FRETR value was 2.4 ± 0.6 (n = 13) (Figure 3B, right), indicating specific FRET emission [25]. Therefore, our results showed that tail-3GFP and Num1-mCherry likely interact at the cell cortex.
Figure 3. Tail-3GFP associates with Num1-mCherry and dynactin at the cell cortex.
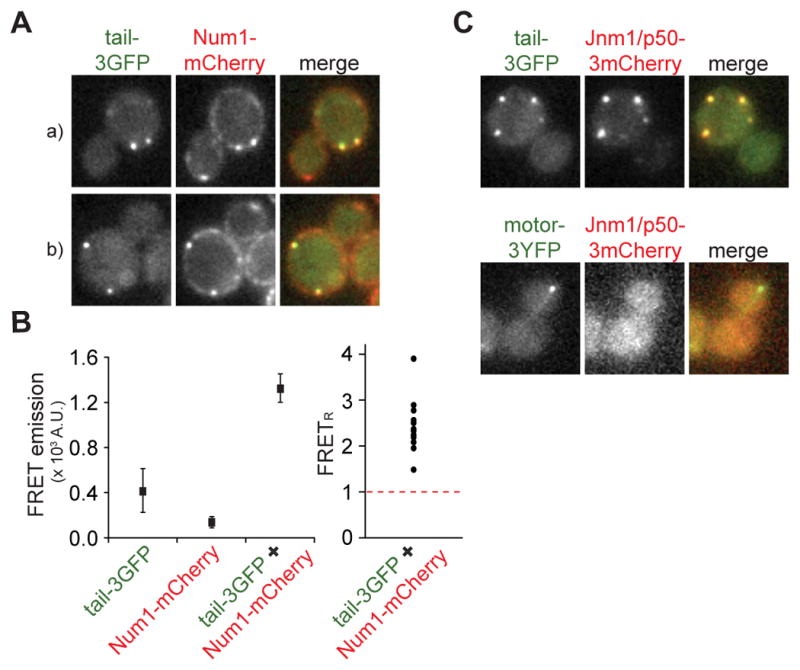
(A) Cells expressing tail-3GFP and Num1-mCherry (two examples are shown, indicated by a and b). Each image is a maximum intensity projection of a 3 μm Z-stack of wide-field images. Merged image on the right depicts colocalization of the two tagged proteins, tail-3GFP in green and Num1-mCherry in red. (B) Left: Mean fluorescence intensity in the FRET channel (excitation λ = 488 nm; emission λ > 560 nm) was plotted for individual foci in strains expressing tail-3GFP only, Num1-mCherry only, or both. Right: Relative FRET values (FRETR) for foci measured in TAIL-3GFP NUM1-mCherry strain (n = 13), calculated by dividing the background-corrected FRET channel fluorescence by the total spillover fluorescence, as described [25]. Tail-3GFP only and Num1-mCherry only strains were imaged to determine the spillover fluorescence into the FRET channel (see Experimental Procedures). FRETR values above 1, indicated by dashed red line, represent significant energy transfer above background. (C) Cells expressing dynactin subunit Jnm1/p50-3mCherry and tail-3GFP (top row) or motor-3YFP (bottom row). Each image is a maximum intensity projection of a 2 μm Z-stack of wide-field images. Merged images on the right depict Jnm1/p50-3mCherry in red, and tail-3GFP or motor-3YFP in green.
We asked whether tail-3GFP colocalizes with Jnm1/p50, the yeast homolog of the dynactin subunit dynamitin, as would be expected if dynactin interacts with the dynein complex via association with the tail domain [26, 27]. We observed that the majority of tail-3GFP foci colocalized with Jnm1/p50-3mCherry foci (Figure 3C, top panel; 76.5% ± 3.0%, n = 204), indicating that the isolated tail domain assembles into a higher order complex at the cell cortex. Together, these results demonstrate that tail-3GFP exhibits properties similar to Dyn1/HC-3GFP at the cell cortex.
