Abstract
Sterile alpha motif (Sam) domains are protein interaction modules that are implicated in many biological processes mainly via homo- and hetero-dimerization. It has been recently reported that the lipid phosphatase Ship2 regulates endocytosis of the EphA2 receptor, a process that has been investigated as a possible route to reduce tumor malignancy. A heterotypic Sam-Sam domain interaction is mediating this process. Here, we report NMR and ITC (Isothermal Titration Calorimetry) studies on the Sam domain of Ship2 revealing its three-dimensional structure and its possible mode of interaction with the Sam domain from the EphA2 receptor. These studies have also resulted in the identification of a minimal peptide region of Ship2 that retains binding affinity for the Sam domain of EphA2 receptor. Hence, this peptide and the detection of key structural elements important for EphA2 receptor endocytosis provide possible ways for the development of novel small molecule antagonists with potential anti-cancer activity.
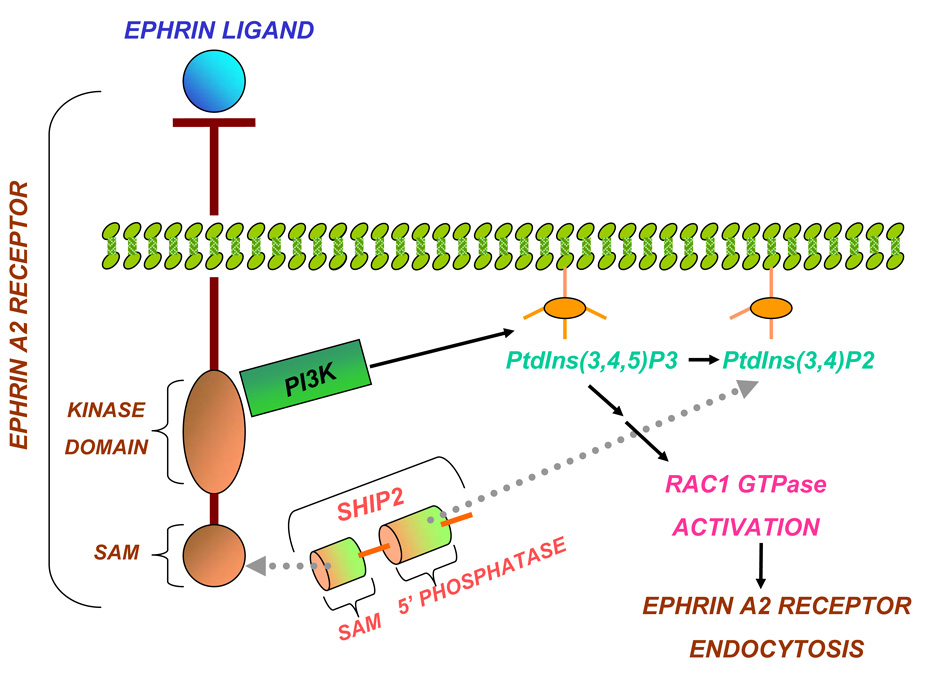
The Src homology 2 domain-containing phosphoinositide-5-phosphatase 2 (Ship2) catalyzes the conversion of phosphatidylinositol(3,4,5)P3 (PtIns(3,4,5)P3) to phosphatidylinositol(3,4)P2 (PtIns(3,4)P2), thus inhibiting processes that are activated by the phospatidylinositol 3 kinase (PI3K) (1, 2) (Figure 1). Specifically, Ship2 plays a role in insulin resistance and obesity by interfering with the phosphoinositide-dependent kinase 1 (PDK-1) activation (3, 4) and hence, it has been proposed as a potential target in drug discovery for type 2 diabetes (5). Less clear is the role of Ship2 in cancer; however, very recently a novel function for the protein has emerged as regulator of the Ephrin A2 (EphA2) receptor endocytosis (6). Since the EphA2 receptor is over-expressed in a variety of cancers (i.e: colon, lung, breast and prostate cancers) (7, 8), the processes of its endocytosis and the consequent degradation have been investigated for their potential correlation to decreased tumor malignancy (8, 9). In vitro studies, have demonstrated that Ship2 over-expression in malignant breast cancer cells reduces EphA2 receptor endocytosis while decreased levels of Ship2 facilitate receptor internalization and subsequent degradation (6), and that a heterotypic Sam-Sam domain interaction is needed to engage Ship2 to the EphA2 receptor site (6) (Figure 1).
Figure 1.
Regulation of EphA2 receptor endocytosis by Ship2. The EphA2 receptor up-regulates PtIns(3,4,5)P3 by interacting with PI3K, and induces activation of the RAC1 GTPase that in turn supports receptor endocytosis. On the contrary, Ship2, that is engaged in this cycle through a heterotypic Sam-Sam association with the EphA2 receptor, may inhibit receptor endocytosis by either reducing PtIns(3,4,5)P3 levels (model proposed by Zhuang et al. (6)) or by direct interaction with the receptor.
Here, we report on the NMR solution structure of the Sam domain of Ship2 (Ship2-Sam) and binding studies with EphA2-Sam. ITC (Isothermal titration calorimetry) experiments indicate that the two domains bind with a dissociation constant of 0.75 ± 0.12 µM and a 1:1 stoichoimetry. Chemical shift perturbation studies reveal the binding interfaces of Ship2-Sam and EphA2-Sam and show that they may adopt a canonical ML (Mid Loop)/EH (End Helix) interaction model that is characteristic of other Sam/Sam complexes (10–12). The identification of key structural elements necessary for the internalization of the EphA2 receptor provides possible routes for the development of novel small molecule antagonists with potential anti-cancer activity.
MATERIALS AND METHODS
Protein expression
Ship2-Sam and EphA2-Sam were expressed as recombinant proteins in E. coli. Synthetic genes coding for residues from 1194 to 1258 of human Ship2 (UniprotKB/TrEMBL code: O15357) and encompassing the Sam domain (residues from 1196 to 1258) were purchased from Retrogen (San Diego, CA). Genes were cloned into the PET15b plasmid and transformed using BL21-Gold (DE3) competent cells (Stratagene).
The PET15b plasmid carrying synthetic genes coding for residues from 901 to 976 of the human EphA2 receptor (Swiss-Prot/TrEMBL: P29317) and encompassing the Sam domain (residues from 904 to 968) was purchased from Celtek (Nashville, TN). PET15b plasmids carrying genes for the EphA2-Sam double mutant (H924N, R950A corresponding to H45N, R71A according to our sequence numbers) and triple mutant (K917A, R957A, Y960S i.e: K38A, R78A, Y81S according to our sequence numbers) were purchased from Celtek (Nashville, TN). All the protein constructs have N-terminal His-tag (See Supplemental data for sequence details).
