Abstract
To determine whether pathogenic mutations in mtDNA are involved in phenotypic expression of Alzheimer’s disease (AD), the transfer of mtDNA from elderly patients with AD into mtDNA-less (ρ0) HeLa cells was carried out by fusion of platelets or synaptosomal fractions of autopsied brain tissues with ρ0 HeLa cells. The results showed that mtDNA in postmortem brain tissue survives for a long time without degradation and could be rescued in ρ0 HeLa cells. Next, the cybrid clones repopulated with exogenously imported mtDNA from patients with AD were used for examination of respiratory enzyme activity and transfer of mtDNA with the pathogenic mutations that induce mitochondrial dysfunction. The presence of the mutated mtDNA was restricted to brain tissues and their cybrid clones that formed with synaptosomes as mtDNA donors, whereas no cybrid clones that isolated with platelets as mtDNA donors had detectable mutated mtDNA. However, biochemical analyses showed that all cybrid clones with mtDNA imported from platelets or brain tissues of patients with AD restored mitochondrial respiration activity to almost the same levels as those of cybrid clones with mtDNA from age-matched normal controls, suggesting functional integrity of mtDNA in both platelets and brain tissues of elderly patients with AD. These observations warrant the reassessment of the conventional concept that the accumulation of pathogenic mutations in mtDNA throughout the aging process is responsible for the decrease of mitochondrial respiration capacity with age and with the development of age-associated neurodegenerative diseases.
There have been many reports suggesting that mammalian mitochondrial respiration capacity decreases with age or the development of age-associated neurodegenerative diseases (1–4). Recently, this age-related decrease in the capacity to produce energy was proposed to be caused by the accumulation of somatic mtDNA mutations, which have been shown to cause various kinds of mitochondrial encephalomyopathies (2, 5–7). In addition to these pathogenic somatic mutations of mtDNA, maternally transferable lesions of mtDNA have also been implicated as causal in patients with neurodegenerative diseases (8–12). For example, the transfer of platelet mtDNA from patients with Parkinson’s and Alzheimer’s disease (AD) to mtDNA-less (ρ0) neuroblastoma cells consistently resulted in depression of the activity of complex I (9) and complex IV cytochrome c oxidase (COX; refs. 10 and 11), respectively. Thus far, however, there is no convincing evidence that maternally transmitted mtDNA mutations and/or accumulated somatic mutations in mtDNA throughout life are causal genetic factors of age-related mitochondrial dysfunction.
Previously, we observed an age-related reduction of respiratory enzyme activity in cultured human skin fibroblasts isolated from donors of various ages (0–97 years; ref. 4). However, the isolation of cybrids by mtDNA transfer from fibroblasts to ρ0 HeLa cells (4) and the isolation of nuclear hybrids by nuclei transfer from ρ0 HeLa cells to fibroblasts provided convincing evidence that the age-related disorders were not caused by mtDNA mutations but by nuclear-recessive mutations of factors involved in the translation in mitochondria (13).
On the other hand, it was uncertain whether our conclusions generated from studies on human fibroblasts also could be extended to other human tissues, particularly brain tissue, which shows much higher oxidative activity than fibroblasts. Recently, we isolated ρ0 lines from mouse cells (14–16) and succeeded in trapping brain mtDNA in dividing cultured cells by fusion of the ρ0 mouse cells with mouse brain synaptosomes (14), which represent synaptic endings isolated from brain tissues. Moreover, we examined how long brain mtDNA could survive and maintain its function after the mice were killed; we found that, even after 1 month, transfer of mouse synaptosomal mtDNA to ρ0 mouse cells resulted in the repopulation of mtDNA and the restoration of normal activity of mitochondrial respiratory enzymes (17). These observations imply that, even 1 month after the mice were killed, mtDNA in brain tissue survives with no change in its functional properties for gene expression. Thus, this procedure could be applied to human samples to examine the influence of aging on mtDNA in brain tissue, even after a prolonged postmortem period before mtDNA transfer from human brain tissues to human ρ0 cells.