Cortical Targeting of Tail-3GFP is Independent of Plus-end Targeting
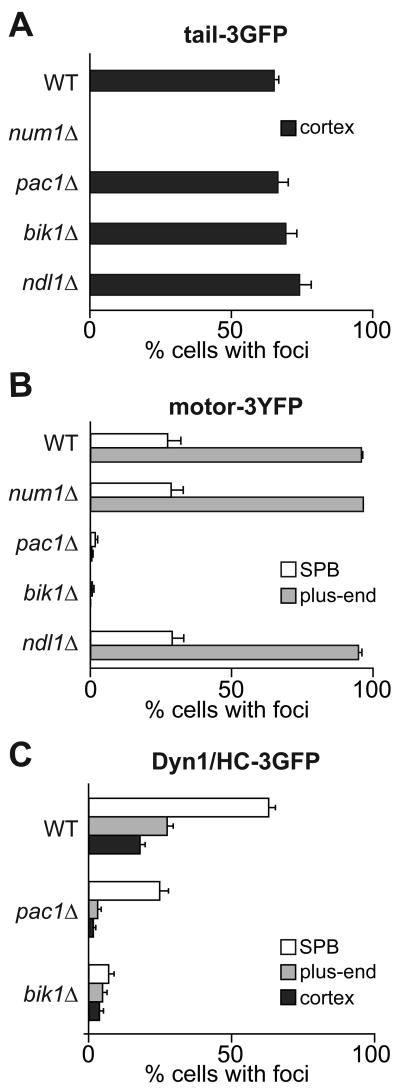
Next, we asked whether cortical tail-3GFP localization depends on components required for plus-end targeting of dynein, as would be expected if tail-3GFP is delivered to the cortex via the off-loading mechanism. The frequency of finding cortical tail-3GFP foci was unaffected in mutants lacking Pac1/LIS1, Bik1/CLIP-170, or Ndl1/NudEL (Figure 4A and S1B), three components required for normal plus-end targeting of Dyn1/HC (Figure 4C) [22, 24, 28, 29]. Additionally, cortical tail-3GFP foci were absent in a num1Δ mutant (Figure 4A and S1B), as previously reported for cortical Dyn1/HC-3GFP foci [17]. Immunoblot analysis revealed that tail-3GFP protein level was unaffected in num1Δ (Figure S1D). Thus, tail-3GFP is recruited to cortical Num1 directly from the cytoplasm, rather than being delivered by the microtubule plus-end.
Figure 4. Localization of tail-3GFP, motor-3YFP and Dyn1/HC-3GFP in null mutants.
The percentage of cells that display (A) tail-3GFP, (B) motor-3YFP, or (C) Dyn1/HC-3GFP foci at a given subcellular site is plotted for different null mutants (n > 100 cells counted for each strain). To identify foci at the cortex, SPB, and plus-end, a 2 μm Z-stack of images was collected at 5 s intervals for each null strain expressing mCherry-Tub1 or CFP-Tub1 and either tail-3GFP, motor-3YFP, or Dyn1/HC-3GFP. Movies were analyzed for foci at indicated locations. Error bars represent standard error of proportion.
Furthermore, the frequency of finding cortical tail-3GFP foci was reduced in mutants lacking dynein accessory chains (Dyn3/LIC or Pac11/IC) or dynactin components (Arp1 or Nip100/p150) (Figure S2A). In dyn3Δ cells, tail-3GFP protein level was reduced, indicating a defect in protein stability (Figure S2C). However, the levels in pac11Δ, arp1Δ and nip100Δ were similar to wild-type (Figure S2C; data not shown). Thus, assembly of tail-3GFP with other dynein and dynactin components may be required for stable association with cortical Num1, a result consistent with that of cortical Dyn1/HC-3GFP [17, 23].
In contrast to tail-3GFP, Dyn1/HC-3GFP depends on plus-end targeting components for localization to the cortex. In pac1Δ or bik1Δ mutants, in which plus-end targeting of Dyn1/HC-3GFP was reduced 8.9- and 5.7-fold (Figure 4C), respectively, we observed an 11.6- and 4.8-fold decrease in the frequency of finding cortical Dyn1/HC-3GFP foci, respectively, compared to wild-type cells (Figure 4C). Thus, cortical targeting of Dyn1/HC depends on plus-end targeting. Unlike tail-3GFP, direct recruitment of Dyn1/HC from the cytoplasm to Num1 does not occur. Our data suggest that the region in the tail domain responsible for interacting with Num1 is masked when Dyn1/HC cannot associate with microtubule plus-ends.
The Motor Domain Targets Dynein to Spindle Pole Bodies and Plus-Ends
The lack of SPB and plus-end localizations for tail-3GFP is in striking contrast to Dyn1/HC-3GFP. To test whether the motor domain is responsible for targeting dynein to these sites, we altered the DYN1 locus to express only the motor domain (amino acids 1364-4092) [30] fused with 3YFP under its native promoter. In haploid cells carrying MOTOR-3YFP as their sole source of dynein, motor-3YFP localized to the SPB and plus-ends (Figure 1C) and was notably absent from the cortex. Motor-3YFP plus-end foci were dynamic (Figure 1D; Movie S4), associating with both growing and shrinking microtubules (Figure 1E; Movie S5). Additionally, motor-3YFP was often detected along the microtubule as speckles, which could be seen moving toward the plus-end (Figure 1F; Movie S6), a behavior that was similarly observed for Dyn1/HC (see below; data not shown) [31]. These localization patterns demonstrate a novel role for the motor domain in SPB and plus-end targeting of dynein.
We compared the SPB and plus-end localizations of Dyn1/HC-3GFP and motor-3YFP. The frequency of finding Dyn1/HC-3GFP at the SPB decreased 1.9-fold from G1 to anaphase (Figure 2B). Motor-3YFP exhibited a similar pattern of decrease at the SPB (Figure 2B), although its frequency of targeting was always 2 to 4-fold lower than Dyn1/HC-3GFP. The frequency of finding Dyn1/HC-3GFP at the plus-end was cell cycle dependent, increasing 5.2-fold from G1 to anaphase (Figure 2C). Dyn1/HC-3GFP was largely absent from plus-ends until pre-anaphase. In contrast, motor-3YFP was found at >99 % (n = 301) of plus-ends irrespective of cell cycle stage (Figure 2C). Thus, the tail domain, which is lacking from motor-3YFP, might play a role in regulating the cell cycle-dependent accumulation of dynein at the plus-end. As expected [23], plus-end foci of motor-3YFP lacked dynactin complex, labeled with Jnm1/p50-3mCherry (Figure 3C, bottom panel). Thus, the tail domain of Dyn1/HC also recruits dynactin to the plus-end.