Unlabeled protein expression was achieved by growing bacteria at 37 °C in LB medium until OD600=0.6. Protein over-expression was then induced by isopropyl β-D-thiogalactopyranoside (IPTG) (1 mM) for 4 hours. Expression of 15N/13C double labeled and 15N labeled proteins was carried out in M9 minimal medium supported with 2 g/l of 13C-Glucose and/or 0.5 g/l of 15NH4Cl. 10% fractional 13C labeling for stereo-specific assignments of Leu-CH3δ1,2/Val-CH3γ1,2 methyl groups (13) was achieved by supporting the M9 medium with 3.96 g of 12C-glucose (natural abundance) and 0.4 g of 13C-glucose. Selective Leu-13CH3δ1,2/Val-13CH3γ1,2 labeling was achieved by adding 70 mg/l of α-ketoisovaleric acid sodium salt (dimethyl-13C) (Cambridge Isotope Laboratories) to the M9 medium right before induction. The expression protocol for uniformly or selectively labeled proteins was identical to that used for unlabeled protein production, but in this case larger expression yields were obtained by growing bacteria overnight, after induction at 25 °C.
After expression, cells pellets were dissolved into the following buffer: 50 mM Tris (pH=8), 500 mM NaCl, 5 mM imidazole, and cells were broken by sonication. The proteins were purified on a nickel column (Amersham) by affinity chromatography with an AKTA prime plus FPLC system.
Resonance assignments of Ship2-Sam and EphA2-Sam
All the experiments for resonance assignments were recorded at 25 °C on a Bruker Avance 600 MHz spectrometer equipped with a TCI cryoprobe. NMR samples consisted of 15N or 15N/13C labeled Sam domains (800 µM) in phosphate buffer saline (PBS, 11.9 mM phosphates, 137 mM NaCl, 2.7 mM KCl) (Fisher) at pH=7.7 with 0.3% NaN3. Sample volumes of 500 µl (95% H2O/5% D2O) were used.
Backbone assignments were achieved by using standard triple resonance experiments (HNCA, HNCACB, HNCO) (14). Carbon side chains were assigned through (H)CC(CO)NH and HCCH-TOCSY experiments. Assignments for proton side chains were obtained by analysis of the HCCH-TOCSY spectrum or by comparing 3D 15N resolved-[1H, 1H] NOESY (100 ms mixing time) and 3D 15N resolved-[1H, 1H] TOCSY (70 ms mixing time) (22).
Backbone assignments for the Ship2-Sam/EphA2-Sam complex were achieved through analysis of HNCA experiments acquired on samples containing either 15N/13C double labeled Ship2-Sam at a concentration of 1 mM and unlabeled EphA2-Sam at a concentration of 2 mM or double labeled EphA2-Sam (1 mM concentration) and unlabeled Ship2-Sam (2 mM concentration) with sample volumes of 500 µl (95% H2O/5% D2O). Stereo-specific assignments for Leu-CH3δ1,2 and Val-CH3γ1,2 methyl groups of Ship2-Sam were achieved by acquiring a [1H, 13C] HSQC experiment of a fractionally 13C labeled Ship2-Sam sample at a concentration of 500 µM (13).
NMR spectra were processed with Bruker software (Topspin version 2.0) and analyzed with NEASY (15) as implemented in Cara (http://www.nmr.ch/).
Relaxation measurements
Backbone 15N nuclear spin relaxation parameters (longitudinal relaxation rates (R1) and transverse relaxation rates (R2)), were measured at 25 °C on a 600 MHz Bruker Avance DRX spectrometer equipped with a TXI probe. Experiments were performed by using a 15N-labeled sample of Ship2-Sam at a concentration of 800 µM and a sample of the Sam-Sam complex containing 15N-labeled Ship2-Sam (100 µM) and unlabeled EphA2-Sam (300 µM).
Gradient-enhanced pulse sequences were used to minimize water saturation (16). R1 and R2 relaxation data were collected as 1D experiments (2 k data points and 512 or 256 transients). Five experiments, with different values of the relaxation delay (0.01, 0.1, 0.3, 0.6, 1.0 s) were performed for R1 measurements; R2 data sets were measured with the following relaxation delays: 0.01, 0.03, 0.05, 0.07, 0.11, 0.15, 0.19 s. Average R1 and R2 values were obtained by the reduction of signal intensity as function of the relaxation delays (17). The rotational correlation time was estimated by the average R2/R1 values with the software tmest (A. G. Palmer III, Columbia University), that is based on the method by Kay et al. (18).
Ship2-Sam structure calculations and analysis
Distance constraints for structure calculations were obtained from a 3D-15N resolved [1H, 1H] NOESY-HSQC spectrum (19) (100 ms mixing time), a 3D-13C resolved [1H, 1H] NOESY-HSQC spectrum (150 ms mixing time) and a 2D [1H, 1H] NOESY (20) (100 ms mixing time), for the aliphatic to aromatic region, that was acquired after dissolving the lyophilized protein sample in 99% D2O. Structure calculations were performed with CYANA version 2.1 (21) and initiated from 100 random conformers; the 20 structures with the lowest CYANA target functions were analyzed with the programs MOLMOL (22) and PROCHECK-NMR (23). Colored figures were generated with MOLMOL (22). Surface representations were generated with MOLCAD (24) as implemented in Sybyl (TRIPOS).
NMR Binding studies
To monitor the protein-protein interactions, NMR titrations were carried out by 2D [1H, 15N]-HSQC or 2D [1H, 13C]-HSQC experiments. First, 15N-labeled Ship2-Sam (100 µM concentration) was titrated with increasing amounts of unlabeled EphA2-Sam (10, 20, 30, 40, 50, 60, 80, 100, 200, 300, 400, 500 µM); a selectively Leu-13CH3δ1,2/Val-13CH3γ1,2 Ship2-Sam sample (50 µM) was also titrated with unlabeled EphA2-Sam (50, 75, 100, 150 µM).
Additional binding studies were performed with a 15N labeled EphA2-Sam sample at a concentration of 200 µM that was titrated with unlabeled Ship2-Sam (concentrations: 100, 200, 300, 400 µM).
The program Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco) was used to generate overlays of 2D spectra.
Isothermal titration calorimetry
ITC measurements were performed on a VP-ITC apparatus (Microcal, Northampton, MA) at 25°C. For the EphA2-Sam/Ship2-Sam interaction, a solution of EphA2-Sam at a concentration of 300 µM was titrated into a solution of Ship2-Sam (25 µM concentration). Proteins were dialyzed against 4 l PBS (pH=7.7) overnight and all further dilutions of proteins and peptide for ITC were made using the leftover external dialyzate.