In this study, mtDNA in platelets and in autopsied brain tissues from elderly patients with AD was rescued in ρ0 HeLa cells by fusion of platelets and brain synaptosomal fractions with ρ0 HeLa cells. This mtDNA transfer resulted in complete restoration of mitochondrial respiratory function, suggesting functional integrity of the mtDNA from the elderly patients with AD.
MATERIALS AND METHODS
Cells and Cell Culture.
ρ0 HeLa cells (18) and mtDNA repopulated cybrids were grown in normal medium: RPMI medium 1640 (Nissui Seiyaku, Tokyo) containing 10% fetal calf serum, 50 μg/ml uridine, and 0.1 mg/ml pyruvate.
mtDNA Donors.
Samples provided with informed consent from four donors with AD and five normal donors were used in this study. The four patients with AD (AD1, AD2, AD3, and AD4) were a 70-year-old female, a 63-year-old female, an 89-year-old male, and an 81-year-old male, respectively. All four were confirmed neuropathologically to have AD. The five normal controls (N1, N2, N3, N4, and N5) were a 67-year-old female, a 76-year-old female, a 43-year-old female, a 74-year-old male, and a 97-year-old female, respectively.
Isolation of Total DNA from Tissues.
Total DNA was isolated as described (19) with slight modifications. Briefly, 0.1-g samples of tissue were frozen in liquid nitrogen, and DNA was extracted from the frozen tissues by the proteinase K/SDS/phenol method.
Isolation of Platelets, Synaptosomal Fractions, and Enucleated Cytoplasts.
Samples of 1 ml of blood were diluted with an equal volume of PBS, and whole cells were removed by centrifugation (1,000 × g for 10 min at 4°C). The resultant supernatant was centrifuged at 1,700 × g for 20 min at 4°C, and the pellet obtained was used as the platelet-rich fraction. Synaptosomal fraction was isolated from autopsied AD4 brain tissue stored at room temperature for 18 h and subsequently at 4°C for 2 h as described (17). Enucleated cytoplasts were isolated from fibroblasts as described (4).
Introduction of mtDNA into ρ0 HeLa Cells.
Platelets, enucleated fibroblasts, and synaptosomes were used as mtDNA donors. Fusion to ρ0 HeLa cells was carried out in the presence of 50% (wt/vol) polyethylene glycol 1500 (Boehringer Mannheim). The fusion mixture was cultivated in selection RPMI medium 1640, in which even cybrids with very low COX activity have been shown to grow, without pyruvate and uridine (13). On days 14–30 after fusion, the cybrid clones growing in the medium were isolated clonally by the cylinder method. In the case of AD4, no selection was used to isolate cybrid clones, and 50 colonies were picked up randomly. Then, cybrid clones with mtDNA were screened by PCR analysis, so that even cybrids with no COX activity could be isolated.
PCR Analysis.
Primers (nucleotides 3,153–3,172 and 3,551–3,531) were used for identification of clones with imported mtDNA, i.e., cybrid clones, and for identification of wild-type mtDNA. For detection of a small amount of a common deletion mutated mtDNA, ΔmtDNA4977, amplification was carried out with 60 ng of total DNA in 10 μl of solution containing 0.5 μM of a primer set and 0.25 units EX-taq polymerase (Takara Shuzo, Kyoto), as described (20) with slight modifications. Briefly, two sets of oligonucleotide primers were used for amplification: (i) F1 (nucleotides 7,901–7,920) on the light strand and R1 (14,220–14,201) on the heavy strand and (ii) F2 (8,282–8,305) on the light strand and R2 (13,650–13,631) on the heavy strand. The amplified products were separated on a 2.5% agarose gel containing ethidium bromide (0.1 μg/ml). For detection of a small amount of mtDNA with the pathogenic mutation tRNALeu(UUR)3243, the amplification refractory-mutation-system method was applied (21). Briefly, a PCR primer with a sequence matched to the mutated mtDNA at the 3′-end was used for selective amplification of the mutated mtDNA. Plasmids with or without the mutation were constructed and used as standards for quantitative analysis. The PCR products were quantified by an automatic sequence detection system (Prism 7700 system; Applied Biosystems) at each amplification step. The primers used were ATTAAAGTCCTACGTGATC (3,048–3,066) and ATGCGATTACCGGGCC (3,258–3,243) for detection of the mutated mtDNA and GCCTTCCCCCGTAAATGATAT (3,163–3,183) and GAAGAGGAATTGAACCTCTGACTG (3,298–3,273) for quantification of mtDNA. The nucleotide sequence of the TaqMan probe (Perkin–Elmer) is TGCCATCTTAACAAACCCTGTTCTTGGGTT (3,241–3,213). The content of mutated mtDNA was normalized by total mtDNA quantified by the same system (S.O., K.N., S. Matsuda, T. Araki, M. Takahashi, and N. Omori, unpublished work).