Motor-3YFP Requires Pac1/LIS1 and Bik1/CLIP-170 for Accumulation at Plus-Ends
We asked whether motor-3YFP targeting depends on its motor activity. Point mutations in the Walker A and Walker B motifs of the AAA3 unit, K2424A and E2488Q, respectively, block Dyn1/HC release from microtubules and inhibit dynein-mediated nuclear segregation [32, 33]. Neither mutation had any effect on motor-3YFP targeting (Figure S2D and S2E). Thus, motor activity is not required for plus-end targeting. Furthermore, the K2424A mutation did not affect full-length Dyn1/HC targeting to SPB, plus-ends and cortex (Figure S2F). Notably, plus-end foci containing Dyn1/HCK2424A-3YFP often remained stably attached to a cortical spot for the entire duration of the movie (Movies S7 and S8). The Dyn1/HCK2424A-3YFP found at the attachment sites may represent off-loaded and/or anchored dynein, which engaged with the plus-end but failed in force production.
We next asked whether motor-3YFP targeting depends on Pac1/LIS1, Bik1/CLIP-170, or Ndl1/NudEL. In pac1Δ and bik1Δ cells, the frequency of finding motor-3YFP foci at the SPB or plus-ends was drastically reduced compared to wild-type cells (Figure 4B and S1C). Immunoblot analysis of motor-3YFP revealed that protein levels were unchanged (Figure S1D). Thus, the dependence on Pac1/LIS1 and Bik1/CLIP-170 for plus-end targeting is similar to that of Dyn1/HC-3GFP (Figure 4C) [22, 24]. In contrast, in ndl1Δ cells, no change in the frequency (Figure 4B and S1C) or intensity (data not shown) of motor-3YFP targeting was observed. Thus, Ndl1/NudEL has no role in the plus-end localization of motor-3YFP, which is different from that of Dyn1/HC-3GFP [29].
Furthermore, the frequency of finding motor-3YFP at the SPB or plus-ends was unaffected in cells lacking dynein accessory components (Dyn3/LIC or Pac11/IC), dynactin components (Arp1 or Nip100/p150), Num1, or Kip2 (a kinesin that transports Bik1/CLIP-170 to the plus-end [28]) (Figure 4B, S1C and S2B). To examine the role of Kip2, we combined kip2Δ with kar3Δ, so that microtubules length was normal [28]. In kip2Δ kar3Δ cells, although the frequency of targeting was unchanged, the intensities of the SPB foci increased (Figure S1E; 133.1% of wild-type) while those of the plus-end foci decreased (73.2% of wild-type). Additionally, we did not find speckles of motor-3YFP traveling along the length of microtubules in kip2Δ kar3Δ cells (Movie S9). Thus, taken together, the Kip2-dependent transport of speckles contributes to normal plus-end targeting of motor-3YFP.
Quantification of Dynein at Plus-ends
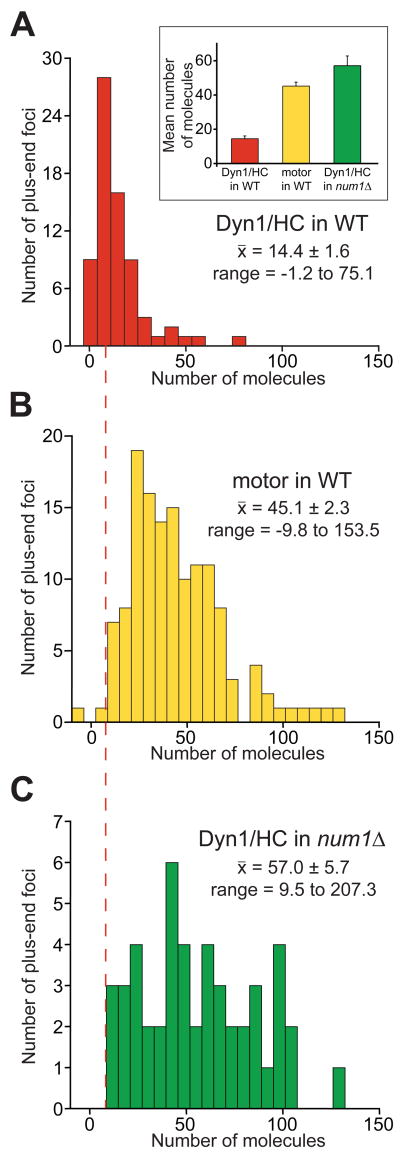
We used quantitative ratiometric methods to determine the number of fluorescent molecules at individual plus-ends. Cse4, a stable kinetochore protein present at two copies per chromosome [34, 35], was used as a standard for the intensity measurements (Figure S3). On average, we found ∼14, 13, and 5 copies of Dyn1/HC-3mCherry per plus-end, SPB, and cortical focus, respectively (Figure 5A and S3D, E, F). Since dynein is a dimer, these numbers represent ∼7, 6.5, and 2.5 molecules of dynein complex, respectively. The intensities of motor-3YFP plus-end foci gave a broader distribution with a mean of ∼45 copies per plus-end (Figure 5B). Motor-3YFP is likely a monomer based on in vitro studies [30]. Thus, plus-end targeting of motor-3YFP is enhanced 6.3-fold compared to dimeric dynein molecules. In num1Δ cells, where Dyn1/HC fails to off-load and is enhanced at the plus-ends [22, 24], we found ∼57 copies of Dyn1/HC, or ∼29 molecules of dynein complex, per plus-end (Figure 5C). Thus, the observed level for motor-3YFP is similar to that for Dyn1/HC in num1Δ (p = 0.02).