For peptide binding studies, the peptide
Ac-EGLVHNGWDDLEFLSDITEEDL-NH2 (Shiptide), was purchased from the Protein/DNA Facility of the Medical College of Wisconsin and dissolved in the same PBS buffer (pH=7.7). Peptide binding was monitored by titrating a 1 mM solution of peptide into a 50 µM solution of EphA2-Sam. To exclude the presence of artifacts due to ligand dilution into the protein buffer, ITC runs were also performed by titrating 300 µM EphA2-Sam into the buffer and 1 mM Shiptide into buffer. To avoid non-specific interactions between the polyHis-tag and the highly negatively charged peptide, the tail was cut from EphA2-Sam by incubation with thrombin.
Details on the ITC measurements as well as the ITC curves for the mutant EphA2-Sam proteins are reported as Supplemental data (Figures S1A, B). Data were analyzed using Microcal Origin software provided by the ITC manufacturer (Microcal, Northampton, MA).
Analytical ultracentrifugation
Sedimentation equilibrium analysis was performed with a Beckman ProteomeLab™ Optima XL-I analytical ultracentrifuge. Three runs were carried out by using samples with concentrations of 0.45 mg/ml, 0.15 mg/ml, and 0.05 mg/ml, respectively. The data were collected at 20 °C at the angular velocity of 30,000 rpm. The software HeteroAnalysis (James L. Cole; http://www.biotech.uconn.edu/auf/) was used to analyze the data.
Docking studies
A model of the Ship2-Sam/EphA2-Sam complex was generated with the program Haddock 1.3 (25) by using the NMR structures number one of both Ship2-Sam and EphA2-Sam. Ambiguous interaction restraints were obtained from the chemical shift perturbation and mutagenesis data. Residues H47, N48, W50, E54, F55, D58 of Ship2-Sam and K38, R67, R71, G74, R77, Y81 of EphA2-Sam were considered active. Active residues were chosen among the ones with the greatest normalized chemical shift deviations (Δδ >0.1 ppm) because they either present high solvent exposure (as determined with MOLMOL (22)) or because they could provide potential interactions at the dimer interface as indicated by analysis of experimental structures of hetero Sam-Sam complexes. For the AIR restraints the upper distance limit default value (i.e: 2 Å) was kept (25). Flexible interfaces were made up of residues 44–71 and 74–81 of Ship2-Sam and EphA2-Sam respectively. In the first iteration (i.e: the rigid body energy minimization) 2000 structures were calculated; in the second iteration the best 200 solutions were subjected to semi-flexible simulated annealing, a final refinement in water was also performed. The final 200 structures were clustered using a rmsd (root-mean-square deviation) cut-off value of 2 Å by using the “cluster_struc” macro of the Haddock program (25). Four clusters were obtained and the best structures of each cluster (according to the Haddock score (25)) were compared. Since the structures resulted very similar to each other, we decided to use the one with the lowest (i.e best) Haddock score as representative of the Ship2-Sam/EphA2-Sam complex.
RESULTS
NMR Solution structure of Ship2-Sam
To investigate the aggregation state of Ship2-Sam in solution, we measured backbone 15N R1 and R2 nuclear spin relaxation rates for the protein in its free and bound forms and performed analytical ultracentrifugation studies.
The rotational correlation time, τc, of Ship2-Sam, estimated by the R2/R1 average value resulted to be 6.7 ± 0.4 ns at a protein concentration of 800 µM. The τc of Ship2-Sam bound to EphA2-Sam increased to 11.2 ± 0.5 ns, as expected for the increased molecular weight of the complex.
Analytical ultracentrifugation measurements showed that the Ship2-Sam tendency to self-associate was very weak in solution (Kd= 2.0 ± 0.7 mM, for the dimerization process). Moreover, no changes were revealed in [1H, 15N]-HSQC spectra of Ship2-Sam by increasing the concentration from 70 to 800 µM. These data indicated that at the concentration used to calculate the Ship2-Sam NMR structure (800 µM), the protein was essentially monomeric in solution.
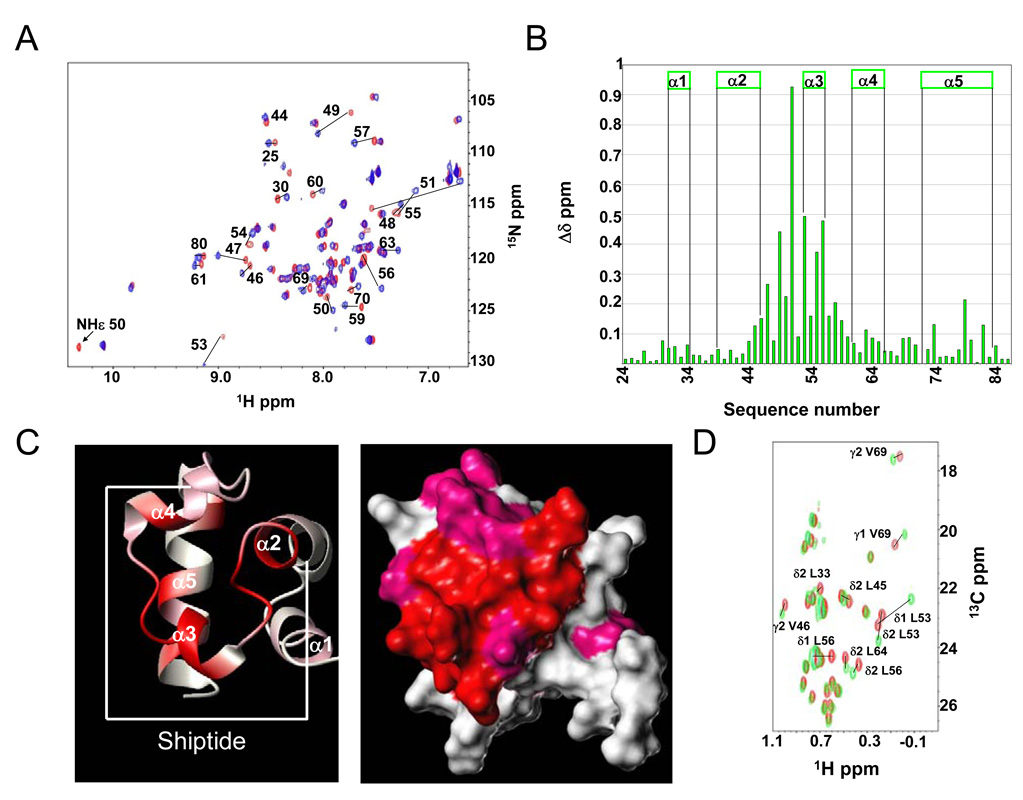
The Ship2-Sam solution structure is shown in Figure 2, while key structural parameters are listed in Table 1. The small number of residual violations (Table 1) indicates that the constraints are well satisfied in the calculated conformers. The low root-mean-square deviation (rmsd) values, calculated for the residues of the Sam domain (Table 1), demonstrate the high precision of the structure determination. The structure represents a canonical Sam domain fold (Figure 2).