Measurement of Oxygen Consumption and COX Activity.
The rate of oxygen consumption was measured by treating cells with trypsin, incubating the suspension in PBS, and recording oxygen consumption in a polarographic cell (1.0 ml) at 37°C with a Clark-type oxygen electrode (Yellow Springs Instruments). For biochemical analysis of COX activity, cells in log-phase growth were harvested, and COX activity was measured as the rate of cyanide-sensitive oxidation of reduced cytochrome c as described above (22).
RESULTS
Comparison of Functional Properties of Platelet mtDNA of Patients with AD and Normal Controls by Using Cybrid Clones.
Recently, it has been proposed that cybrids with platelet mtDNA from patients with AD consistently showed progressively reduced COX activity compared with those with mtDNA from normal controls (10, 11). In the case of mtDNA transfer from platelets, mitochondrial dysfunction in cybrids should not be caused by somatic mutations but could be caused by maternally transmitted mutations in mtDNA, because mitotic tissue usually does not accumulate somatic mtDNA mutations (13). However, there has been no report that this disease is consistently inherited maternally. To study this problem, we examined whether maternally transmitted mtDNA lesions that could induce remarkable reduction of COX activity are present in all patients with AD by developing a simple procedure for introducing platelet mtDNA into ρ0 HeLa cells by using only 1 ml of peripheral blood (see Materials and Methods). Cybrid clones were isolated by the fusion of ρ0 HeLa cells with platelets from three patients with AD and three age-matched normal controls (Table 1). As normal controls, we also used two cybrid clones isolated by the fusion of ρ0 HeLa cells with enucleated fibroblasts from two elderly controls. Results showed that the COX activity of all the cybrid clones we examined were similar irrespective of whether the ρ0 HeLa cells were repopulated with mtDNA from platelets of patients with AD, platelets of age-matched normal controls, or enucleated fibroblasts of normal controls (Fig. 1). These observations suggest that platelet mtDNA, at least the mtDNA from those patients with AD that we tested, was functionally intact, contrary to the observations of Davis et al. (10, 11).
Table 1.
Characterization of cybrids with mtDNA from mitotic tissues of AD patients
| Parents and cybrids | Fusion combination |
|---|---|
| Parents | |
| mtDNA recipients | |
| ρ0 HeLa cells | |
| mtDNA donors (blood) | |
| AD1 | |
| AD2 | |
| AD3 | |
| N1 | |
| N2 | |
| N3 | |
| mtDNA donors (fibroblasts) | |
| N4 | |
| N5 | |
| Cybrids | |
| CyAD1-B | ρ0 HeLa cells × AD1 |
| CyAD2-B | ρ0 HeLa cells × AD2 |
| CyAD3-B | ρ0 HeLa cells × AD3 |
| CyN1-B | ρ0 HeLa cells × N1 |
| CyN2-B | ρ0 HeLa cells × N2 |
| CyN3-B | ρ0 HeLa cells × N3 |
| CyN4-F | ρ0 HeLa cells × N4 |
| CyN5-F | ρ0 HeLa cells × N5 |
Figure 1.