Figure 5. Quantitative ratiometric measurements of plus-end associated Dyn1/HC and isolated motor domain.
(A) Histogram of the number of molecules of Dyn1/HC-3mCherry per plus-end focus in wild-type cells (n = 71). Fields containing a mixed population of strains expressing Cse4-3mCherry only or Dyn1/HC-3mCherry with CFP-Tub1 were imaged simultaneously to ensure identical imaging conditions. The mean fluorescence intensity of Cse4-3mCherry (Figure S3A) was assigned a value of 32 molecules of 3mCherry [34, 35] and was used to normalize Dyn1/HC-3mCherry fluorescence intensity (A.U.) to number of molecules. Inset graph depicts the mean number of molecules for (A), (B) and (C) with standard error. Vertical dashed red line indicates mean value for Dyn1/HC in wild-type cells. (B) Histogram of the number of molecules of motor-3YFP per plus-end focus (n = 137). For normalizing motor-3YFP intensity, we used the intensity distribution of Dyn1/HC-3YFP as a standard (Figure S3B), which was normalized using the mean value of 14.4 copies of Dyn1/HC per plus-end, as determined for Dyn1/HC-3mCherry in (A). (C) Histogram of the number of molecules of Dyn1/HC-3GFP per plus-end in num1Δ cells (n = 52). Cse4-3GFP (Figure S3C) was used as a standard, as in (A). Values less than zero are a result of background subtraction. Plus-end foci were identified with either CFP-Tub1 or mCherry-Tub1.
Motor-3YFP Blocks Plus-end Targeting and Function of Dyn1/HC
We next asked whether the increased frequency and levels of motor-3YFP at plus-ends would inhibit Dyn1/HC function and localization, as predicted if motor-3YFP competes with Dyn1/HC for plus-end targeting. We constructed a diploid strain carrying MOTOR-3YFP and DYN1-2mCherry at the two DYN1 chromosomal loci, both under the control of the native promoter. The resulting diploid strain had a level of spindle misorientation similar to that of a dynein null strain, dyn1Δ/3GFP (Figure 6A), indicating a dominant negative effect of MOTOR-3YFP on dynein pathway function [17, 22]. Strains carrying MOTOR-3YFP with the AAA3 point mutations (K2424A or E2488Q) also exhibited similar levels of spindle misorientation (Figure 6A). In contrast, strains carrying TAIL-3GFP or 3GFP with DYN1/HC-2mCherry did not exhibit any spindle misorientation defects (Figure 6A). Our findings are consistent with previous overexpression studies of dynein motor domain in Dictyostelium [36].
Figure 6. Dominant negative effects of motor-3YFP can be rescued by overexpression of Pac1/LIS, but not Bik1/CLIP-170.