Figure 2.
Left panel. Overlay on the backbone atoms (residues 29–85) of 20 Ship2-Sam NMR conformers. The final structure calculation includes 989 non redundant distance constraints (263 intra-residue, 240 short-range, 242 medium-range, 244 long-range) and 282 angle constraints. Right panel. Ribbon drawing of one Ship2-Sam NMR conformer (number one), it contains the following secondary structure elements: α1 (residues 30–34), α2 (residues 39–46), α3 (residues 53–56), α4 (residues 61–66), α5 (residues 72–84).
Table 1.
Structure statistics for the NMR ensemble of Ship2-Sam
| NOE upper distance limits | 989 |
| Angle constraints | 282 |
| Residual target function, Å2 | 1.08±0.13 |
| Residual NOE violations | |
| Number > 0.1 Å# | 3 |
| Maximum, Å | 0.29±0.07 |
| Residual angle violations | |
| Number | 0 |
| Atomic pairwise RMSD, Å | |
| Backbone atoms (aa 29–85) |
0.25±0.06 |
| Heavy atoms (aa 29–85) |
0.77±0.08 |
| Ramachandran analysis@ | |
| Residues in core regions | 85.4% |
| Residues in allowed regions | 14.4% |
| Residues in generous regions | 0.1% |
| Residues in disallowed regions | 0.1% |
Ship2-Sam/EphA2-Sam interaction studies
Binding of Ship2-Sam to EphA2-Sam was monitored by means of chemical shift perturbation studies (26, 27). First, 2D [1H, 15N]-HSQC experiments were recorded for a 15N uniformly labeled Ship2-Sam sample in presence and absence of unlabeled EphA2-Sam (Figure 3A). Normalized chemical shifts variations were then calculated according to the equation Δδ = [(ΔHN)2 + (0.17 * Δ15N)2]1/2 (Figure 3B) (28). The greatest Δδ (values > 0.1 ppm) were found in the middle region of the protein, including the α3 helix, the C-terminal portion of α2 helix and N-terminal part of the α4 helix (Figures 3B, C). To confirm these mapping data, 2D [1H, 13C]-HSQC spectra of a selectively Leu-13CH3δ1,2/Val-13CH3γ1,2 labeled Ship2-Sam sample, were also recorded for the apo protein and after addition of unlabeled EphA2-Sam (Figure 3D). In agreement with what we had previously observed in the experiments with 15N labeled protein, the side chains of leucine and valine residues localized in the middle part of Ship2-Sam (i.e α3 helix and nearby regions) induced largest chemical shifts variations upon complex formation (Figure 3D).
Figure 3.
A. Comparison of [1H, 15N]-HSQC spectra of Ship2-Sam (100 µM) in its apo form (red) and after addition of EphA2-Sam (400 µM) (blue). B. Histogram showing normalized chemical shift deviations (Δδ=[(ΔHN)2 + (0.17 * Δ15N)2]1/2) as function of the residue number. Residues L45, V46, H47, G49, W50, D51, L53, E54, F55, L56, S57, D58, I59, D63, H74, L79, L82 present normalized deviations with values higher that 0.1 ppm. Δδ values in between 0.05 and 0.1 ppm are observed for the following residues: S30, A31, W32, R34, G44, N48, D52, T60, E61, L64, E65, E66, A67, V69, Q70, D71, D80. C. Residues with normalized chemical shifts deviations (Δδ values) greater than 0.1 ppm and included in between 0.05 and 0.1 are colored in red and pink respectively on the 3D solution structure of Ship2-Sam (conformer number 1) in its ribbon (left panel) and surface (right panel) representations. The peptide region used for our studies is highlighted in the left panel. D. [1H, 13C]-HSQC spectra of selective Leu-13CH3δ1,2/Val-13CH3γ1,2 labeled Ship2-Sam (50 µM) in absence (red) and presence (green) of EphA2-Sam (150 µM).
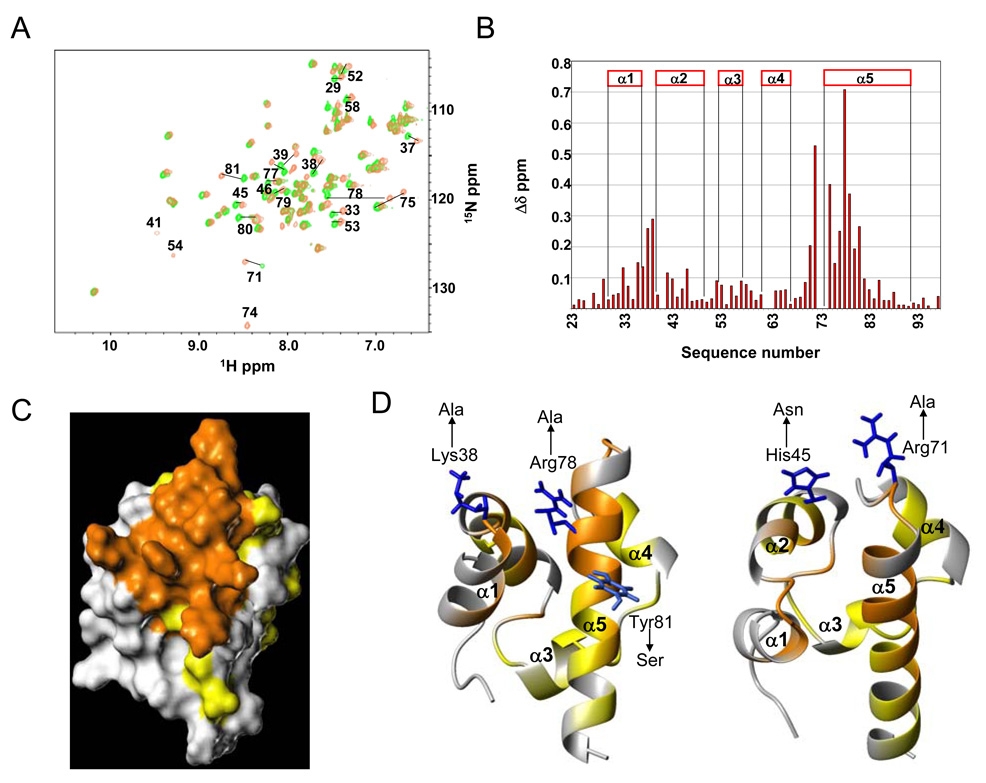
To map the binding surface of EphA2-Sam for Ship2-Sam, similar titration experiments were performed with 15N labeled EphA2-Sam and unlabeled Ship2-Sam (Figure 4A). Upon binding to Ship2-Sam, several changes occurred in the spectrum of EphA2-Sam (Figure 4A). We evaluated normalized chemical shift deviations to identify the residues of the receptor that were most affected by the interaction (Figure 4B). The largest deviations were localized at the interface between the N-terminal segment of the α5 helix and the adjacent α1α2 and α4α5 loop regions (Figures 4C, D).