Comparison of COX activity in cybrids with mtDNA from mitotic tissues of patients with AD and normal controls. He, HeLa cells; ρ0, ρ0 HeLa cells; CyAD1-B, 2-B, and 3-B, cybrid clones with platelet mtDNA from AD1, AD2, and AD3, respectively; CyN1-B, 2-B, and 3-B, cybrid clones with platelet mtDNA from N1, N2, and N3, respectively; CyN4-F and 5-F, cybrid clones with fibroblast mtDNA from N4 and N5, respectively.
Isolation of Cybrids by Introduction of mtDNA from Autopsied Brain Tissues to ρ0 HeLa Cells.
Next, we examined whether somatic mutations of mtDNA were accumulated sufficiently in various autopsied brain tissues from an elderly patient with AD to induce progressive reduction of mitochondrial respiratory activity, because postmitotic and highly oxidative tissues, such as brain tissue, are likely to accumulate somatic mutated mtDNA throughout life and/or during the development of neurodegenerative diseases (2).
We recently showed that mouse mtDNA in postmortem brain tissue that was stored for up to 1 month still survived and retained functional integrity, causing complete recovery of mitochondrial respiratory function when transferred to ρ0 mouse cells (17). We applied this procedure to autopsied human brain tissues for examination of accumulation of heritable lesions in mtDNA in postmitotic and highly oxidative tissues from elderly patients with AD. Platelets and the synaptosomal fractions isolated from autopsied brain tissues of AD4 were fused with ρ0 HeLa cells for isolation of cybrid clones (Table 2). We did not use a selection medium for excluding unfused ρ0 HeLa cells, so that respiration-deficient cybrids were not excluded. We randomly picked 50 colonies from each tissue and examined them for the presence of mtDNA by PCR analysis. Results showed that most of the clones did not possess mtDNA, suggesting that they were derived from unfused parental ρ0 HeLa cells. mtDNA was observed in only one clone obtained from substantia nigra as the mtDNA donor (CyAD4-SN), two clones obtained from globus pallidus as the mtDNA donor (CyAD4-GP1 and CyAD4-GP2), and two clones obtained from blood as the mtDNA donor (CyAD4-B1 and CyAD4-B2) (Table 2).
Table 2.
Characterization of cybrids with mtDNA from autopsied brain tissues of patient AD4
| Parents and cybrids | Fusion combination | tRNALeu(UUR)3243 |
|---|---|---|
| Parents | ||
| mtDNA recipients | ||
| ρ0 HeLa cells | − | |
| mtDNA donor AD4 | ||
| Substantial nigra | + (0.05%) | |
| Globus pallidus | + (0.04%) | |
| Blood | − | |
| Cybrids | ||
| CyAD4-SN | ρ0 HeLa cells × substantia nigra | + (0.04%) |
| CyAD4-GP1 | ρ0 HeLa cells × globus pallidus | + (0.05%) |
| CyAD4-GP2 | ρ0 HeLa cells × globus pallidus | + (0.05%) |
| CyAD4-B1 | ρ0 HeLa cells × blood | − |
| CyAD4-B2 | ρ0 HeLa cells × blood | − |
Characterization of Cybrids with Respect to Transfer of Neuronal mtDNA and Respiratory Activity.
The presence of the common deletion mutated ΔmtDNA4977, which is responsible for mitochondrial diseases (23), was examined by using blood and autopsied brain samples from AD4. The 392-bp fragment amplified from ΔmtDNA4977 was observed in brain tissues, the substantia nigra, and globus pallidus, but no fragment was amplified from a blood sample of the same patient (Fig. 2). These results are consistent with previous observations that age-associated accumulation of somatic mtDNA mutations was observed in highly oxidative and postmitotic tissues (5, 6) but not in mitotic tissues, such as fibroblasts (13). Similar results were obtained when another pathogenic mtDNA mutation, tRNALeu(UUR)3243 (24), was examined by applying quantitative PCR amplification: brain tissues accumulated up to 0.05% tRNALeu(UUR)3243 mtDNA, but tRNALeu(UUR)3243 mtDNA was not detectable in blood samples (Table 2).