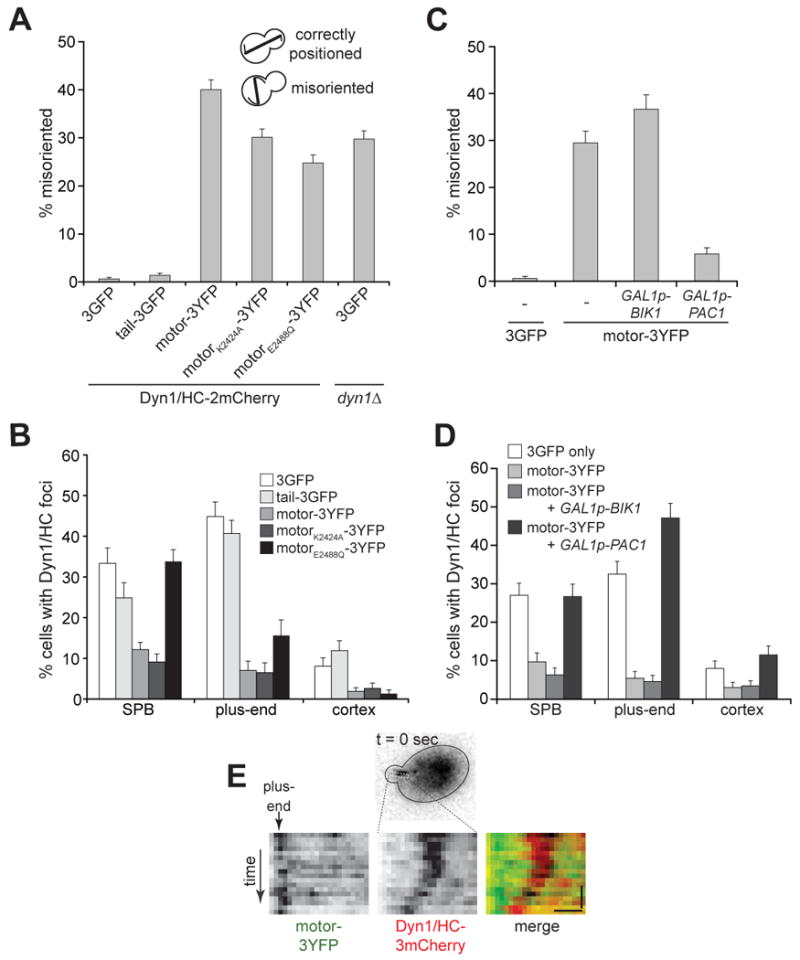
(A) The percentage of cells with a misoriented mitotic spindle in a cold (16°C) spindle position assay [24, 29] is plotted for diploid strains carrying DYN1/HC-2mCherry at one DYN1 locus, and either 3GFP, TAIL-3GFP, or MOTOR-3YFP (wild-type or AAA3 mutants) at the other DYN1 locus (n > 570 cells for each strain). The spindle was visualized using CFP-Tub1. A control diploid strain carrying no functional DYN1 is indicated on the right. (B) The percentage of cells that display Dyn1/HC-3mCherry foci at a given location is plotted for diploid strains similar to those in (A), except that the DYN1/HC-2mCherry allele was replaced with DYN1/HC-3mCherry for brighter fluorescence (n > 150 cells for each strain). Movies of Dyn1/HC-3mCherry and CFP-Tub1 were collected to distinguish foci at plus-ends from SPB and the cell cortex. Strains were imaged after growth under the same conditions as in (A). (C) The percentage of cells with a misoriented mitotic spindle in a cold spindle position assay is plotted for diploid strains carrying DYN1/HC-3mCherry and either 3GFP or MOTOR-3YFP, as indicated. The GAL1 promoter was inserted at the 5′ end of one of the chromosomal BIK1 or PAC1 loci. Strains were imaged after growth at 16°C to mid-log in synthetic defined media supplemented with 2% galactose (n > 250 cells for each strain). (D) Percentage of cells that display Dyn1/HC-3mCherry foci at a given location is plotted for the same strains used in (C), and imaged after growth under the same conditions as in (C) (n > 160 cells for each strain). (E) Kymograph depicting Dyn1/HC-3mCherry migrating along cytoplasmic microtubule toward a plus-end in a Pac1/LIS overexpressing cell. The motor-3YFP signal was used as reference point to identify the plus-end. Vertical bar represents 10 s; horizontal bar represents 1 μm. See Movie S10.
We further tested the effects of motor-3YFP on dynein pathway by assaying for synthetic growth defects with the Kar9 pathway. Budding yeast need either the dynein or Kar9 pathway for normal growth [17, 22]. Overexpression of motor-3YFP from a plasmid caused synthetic growth defects in kar9Δ and bim1Δ, two Kar9 pathway mutants, but not in dyn1Δ (Figure S4A). Overexpression of tail-3GFP or 3GFP did not cause synthetic growth defects (Figure S4A). Taken together, the spindle orientation and growth assays demonstrate that motor-3YFP, but not tail-3GFP, disrupt Dyn1/HC function.
We next asked whether motor-3YFP blocked Dyn1/HC function by inhibiting plus-end targeting of Dyn1/HC. Motor-3YFP (wild-type or AAA3 mutants), but not tail-3GFP, significantly reduced the frequencies of finding Dyn1/HC-3mCherry foci at plus-ends and the cell cortex (Figure 6B). Loss of cortical Dyn1/HC-3mCherry caused by motor-3YFP further demonstrates that plus-end targeting of Dyn1/HC is a prerequisite for off-loading and anchoring at the cortex. We conclude that the dominant negative capacity of motor-3YFP can be specifically attributed to its ability to block plus-end targeting of Dyn1/HC.