Figure 4.
A. Superposition of [1H, 15N]-HSQC spectra of EphA2-Sam (200 µM concentration) in its unbound form (green) and after complex formation with Ship2-Sam (400 µM) (brown). B. Plot of normalized chemical shift deviations (Δδ=[(ΔHN)2 + (0.17 * Δ15N)2]1/2) versus residue number. The highest normalized chemical shift deviations (Δδ values > 0.1 ppm) are observed for residues W33, I37, K38, M39, Y42, F46, R71, L72, H75, Q76, K77, R78, I79, A80, Y81. Minor changes (0.05 < Δδ values < 0.1 ppm) affect amino acids T29, L34, T43, H45, T52, A53, E55, V57, V58, Q59, T61, D64, I65, K66, S82, L83, G85, D88. Data are not shown for residues N62 and D63 (unassigned); G74, Q41 (their peaks only appear in the spectrum of the complex). C. Surface representation of EphA2-Sam (conformer number 1, pdb i.d: 2E8N, from RIKEN Structural Genomics Initiative) (in the same orientation as Figure 4D-left panel), residues exhibiting the greatest normalized chemical shift deviations (Δδ values > 0.1 ppm) are colored in orange, those presenting minor changes (0.05 < Δδ values < 0.1 ppm) are reported in yellow. D. EphA2-Sam mutants. Ribbon representation of EphA2-Sam (colored as in the 4C panel). The side chains of the mutated residues are also shown together with the corresponding mutations.
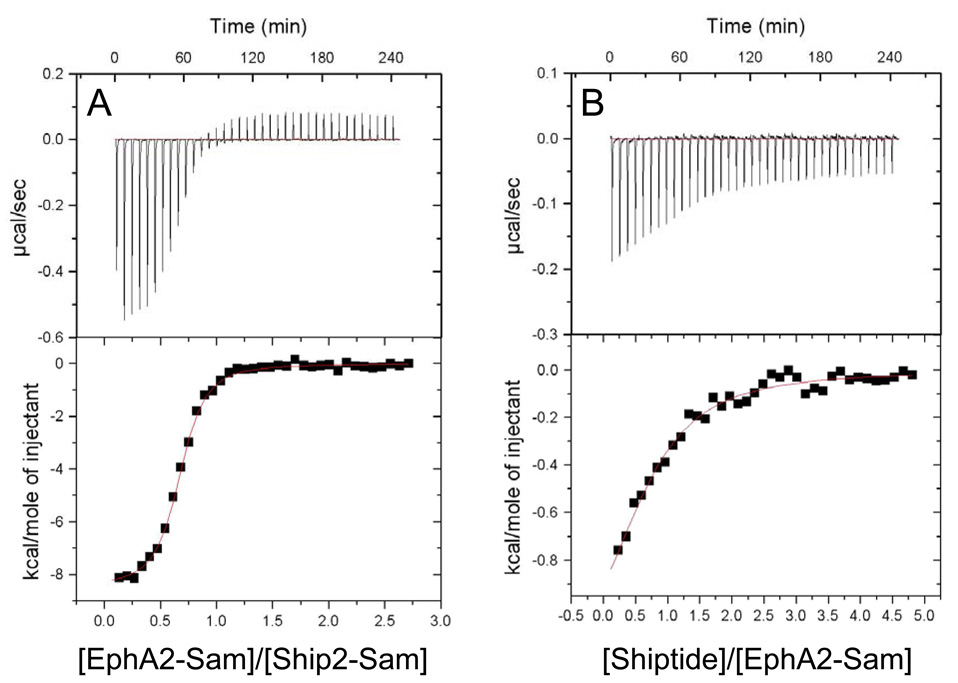
Finally, ITC experiments were performed to determine the dissociation constant of the complex. Several ITC runs were carried and indicated that the two Sam domains interact with a Kd value of 0.75 ± 0.12 µM and a 1:1 binding stoichoimetry (Figures 5A).
Figure 5.
ITC studies. A. The raw and integrated data are shown in the upper and lower panels respectively. In the lower panel, solid squares represent data from the EphA2-Sam (300 µM) titration into Ship2-Sam solution (25 µM); the solid red line represents the fit to a single binding site model. B. Calorimetric curve representing the EphA2-Sam (50 µM) titration with the Shiptide peptide (1 mM).
In order to further validate the chemical shift mapping studies, we investigated whether a minimal peptide region of Ship2-Sam (Ac-EGLVHNGWDDLEFLSDITEEDL-NH2; Shiptide) could still interact with EphA2-Sam. This peptide encompasses the region of Ship2-Sam that includes the α3 helix together with part of the C-terminus of α2 and N-terminus of α4 (aa 43–64) (Figure 3C, left panel). ITC measurements showed peptide binding to EphA2-Sam with a Kd= 19 ± 3 µM and a single binding site model (Figure 5B).
Additional insights on the binding mode of EphA2-Sam to Ship2-Sam were derived by further NMR and ITC studies with EphA2-Sam mutants. Two mutants were designed based on the chemical shift mapping studies (Figure 4D) and sequence alignment data with other Sam domains (Figure S2). The triple mutant (K38A, R78A, Y81S) failed to bind Ship2-Sam whereas the double mutant (H45N, R71A) retained its ability to bind Ship2-Sam with a Kd= 1.8 ± 0.6 µM (Figures S1A, B). We chose to mutate K38, R71, R78 to alanine in order to perturb crucial electrostatic interactions at the dimer interface; however, we also wanted to avoid introducing perturbations that could disrupt the fold of the Sam domain. Hence, residues Y81 and H45 were mutated to serine and asparagine respectively since these substitutions seem allowed in Sam domains from other Ephrin receptors.
Finally, a model of the Ship2-Sam/EphA2-Sam complex was obtained by docking studies with the software Haddock 1.3 (25) (See Materials and Methods section). A representative solution is shown in Figure 6. In this model, acidic residues of Ship2-Sam and basic residues of EphA2-Sam are providing a dense network of hydrogen-bond and electrostatic interactions at the dimer interface (Figure 6).