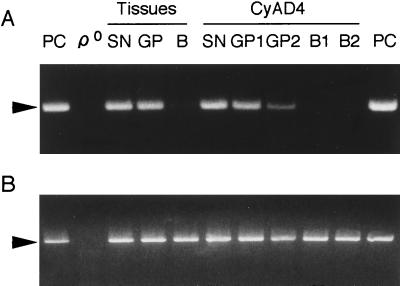
Figure 2.
PCR analysis of ΔmtDNA4977 in autopsied tissues from AD4 and their cybrid clones. (A) Amplification of ΔmtDNA4977. (B) Amplification of wild-type mtDNA. PCR analysis was carried out with total DNA prepared from ρ0 HeLa cells as a negative control (ρ0) and ΔmtDNA4977 prepared from a patient with Kearns–Sayre syndrome (provided by Y.-i. Goto and I. Nonaka, National Center of Neurology and Psychiatry, Kodaira, Tokyo, Japan) as a positive control (PC). SN, substantia nigra; GP, globus pallidus; B, blood; CyAD4-SN, a cybrid clone with mtDNA from the substantia nigra; CyAD4-GP1 and CyAD4-GP2, cybrid clones with mtDNA from the globus pallidus; CyAD4-B1 and CyAD4-B2, cybrid clones with mtDNA from platelets of AD4. Arrowheads indicate a fragment of 392 bp amplified from ΔmtDNA4977 (A) and a fragment of 399 bp amplified from wild-type mtDNA (B).
By using cybrid clones repopulated with exogenously imported mtDNA, we examined transfer of the mutated mtDNA from brain tissue. The mutated ΔmtDNA4977 and mtDNA with a tRNALeu(UUR)3243 mutation were detected in cybrid clones only when synaptosomes prepared from the substantia nigra and globus pallidus were used as mtDNA donors (Fig. 2 and Table 2). Cybrid clones isolated from blood samples did not have any detectable amounts of the pathogenic mutated mtDNA. Moreover, the amounts of mutated tRNALeu(UUR)3243 mtDNA in cybrid clones were almost the same as those in mtDNA donor brain tissue (Table 2). Therefore, the mtDNA in the cybrid clones CyAD4-SN, CyAD4-GP1, and CyAD4-GP2 should be derived exclusively from brain tissue.
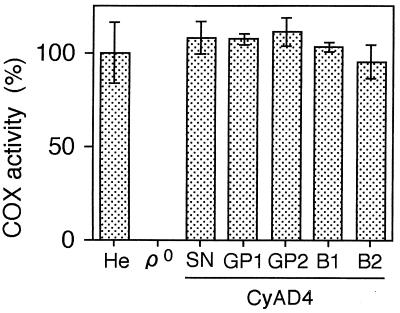
Next, we tested COX activity and found that all the cybrid clones with imported mtDNA from brain tissue of a patient with AD had COX activity as high as that of cybrid clones with mtDNA from platelets of the same patient or with mtDNA from platelets and fibroblasts of age-matched normal controls (Figs. 1 and 3). To examine the possibility of impairment of other mitochondrial respiration complexes, we measured the O2 consumption rate and observed its complete recovery in cybrid clones with AD brain mtDNA (data not shown), suggesting that mtDNA in autopsied brain tissue from the patient with AD is functionally intact with respect to replication and gene expression providing a normal oxidative phosphorylation capacity in ρ0 HeLa cells. Therefore, the amounts of accumulated somatic mutations in mtDNA populations of brain tissue are insufficient to affect mitochondrial respiratory functions.
Figure 3.
Comparison of COX activity in cybrids with mtDNA from brain tissues and platelets of AD4. He, HeLa cells; ρ0; ρ0 HeLa cells; CyAD4-SN, a cybrid clone with mtDNA from the substantia nigra; CyAD4-GP1 and CyAD4-GP2, cybrid clones with mtDNA from the globus pallidus; CyAD4-B1 and CyAD4-B2, cybrid clones with mtDNA from platelets of AD4.