Overexpression of Pac1/LIS1 Rescues Motor-3YFP Dominant Negative Effect at Plus-Ends
Since Pac1/LIS1 and Bik1/CLIP-170 are required for plus-end targeting of motor-3YFP, it is possible that either one of them may be tightly sequestered by motor-3YFP, such that they are not free to recruit Dyn1/HC to plus-ends. We asked whether we could relieve the dominant negative effect of motor-3YFP by overexpressing Pac1/LIS1 or Bik1/CLIP-170. We used an inducible GAL1 promoter to drive high expression of chromosomal PAC1 or BIK1 in the diploid strain carrying MOTOR-3YFP and DYN1/HC-3mCherry. Overexpression of Pac1/LIS1, but not Bik1/CLIP-170, rescued the dominant negative effect of motor-3YFP on dynein pathway function, based on spindle misorientation assays (Figure 6C). Additionally, overexpression of Pac1/LIS1, but not Bik1/CLIP-170, restored targeting of Dyn1/HC-3mCherry to plus-ends, SPB and cell cortex to levels similar to those in a control strain without motor-3YFP (Figure 6D and S4B). Movies of Pac1/LIS1-overexpressing cells showed speckles of Dyn1/HC-3mCherry moving along the cytoplasmic microtubules toward the plus-end (Figure 6E and Movie S10). In contrast, overexpression of Bik1/CLIP-170 did not have an effect on Dyn1/HC-3mCherry targeting when motor-3YFP was present (Figure 6D and S4B). We conclude that motor-3YFP inhibits dynein pathway function by sequestering Pac1/LIS1 from Dyn1/HC.
Discussion
In summary, we found that the tail domain targets Dyn1/HC to the cell cortex but not microtubule plus-ends. We discovered that the motor domain is responsible for plus-end targeting of Dyn1/HC (Figure 7A). Our analysis reveals a new regulatory step that may involve an unmasking of the cortical association domain in Dyn1/HC (Figure 7B). Furthermore, we discovered that the isolated motor domain exhibits dominant negative effects on Dyn1/HC targeting and function by tightly sequestering Pac1/LIS1.
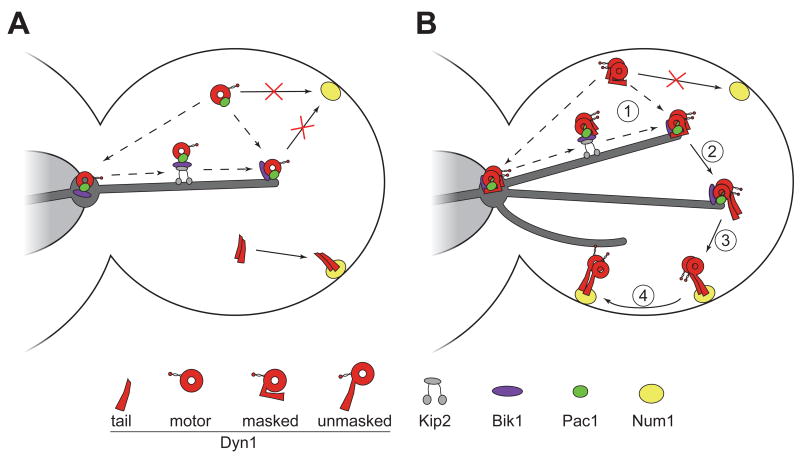
Figure 7. A model for Dyn1/HC unmasking at the plus-end.
(A) Summary of results for TAIL-3GFP and MOTOR-3YFP strains. Isolated motor and tail domains are illustrated in the same cell. (B) Prior to association with the cell cortex, our data indicates that the masked full-length Dyn1/HC molecule must first be targeted to microtubule plus-ends (Step 1). Our observations that Dyn1/HC and the isolated motor domain track along microtubules toward plus-ends in a Kip2-dependent manner suggest that Dyn1/HC is actively transported in a manner similar to Bik1/CLIP-170 [28]. However, Dyn1/HC also appears to be recruited to plus-ends directly from the cytoplasm. Upon association with plus-ends, we propose that Dyn1/HC undergoes a conformational change (Step 2), resulting in the unmasking of the N-terminal tail cortical association domain. Once unmasked, Dyn1/HC is primed for off-loading to the cell cortex (Step 3), where it functions to power the sliding of astral microtubules (Step 4). For simplicity, dynactin complex and dynein accessory chains were omitted in the illustration.
The Role of Tail Domain in Dynein Function
We observed that tail-3GFP associates with the cortex more often than full-length Dyn1/HC. One possible explanation for this difference is that Dyn1/HC has an affinity for (and is sequestered by) microtubules, either along their length or at their plus-ends, and SPBs. Thus the concentration of free Dyn1/HC available to bind to the cortex is lower than that of tail-3GFP. Another possible explanation is that the cortical association domain is masked in the full-length Dyn1/HC molecule, preventing it from binding to the cortex when it is free. Our findings did not support the former explanation, since free Dyn1/HC did not appear at the cortex when SPB and plus-end targeting were inhibited by deletion of Pac1/LIS1 or Bik1/CLIP-170 (Figure 4C), or when astral microtubules were disrupted by nocodazole treatment (Figure S1F). Instead, cortical Dyn1/HC foci were reduced compared to wild-type. Given that Dyn1/HC protein has been shown to be stable in bik1Δ and pac1Δ cells [22, 24], we conclude that free Dyn1/HC fails to bind to the cortex in these cells because its cortical-association domain is masked. The motor domain, or a motor domain-associated factor, likely performs this masking function.