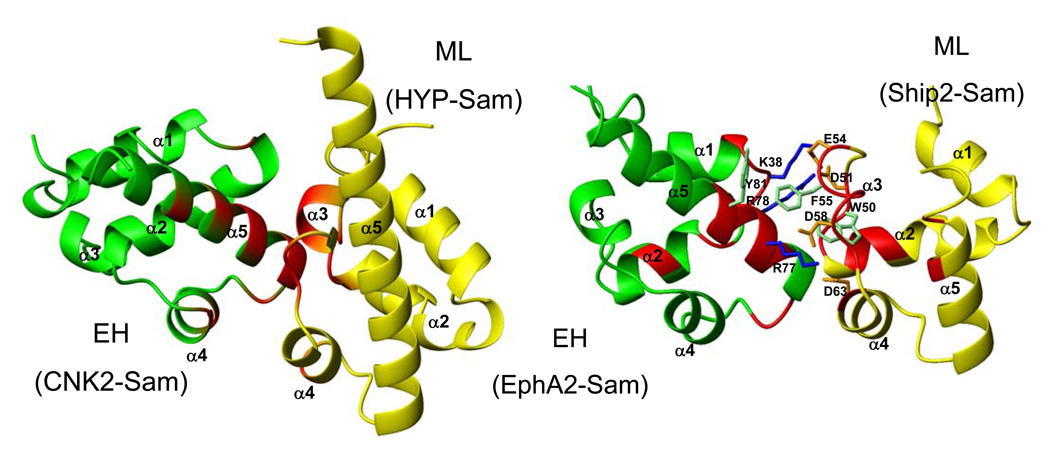
Figure 6.
Left panel. Crystal structure of the complex between CNK2-Sam (green) and HYP-Sam (yellow) (pdb code: 3BS5 (11)). Residues contributing to the ML and EH interfaces (11) have been colored in red on the ribbon representations of the two molecules. Right panel. Model, generated with Haddock 1.3 (25) of the Ship2-Sam/EphA2-Sam complex (solution with the lowest Haddock score). Residues with the largest chemical shift perturbations, according to our NMR binding studies, have been reported in red. The side chains of amino acids which are involved in interactions at the dimer interface are also shown.
DISCUSSION
The EphA2 receptor has recently gained much attention by both academic and industrial laboratories as a potential target for drug discovery in cancer. The processes of receptor endocytosis and consequent degradation are being investigated as potential routes to reduce cancer progression (9). Previous studies have reported on the regulatory influence of the lipid phosphatase Ship2 in receptor endocytosis (Figure 1) (6). Ship2 exerts its function through recruitment at the EphA2 receptor site by means of a heterotypic Sam-Sam association (6). Due to the lack of structural information on the Ship2-Sam domain and its mode of binding to EphA2-Sam, we have carried out structural and binding studies by means of NMR spectroscopy and ITC experiments.
Structural details of the Ship2-Sam/EphA2-Sam interaction
Solution and X-ray structures of several Sam domains have been reported thus far (29–33). A “blastp” search (34) of the Ship2-Sam sequence against the PDB database reveals highest identities (34) with EphB4-Sam (pdb id: 2QKQ, Structural genomics Consortium) and EphA4-Sam (pdb id: 1B0X, (35)) which share with Ship2-Sam 38% and 28% sequence identities, respectively. A similar blastp search for the EphA2-Sam sequence indicates that it presents high homology with Sam domains from other ephrin receptors. Accordingly, the 3D structure of Ship2-Sam and EphA2-Sam (pdb i.d: 2E8N, Riken Structural Genomics Initiative) consist of canonical Sam domains.
Sam domains may self-associate very weakly in solution, as it has been previously demonstrated by analytical ultracentrifugation studies carried out for Sam domains from some of the ephrin receptors, including EphA2-Sam (36).
Gel filtration and GST pull down assays have already pointed out that Ship2-Sam shows a preference for heterotypic associations (37). Our 15N relaxation measurements indicate for Ship2-Sam a τc equal to 6.7 ns at a protein concentration of 800 µM that is indeed similar to the τc value reported for other Sam domains which have been shown to be monomeric in solution, such as Ets-1-Sam (6.9 ns) (38), DLC2-Sam (7.2 ns) (32), and EphB2-Sam (6 ns) (39). Thus, in agreement with these previous findings on other Sam domains, our results indicate that Ship2-Sam is mainly monomeric in solution as further confirmed by analytical ultracentrifugation data.
The correlation time τc for Ship2-Sam in complex with EphA2-Sam is 11.2 ± 0.5 ns that is a value compatible with that of proteins with a molecular weight of about 15 kDa and close to that measured for the dimeric Ste11-Sam (i.e: 10 ± 0.2 ns) (40), thus suggesting that EphA2-Sam and Ship2-Sam may form a dimer at the condition used in our NMR experiments; the presence of high order oligomers can be excluded also because of the very high quality of the NMR spectra for the complex.
The interaction between EphA2-Sam and Ship2-Sam has been investigated by means of ITC measurements and chemical shift perturbation studies. ITC data show that the Sam domain of Ship2 binds to the Sam domain of the EphA2 receptor with high affinity (Kd= 0.75 ± 0.12 µM) with a single binding site model (Figure 5A).
Recent studies have shown that the Sam domain of Ship2 is also able to interact with the Sam domain of the PI3K effector protein Arap3 (37). Recently reported ITC measurements show that Arap3-Sam binds to Ship2-Sam by forming a dimer (Kd= 100 nM and a binding stoichoimetry of 1:1) (37). The comparison between these data and our results demonstrates that Ship2-Sam is able to interact with both Arap3-Sam and EphA2-Sam with similar binding affinities and the same binding stoichoimetry.
By having NMR assignments and 3D structures of both EphA2-Sam (i.d: 2E8N, from RIKEN Structural Genomics Initiative) and Ship2-Sam, it has been possible to carry out more detailed NMR binding studies.
The binding region on Ship2-Sam for EphA2-Sam is located in the middle part of the molecule and contains mainly negatively charged and hydrophobic residues (Figure 6 and Figure S2) including a tryptophan, whose side chains is affected by major chemical shift changes upon complex formation (Figure 3A), and a solvent-exposed phenylalanine (Figure 6).
The putative EphA2-Sam binding interface for Ship2-Sam is presented in Figure 4 and includes the N-terminal part of α5 and the closest α1α2 and α4α5 loops. This binding region on EphA2-Sam has been further confirmed by mutagenesis studies, indicating that the triple mutant K38A, R78A and Y81S is not effective in binding Ship2-Sam (Figure 4D and Figure S1A).