DISCUSSION
In this study, mtDNA transfer from platelets and autopsied brain tissues of elderly patients with AD to ρ0 HeLa cells was carried out by the fusion of platelets and synaptosomal fractions with ρ0 HeLa cells, respectively. The results showed that mtDNA in postmortem brain tissue survives for a long time without degradation, as is the case with mouse brain tissue (17), and mtDNA could be rescued in ρ0 HeLa cells. Moreover, this mtDNA transfer resulted in complete restoration of mitochondrial respiratory function, suggesting the functional integrity of mtDNA in both platelets and brain tissue of patients with AD.
These observations are not consistent with the report by Davis et al. (10) that claims that specific mtDNA mutations in COX genes are responsible for the COX deficiency observed in platelets and the brain of patients with AD. Recently, it was concluded that these COX mutations proposed to cause AD (10) were not derived from mtDNA but from mtDNA sequences located in nuclear DNA-coded pseudogenes, owing to the presence of these sequences even in ρ0 human cells (25, 26). However, the results did not exclude the possibility of involvement of other mtDNA mutations in the expression of COX deficiency, because cybrid clones with platelet mtDNA from all patients with AD examined consistently showed reduced COX activity (10, 11). Nonetheless, it is unlikely that all patients with AD possess sufficient pathogenic mtDNA mutations for inducing COX deficiency for the following reasons. First, if these putative pathogenic mtDNA mutations are causal genetic factors, then most late-onset AD must be consistently inherited maternally, or patients with mitochondrial diseases caused by pathogenic mtDNA mutations must express phenotypes of AD much more frequently than normal controls, which is not the case. Second, even if mutated mtDNA is not transmitted maternally but derived from newly formed somatic mutations during the aging process in the patients with AD, then it is unlikely that platelets possess them because mitotic tissue does not accumulate such pathogenic mutated mtDNA because of mitotic segregation (13). Finally, our present observations suggest that platelet mtDNA from patients with AD, as well as from normal elderly controls, does not contain sufficient mtDNA with pathogenic mutations to reduce mitochondrial respiratory function.
In contrast to mitotic tissue, postmitotic tissue requires much higher energy-producing activity, and the resultant progressive oxidative stress in its mitochondria is proposed to be responsible for preferential accumulation of somatic mtDNA mutations, leading to reduced respiration activity in the brain tissue of elderly subjects and patients with age-associated neurodegenerative diseases such as AD and Parkinson’s disease (2). For testing this hypothesis, mtDNA in human brain tissue must be transferred to ρ0 human cells, and the resulting cybrid clones must be examined for cotransmission of reduced respiratory activity. However, there have been no reports on the introduction of human neuronal mtDNA into ρ0 cells. By using mouse tissue and ρ0 mouse cells, we recently showed that even more than 1 month after mice were killed, the mtDNA in their brain tissues survived and could be rescued in ρ0 mouse cells without change in its functional properties (17). In this study, we applied this procedure to human samples and showed that mtDNA in autopsied brain tissue from an elderly patient with AD could be rescued by fusion of synaptosomal fractions with ρ0 HeLa cells. This mtDNA transfer resulted in complete restoration of mitochondrial respiratory function, suggesting functional integrity of the mtDNA in brain tissues of the elderly patients with AD.
It is unlikely that we selected only respiration-competent cybrid clones, because we used no selection procedure, thus allowing growth of respiration-deficient cybrids. It is also unlikely that mtDNA of platelets in autopsied brain tissue was transferred to ρ0 cells for the following two reasons. First, these cybrid clones possessed pathogenic mutated mtDNA, whereas platelets and cybrid clones isolated by using platelets as mtDNA donors did not (Fig. 2 and Table 2). Moreover, the amounts of the tRNALeu(UUR)3243 mutation in CyAD4-SN, CyAD4-GP1, and CyAD4-GP2 were comparable to those in donor brain tissues (Table 2). Therefore, the mtDNA in the cybrid clones should be derived from brain tissue. Second, we previously found that success in transfer of mtDNA from various mouse tissues to ρ0 mouse cells was limited to brain tissue, and no cybrid colonies were obtained when other mouse tissues were used as mtDNA donors (17). As all tissues contain platelets, this observation also excludes the possibility that platelet-derived mtDNA was transferred to ρ0 cells.