While targeting of tail-3GFP to the cell cortex is very robust, its presence does not interfere with the normal targeting and function of Dyn1/HC. This is not surprising given the large number of cortical Num1 foci (Figure 3A). This also indicates that receptor sites at the cell cortex are not limiting in the dynein pathway.
The Role of Motor Domain in Dynein Function
Molecule-counting measurements of Dyn1/HC and motor at the plus-end reveal that, in a wild-type cell, Dyn1/HC is well below saturation. Our data indicate that there are approximately 7 dynein complexes per microtubule plus-end. While this number is of similar magnitude to that previously reported for the dynactin complex at the plus-end (mean ≈ 1; ≤ 5 dynactin complexes per plus-end) [23], it is clear that Dyn1/HC is present at higher levels (Figure 5A; up to 38 dynein complexes per plus-end). Thus, relative to dynein, dynactin at microtubule plus-ends is limiting.
In contrast to tail, the motor inhibits dynein pathway activity by blocking Dyn1/HC targeting. The fact that Pac1/LIS1 overexpression is capable of rescuing this phenotype indicates that Dyn1/HC is unable to compete with the motor for Pac1/LIS1 binding. If the association between motor and Pac1/LIS1 was transient, upon their dissociation, Dyn1/HC and motor would have equal opportunity to compete for Pac1/LIS1 binding. Dyn1/HC would therefore be found at plus-ends in equal proportion to motor; however, this was not observed (Figures 6B and 6D). Therefore, motor likely binds Pac1/LIS1 with much higher affinity than the full-length Dyn1/HC. This is consistent with in vitro studies by Reck-Peterson et al. [30], who observed Pac1/LIS1 copurifying with the isolated motor but not the full-length molecule following microtubule affinity purification. The differential affinity of Pac1/LIS1 for the motor versus Dyn1/HC indicates that in the context of the full-length molecule, the tail domain plays a role in modulating Pac1/LIS1 binding to the motor domain. Previous studies in mammalian cells have demonstrated that LIS1 interacts with two distinct regions of the dynein heavy chain: one in the AAA1 unit of the motor domain and another in the tail domain [19]. Whether these interactions are conserved in yeast remains to be determined. The strong Pac1/LIS1 affinity state exhibited by the motor may represent a particular conformation adopted by Dyn1/HC throughout its life cycle; however this phenomenon and the role of the tail domain in affecting this interaction requires further study.
In contrast to Dyn1/HC, motor does not require Ndl1/NudEL for plus-end targeting. Ndl1/NudEL has been proposed to affect plus-end targeting of Dyn1/HC by stabilizing the association between Dyn1/HC and Pac1/LIS1 [29]. The fact that Ndl1/NudEL is dispensable for plus-end targeting of motor is consistent with a stronger association of Pac1/LIS1 with the motor than with Dyn1/HC.
Although overexpression of Bik1/CLIP-170 did not rescue the dominant negative effect of motor on dynein function, it altered the interaction of motor with astral microtubules. In Bik1/CLIP-170 overexpressing cells, the motor (likely in complex with Pac1/LIS1) decorated the entire length of astral microtubules, and was less apparent as punctate foci (Figure S4B; compare control or GAL1p-PAC1 to GAL1p-BIK1). Since Dyn1/HC targeting to microtubules was not rescued by Bik1/CLIP-170 overexpression (Figure 6D), and Pac1/LIS1 was limiting in the cell, we reason that binding of Dyn1/HC to Pac1/LIS1 represents a first step in the assembly of a complex competent for plus-end targeting. Assembly of this complex with Bik1/CLIP-170 likely occurs subsequently.
Model for Dynein Unmasking at the Plus-End
Previously, we proposed a model whereby dynein is off-loaded to the cell cortex from microtubule plus-ends. Our data here support this model, and reveal new insights into the mechanism underlying the spatial regulation of dynein. First, Dyn1/HC is targeted to plus-ends by one of two mechanisms (Figure 7B, step 1, dashed lines): in a Bik1/CLIP-170 and Kip2-dependent manner, Dyn1/HC is transported along astral microtubules to the plus-end; alternatively, Dyn1/HC is directly recruited to plus-ends from the cytoplasm. This study and work from other labs [28, 29, 31] support both mechanisms of plus-end targeting. The former Kip2-dependent mechanism may account for ∼30% of plus-end targeting (Figure S1E), while the latter accounts for ∼70%. We propose that association of Dyn1/HC with the plus-end triggers an unmasking of the tail domain (Figure 7B, Step 2), which results in the consequent ability of Dyn1/HC to be off-loaded to Num1 sites at the cortex, its site of action (Figure 7B, Step 3). Subsequently, cortically anchored dynein powers the sliding of astral microtubules (Step 4), pulling the mitotic spindle toward the anchored site. Our analysis of full-length Dyn1/HCK2424A-3YFP mutant (Figure S2F and Movies S7 and S8) demonstrates that cortically anchored Dyn1/HC is capable of capturing astral microtubule plus-ends.