The role of the N-terminal arm and the C-terminal α5 helix in directing homotypic Sam-Sam interactions has already been pointed out for both the dimeric crystal structure of EphA4-Sam (35) and the oligomeric crystal structure of EphB2-Sam (41). Besides, studies on different Sam domains have confirmed the important role of the C-terminal α5 helix which seems to be a common motif used by Sam domains to associate in different manners (40). In fact, for several homotypic (12, 41, 42, 43) and heterotypic (11, 44) Sam-Sam complexes, the interactions in between monomers occur through the Mid-Loop (ML) surface, where residues from the regions close to the middle portion of the protein are providing the contact surface of one subunit, and the End-Helix (EH) surface which involves mainly the α5 helix of another subunit and adjacent loop regions. Analysis of chemical shift mapping data indicates indeed that Ship2-Sam and EphA2-Sam may adopt the ML/EH binding topology. A model of the ML/EH interaction for Ship2-Sam/EphA2-Sam (Figure 6) has been generated by molecular docking and is indeed in good agreement with our experimental data. In fact, the Ship2-Sam sequence encompassing the Shiptide region (Figure 3C) is mainly providing the ML interface, while the α5 helix and nearby loop regions are proving the EH interface of EphA2-Sam (Figure 6). Phe55 of Ship2-Sam provides interactions with Tyr81 of EphA2-Sam, while Lys38 and Arg78 on the receptor are involved in electrostatic interactions with Asp51 and Glu54 (Figure 6), thus explaining our results with the EphA2-Sam triple mutant.
This model resembles closely the structures of other Sam-Sam complexes which adopt the ML/EH binding mode (Figure 6).
In conclusion, the structural information we have obtained (i.e: the 3D solution structure of Ship2-Sam, the identification of its putative binding mode to EphA2-Sam and the detection of a minimal Ship2-Sam binding peptide) paves the way for future studies aiming at the development of small-molecules inhibitors of this interaction which could be further investigated for their effects in tumor cells over-expressing the EphA2 receptor.
Supplementary Material
Details about chemical shift perturbation and ITC studies with EphA2-Sam mutants; sequence alignment of Sam domains from Ship2 and from different Ephrin receptors; protein sequences used in this study. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENT
We thank J. Ryan for cloning the Sam domain of Ship2 into the PET15b plasmid; Dr. Jinghua Yu and Dr. Andrey Bobkov of the Burnham Institute NMR and protein Facilities for technical assistance. We also thank Dr. A. M. J. J Bonvin for providing us with Haddock 1.3.
Abbreviations
- DLC2
deleted in Liver Cancer 2
- EH
End-Helix
- EphA2
Ephrin A2
- EphA4
Ephrin A4
- EphB2
Ephrin B2
- EphA2-Sam
Sam domain of the EphA2 receptor
- FPLC
Fast Performance Liquid Chromatography
- HSQC
Heteronuclear Single Quantum Coherence Spectroscopy
- IPTG
isopropyl β-D-thiogalactopyranoside
- ITC
isothermal titration calorimetry
- MD
Mid-Loop
- NOESY
Nuclear Overhauser Enhancement Spectroscopy
- PBS
phosphate buffer saline
- PDK1
3-phosphoinositide-dependent protein kinase-1
- PI3K
phospatidylinositol 3 kinase
- PtIns(3,4,5)P3
phosphatidylinositol(3,4,5)P3
- rmsd
root-mean-square deviation
- Sam
Sterile Alpha Motif
- Ship2
Src homology 2 domain-containing phosphoinositide-5-phosphatase 2
- Ship2-Sam
Sam domain of Ship2
- TOCSY
Total Correlation Spectroscopy.
Footnotes
Financial support was provided by NIH grant CA102583
NMR structures have been deposited in the Protein Data Bank under accession code 2K4P.
REFERENCES
- 1.Pesesse X, Moreau C, Drayer AL, Woscholski R, Parker P, Erneux C. The SH2 domain containing inositol 5-phosphatase SHIP2 displays phosphatidylinositol 3,4,5-trisphosphate and inositol 1,3,4,5-tetrakisphosphate 5-phosphatase activity. FEBS Lett. 1998;437(3):301–303. doi: 10.1016/s0014-5793(98)01255-1. [DOI] [PubMed] [Google Scholar]
- 2.Pesesse X, Deleu S, De Smedt F, Drayer L, Erneux C. Identification of a second SH2-domain-containing protein closely related to the phosphatidylinositol polyphosphate 5-phosphatase SHIP. Biochem Biophys Res Commun. 1997;239(3):697–700. doi: 10.1006/bbrc.1997.7538. [DOI] [PubMed] [Google Scholar]
- 3.Clement S, Krause U, Desmedt F, Tanti JF, Behrends J, Pesesse X, Sasaki T, Penninger J, Doherty M, Malaisse W, Dumont JE, Le Marchand-Brustel Y, Erneux C, Hue L, Schurmans S. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature. 2001;409(6816):92–97. doi: 10.1038/35051094. [DOI] [PubMed] [Google Scholar]
- 4.Sleeman MW, Wortley KE, Lai KM, Gowen LC, Kintner J, Kline WO, Garcia K, Stitt TN, Yancopoulos GD, Wiegand SJ, Glass DJ. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat Med. 2005;11(2):199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]
- 5.Lazar DF, Saltiel AR. Lipid phosphatases as drug discovery targets for type 2 diabetes. Nat Rev Drug Discov. 2006;5(4):333–342. doi: 10.1038/nrd2007. [DOI] [PubMed] [Google Scholar]
- 6.Zhuang G, Hunter S, Hwang Y, Chen J. Regulation of EphA2 receptor endocytosis by SHIP2 lipid phosphatase via phosphatidylinositol 3-Kinase-dependent Rac1 activation. J Biol Chem. 2007;282(4):2683–2694. doi: 10.1074/jbc.M608509200. [DOI] [PubMed] [Google Scholar]
- 7.Walker-Daniels J, Hess AR, Hendrix MJ, Kinch MS. Differential regulation of EphA2 in normal and malignant cells. Am J Pathol. 2003;162(4):1037–1042. doi: 10.1016/S0002-9440(10)63899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ireton RC, Chen J. EphA2 receptor tyrosine kinase as a promising target for cancer therapeutics. Curr Cancer Drug Targets. 2005;5(3):149–157. doi: 10.2174/1568009053765780. [DOI] [PubMed] [Google Scholar]
- 9.Carles-Kinch K, Kilpatrick KE, Stewart JC, Kinch MS. Antibody targeting of the EphA2 tyrosine kinase inhibits malignant cell behavior. Cancer Res. 2002;62(10):2840–2847. [PubMed] [Google Scholar]