In this study, synaptosomal fractions were prepared from about 0.5 g of brain tissue. Therefore, we could not exclude the possibility that age- and/or disease-associated accumulation of mtDNA with pathogenic somatic mutations occurred in limited cell types or subregions of the brain tissue. It is also possible that the amount of the pathogenic mutated mtDNA in the cybrid clones was diluted with a large amount of wild-type mtDNA from normal regions of the tissues. However, age- and/or disease-associated mitochondrial respiratory abnormalities were recognized by biochemical analysis of homogenates of the brain tissues (27, 28). Therefore, our present findings suggest that these biochemical mitochondrial abnormalities observed in human brain tissue could not be explained by accumulation of mtDNA with somatic pathogenic mutations.
By using mitochondrial- (4) and nuclear-genome transfer (13), we previously showed that mtDNA mutations were not involved in the age-associated mitochondrial dysfunction observed in human fibroblasts (4). In this study, we extended this finding to postmitotic, highly oxidative human brain tissue. However, small amounts of ΔmtDNA4977 and mtDNA with the tRNALeu(UUR)3243 mutation were present in human brain tissue, and probably mtDNA with other pathogenic mutations also could be accumulated. This discrepancy may be explained by the occurrence of transcomplementation between mtDNAs (18, 29, 30). In fact, when two cells that were respiration-deficient because of a predominance of different types of pathogenic mtDNA mutations were fused, normal respiratory function was restored in the fused cells (31). Therefore, presence of transcomplementation between mitochondria seems to be very important for the cells to maintain normal respiration function throughout the aging process, and mitochondrial respiratory function would not be affected significantly when many kinds of mtDNA with different types of somatic mutations accumulate throughout the aging process.
Acknowledgments
This work was supported in part by grants from the Tsukuba Advanced Research Alliance to J.-I.H. and S.-y.K., by a University of Tsukuba Special Research Grant (Superior) to J.-I.H., and by grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan to J.-I.H.
ABBREVIATIONS
- AD
Alzheimer’s disease
- COX
cytochrome c oxidase
- ρ0
mtDNA-less
References
- 1.Linnane A W, Marzuki S, Ozawa T, Tanaka M. Lancet. 1989;i:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- 2.Wallace D C. Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 3.Ames B N, Shigenaga M K, Hagen T M. Biochim Biophys Acta. 1995;1271:165–170. doi: 10.1016/0925-4439(95)00024-x. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi J-I, Ohta S, Kagawa Y, Kondo H, Kaneda H, Yonekawa H, Takai D, Miyabayashi S. J Biol Chem. 1994;269:6878–6883. [PubMed] [Google Scholar]
- 5.Soong N W, Hinton D R, Cortopassi G, Arnheim N. Nat Genet. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 6.Corral-Debrinski M, Horton T, Lott M T, Shoffner J M, Flint Beal M, Wallace D C. Nat Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 7.Larson N G, Clayton D A. Annu Rev Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 8.Hutchin T, Cortopassi G. Proc Natl Acad Sci USA. 1995;92:6892–6895. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swerdlow R H, Parks J K, Miller S W, Tuttle J B, Trimmer P A, Sheehan J P, Bennett J P, Jr, Davis R E, Parker W D., Jr Ann Neurol. 1996;40:663–671. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- 10.Davis R E, Miller S, Herrnstadt C, Ghosh S S, Fahy E, Shinobu L A, Galasko D, Thal L J, Beal M F, Howell N, et al. Proc Natl Acad Sci USA. 1997;94:4526–4531. doi: 10.1073/pnas.94.9.4526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Sheehan J P, Swerdlow R H, Miller S W, Davis R E, Parks J K, Parker W D, Tuttle J B. J Neurosci. 1997;17:4612–4622. doi: 10.1523/JNEUROSCI.17-12-04612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattson M P. Trends Neurosci. 1997;20:373–375. doi: 10.1016/s0166-2236(97)01114-4. [DOI] [PubMed] [Google Scholar]
- 13.Isobe K, Ito S, Hosaka H, Iwamura Y, Kondo H, Kagawa Y, Hayashi J-I. J Biol Chem. 1998;273:4601–4606. doi: 10.1074/jbc.273.8.4601. [DOI] [PubMed] [Google Scholar]
- 14.Inoue K, Ito S, Takai D, Soejima A, Shisa H, LePecq J-B, Segal-Bendirdjian E, Kagawa Y, Hayashi J-I. J Biol Chem. 1997;272:15510–15515. doi: 10.1074/jbc.272.24.15510. [DOI] [PubMed] [Google Scholar]
- 15.Soejima A, Inoue K, Takai D, Kaneko M, Ishihara H, Oka Y, Hayashi J-I. J Biol Chem. 1996;271:26194–26199. doi: 10.1074/jbc.271.42.26194. [DOI] [PubMed] [Google Scholar]
- 16.Inoue K, Takai D, Hosaka H, Ito S, Shitara H, Isobe K, LePecq J-B, Segal-Bendirdjian E, Hayashi J-I. Biochem Biophys Res Commun. 1997;239:257–260. doi: 10.1006/bbrc.1997.7446. [DOI] [PubMed] [Google Scholar]
- 17.Ito S, Inoue K, Yanagisawa N, Kaneko M, Hayashi J-I. Biochem Biophys Res Commun. 1998;247:432–435. doi: 10.1006/bbrc.1998.8800. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi J-I, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. Proc Natl Acad Sci USA. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shitara H, Hayashi J-I, Takahama S, Kaneda H, Yonekawa H. Genetics. 1998;148:851–857. doi: 10.1093/genetics/148.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubota N, Hayashi J-I, Inada T, Iwamura Y. Radiat Res. 1997;148:395–398. [PubMed] [Google Scholar]
- 21.Newton C R, Graham A, Heptinstall L E, Powell S J, Summers C, Kalsheker N, Smith J C, Markham A F. Nucleic Acids Res. 1989;17:2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyabayashi S, Narisawa K, Iinuma K, Tada K, Sakai K, Kobayashi K, Kobayashi Y, Morinaga S. Brain Dev. 1984;6:362–372. doi: 10.1016/s0387-7604(84)80112-6. [DOI] [PubMed] [Google Scholar]
- 23.Schon E A, Rizzuto R, Moraes C T, Nakase H, Zeviani M, DiMauro S. Science. 1989;244:346–349. doi: 10.1126/science.2711184. [DOI] [PubMed] [Google Scholar]
- 24.Goto Y, Nonaka I, Horai S. Nature (London) 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 25.Hirano M, Shtilbans A, Mayeux R, Davidson M M, DiMauro S, Knowles J A, Schon E A. Proc Natl Acad Sci USA. 1997;94:14894–14899. doi: 10.1073/pnas.94.26.14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace D C, Stugard C, Murdock D, Schurr T, Brown M D. Proc Natl Acad Sci USA. 1997;94:14900–14905. doi: 10.1073/pnas.94.26.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mutisya E M, Bowling A C, Beal M F. J Neurochem. 1994;63:2179–2184. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- 28.Parker W D, Jr, Parks J K, Filley C M, Kleinschmidt-DeMasters B K. Neurology. 1994;44:1090–1096. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi J-I, Takemitsu M, Goto Y-i, Nonaka I. J Cell Biol. 1994;125:43–50. doi: 10.1083/jcb.125.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takai D, Inoue K, Goto Y-i, Nonaka I, Hayashi J-I. J Biol Chem. 1997;272:6028–6033. doi: 10.1074/jbc.272.9.6028. [DOI] [PubMed] [Google Scholar]
- 31.Takai, D., Isobe, K. & Hayashi, J.-I. (1999) J. Biol. Chem., in press. [DOI] [PubMed]