Dyn3/LIC and components of the dynactin complex have been implicated in the off-loading process, since their absence leads to the same phenotype as loss of Num1, namely loss of cortical Dyn1/HC foci and an enhancement of Dyn1/HC at the plus-ends [17, 22, 23]. Because dynactin appears to be limiting at the plus-end relative to Dyn1/HC, association of dynactin with Dyn1/HC may be the crucial step in the regulation of the off-loading process, possibly through mediating the unmasking of the tail domain. This regulatory step may ensure that only fully assembled dynein-dynactin complexes are delivered to cortical sites.
Supplementary Material
Acknowledgments
We thank Roy Kinoshita and Sean Christie for arranging Nikon microscope and cameras for use at MBL. We thank Dr. Moore and Dr. Schiebel for sending plasmids containing MET3 promoter and mCherry-TUB1. We are grateful to Drs. Xianying Tang, Patricia Wadsworth and Jennifer Ross for valuable discussions throughout this study. This work was supported by an HHMI Academic Research Internship and a University of Massachusetts Amherst Biology Department Junior Fellowship to J. P., and an NIH/NIGMS grant (1R01GM076094) and a Laura and Arthur Colwin Endowed Summer Research Fellowship at MBL to W.-L. L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data: Additional materials include Experimental Procedures, four figures, ten movies, and one table.
References
- 1.Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt DJ, Rose DJ, Saxton WM, Strome S. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol Biol Cell. 2005;16:1200–1212. doi: 10.1091/mbc.E04-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rusan NM, Tulu US, Fagerstrom C, Wadsworth P. Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. J Cell Biol. 2002;158:997–1003. doi: 10.1083/jcb.200204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- 7.O'Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wojcik E, Basto R, Serr M, Scaerou F, Karess R, Hays T. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- 10.Koonce MP, Samso M. Of rings and levers: the dynein motor comes of age. Trends Cell Biol. 2004;14:612–619. doi: 10.1016/j.tcb.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Mocz G, Gibbons IR. Model for the motor component of dynein heavy chain based on homology to the AAA family of oligomeric ATPases. Structure. 2001;9:93–103. doi: 10.1016/s0969-2126(00)00557-8. [DOI] [PubMed] [Google Scholar]
- 12.Hook P, Vallee RB. The dynein family at a glance. J Cell Sci. 2006;119:4369–4371. doi: 10.1242/jcs.03176. [DOI] [PubMed] [Google Scholar]
- 13.King SJ, Bonilla M, Rodgers ME, Schroer TA. Subunit organization in cytoplasmic dynein subcomplexes. Protein Sci. 2002;11:1239–1250. doi: 10.1110/ps.2520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King SM. The dynein microtubule motor. Biochim Biophys Acta. 2000;1496:60–75. doi: 10.1016/s0167-4889(00)00009-4. [DOI] [PubMed] [Google Scholar]
- 15.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 16.Busson S, Dujardin D, Moreau A, Dompierre J, De Mey JR. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol. 1998;8:541–544. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- 17.Lee WL, Kaiser MA, Cooper JA. The offloading model for dynein function: differential function of motor subunits. J Cell Biol. 2005;168:201–207. doi: 10.1083/jcb.200407036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purohit A, Tynan SH, Vallee R, Doxsey SJ. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J Cell Biol. 1999;147:481–492. doi: 10.1083/jcb.147.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang X, Han G, Winkelmann DA, Zuo W, Morris NR. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr Biol. 2000;10:603–606. doi: 10.1016/s0960-9822(00)00488-7. [DOI] [PubMed] [Google Scholar]
- 21.Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell. 2006;10:425–439. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 25.Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, O'Toole ET, Giddings TH, Jr, Sundin BA, Davis TN. The organization of the core proteins of the yeast spindle pole body. Mol Biol Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tynan SH, Gee MA, Vallee RB. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J Biol Chem. 2000;275:32769–32774. doi: 10.1074/jbc.M001537200. [DOI] [PubMed] [Google Scholar]
- 27.Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalho P, Gupta ML, Jr, Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caudron F, Andrieux A, Job D, Boscheron C. A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. J Cell Sci. 2008;121:1506–1513. doi: 10.1242/jcs.023374. [DOI] [PubMed] [Google Scholar]
- 32.Reck-Peterson SL, Vale RD. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein's AAA domains in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:1491–1495. doi: 10.1073/pnas.2637011100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Reck-Peterson SL, Vale RD. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein's AAA domains in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:14305. doi: 10.1073/pnas.0404506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 35.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koonce MP, Samso M. Overexpression of cytoplasmic dynein's globular head causes a collapse of the interphase microtubule network in Dictyostelium. Mol Biol Cell. 1996;7:935–948. doi: 10.1091/mbc.7.6.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner MP, Costa GL, Schoettlin W, Cline J, Mathur E, Bauer JC. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene. 1994;151:119–123. doi: 10.1016/0378-1119(94)90641-6. [DOI] [PubMed] [Google Scholar]
- 39.Vorvis C, Markus SM, Lee WL. Photoactivatable GFP tagging cassettes for protein-tracking studies in the budding yeast Saccharomyces cerevisiae. Yeast. 2008;25:651–659. doi: 10.1002/yea.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 41.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 42.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.