- 10.Ramachander R, Bowie JU. SAM domains can utilize similar surfaces for the formation of polymers and closed oligomers. J Mol Biol. 2004;342(5):1353–1358. doi: 10.1016/j.jmb.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Rajakulendran T, Sahmi M, Kurinov I, Tyers M, Therrien M, Sicheri F. CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling. Proc Natl Acad Sci U S A. 2008;105(8):2836–2841. doi: 10.1073/pnas.0709705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada BT, Knight MJ, Imai S, Qiao F, Ramachander R, Sawaya MR, Gingery M, Sakane F, Bowie JU. Regulation of enzyme localization by polymerization: polymer formation by the SAM domain of diacylglycerol kinase delta1. Structure. 2008;16(3):380–387. doi: 10.1016/j.str.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28(19):7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 14.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR. 1993;3(2):185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 15.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 16.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33(19):5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 17.Viles JH, Duggan BM, Zaborowski E, Schwarzinger S, Huntley JJ, Kroon GJ, Dyson HJ, Wright PE. Potential bias in NMR relaxation data introduced by peak intensity analysis and curve fitting methods. J Biomol NMR. 2001;21(1):1–9. doi: 10.1023/a:1011966718826. [DOI] [PubMed] [Google Scholar]
- 18.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28(23):8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 19.Talluri S, Wagner G. An optimized 3D NOESY-HSQC. J Magn Reson B. 1996;112(2):200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A, Ernst RR, Wuthrich K. A 2D nuclear Overhauser enhancement (2D NOE) experiment for elucidation of complete proton-proton cross relaxation networks in biological macromolecules. Biochm. Biophys. Res. Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319(1):209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 22.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(1):51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 24.Teschner M, Henn C, Vollhardt H, Reiling S, Brickmann J. Texture mapping: a new tool for molecular graphics. J Mol Graph. 1994;12(2):98–105. doi: 10.1016/0263-7855(94)80074-x. [DOI] [PubMed] [Google Scholar]
- 25.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 26.Pellecchia M. Solution nuclear magnetic resonance spectroscopy techniques for probing intermolecular interactions. Chem Biol. 2005;12(9):961–971. doi: 10.1016/j.chembiol.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 27.Pellecchia M, Sem DS, Wuthrich K. NMR in drug discovery. Nat Rev Drug Discov. 2002;1(3):211–219. doi: 10.1038/nrd748. [DOI] [PubMed] [Google Scholar]
- 28.Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Biol. 1996;3(12):995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 29.Kwan JJ, Donaldson LW. The NMR structure of the murine DLC2 SAM domain reveals a variant fold that is similar to a four-helix bundle. BMC Struct Biol. 2007;7(1):34. doi: 10.1186/1472-6807-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cicero DO, Falconi M, Candi E, Mele S, Cadot B, Di Venere A, Rufini S, Melino G, Desideri A. NMR structure of the p63 SAM domain and dynamical properties of G534V and T537P pathological mutants, identified in the AEC syndrome. Cell Biochem Biophys. 2006;44(3):475–489. doi: 10.1385/CBB:44:3:475. [DOI] [PubMed] [Google Scholar]
- 31.Edwards TA, Butterwick JA, Zeng L, Gupta YK, Wang X, Wharton RP, Palmer AG, 3rd, Aggarwal AK. Solution structure of the Vts1 SAM domain in the presence of RNA. J Mol Biol. 2006;356(5):1065–1072. doi: 10.1016/j.jmb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Fung KL, Jin DY, Chung SS, Ching YP, Ng IO, Sze KH, Ko BC, Sun H. Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2. Proteins. 2007;67(4):1154–1166. doi: 10.1002/prot.21361. [DOI] [PubMed] [Google Scholar]
- 33.Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat Struct Mol Biol. 2006;13(2):160–167. doi: 10.1038/nsmb1038. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stapleton D, Balan I, Pawson T, Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat Struct Biol. 1999;6(1):44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- 36.Thanos CD, Faham S, Goodwill KE, Cascio D, Phillips M, Bowie JU. Monomeric structure of the human EphB2 sterile alpha motif domain. J Biol Chem. 1999;274(52):37301–37306. doi: 10.1074/jbc.274.52.37301. [DOI] [PubMed] [Google Scholar]
- 37.Raaijmakers JH, Deneubourg L, Rehmann H, de Koning J, Zhang Z, Krugmann S, Erneux C, Bos JL. The PI3K effector Arap3 interacts with the PI(3,4,5)P(3) phosphatase SHIP2 in a SAM domain-dependent manner. Cell Signal. 2007;19(6):1249–1257. doi: 10.1016/j.cellsig.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 38.Slupsky CM, Gentile LN, Donaldson LW, Mackereth CD, Seidel JJ, Graves BJ, McIntosh LP. Structure of the Ets-1 pointed domain and mitogen-activated protein kinase phosphorylation site. Proc Natl Acad Sci U S A. 1998;95(21):12129–12134. doi: 10.1073/pnas.95.21.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smalla M, Schmieder P, Kelly M, Ter Laak A, Krause G, Ball L, Wahl M, Bork P, Oschkinat H. Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites. Protein Sci. 1999;8(10):1954–1961. doi: 10.1110/ps.8.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharjya S, Xu P, Gingras R, Shaykhutdinov R, Wu C, Whiteway M, Ni F. Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: implications for Ste11 activation and signal transmission through the Ste50-Ste11 complex. J Mol Biol. 2004;344(4):1071–1087. doi: 10.1016/j.jmb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Thanos CD, Goodwill KE, Bowie JU. Oligomeric structure of the human EphB2 receptor SAM domain. Science. 1999;283(5403):833–836. doi: 10.1126/science.283.5403.833. [DOI] [PubMed] [Google Scholar]
- 42.Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, Faham S, Bowie JU. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20(15):4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim CA, Gingery M, Pilpa RM, Bowie JU. The SAM domain of polyhomeotic forms a helical polymer. Nat Struct Biol. 2002;9(6):453–457. doi: 10.1038/nsb802. [DOI] [PubMed] [Google Scholar]
- 44.Qiao F, Song H, Kim CA, Sawaya MR, Hunter JB, Gingery M, Rebay I, Courey AJ, Bowie JU. Derepression by depolymerization; structural insights into the regulation of Yan by Mae. Cell. 2004;118(2):163–173. doi: 10.1016/j.cell.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 45.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details about chemical shift perturbation and ITC studies with EphA2-Sam mutants; sequence alignment of Sam domains from Ship2 and from different Ephrin receptors; protein sequences used in this study. This material is available free of charge via the Internet at http://pubs.acs.org.