Abstract
Recently, a genomic distance-based regression for multilocus associations was proposed (Wessel and Schork [2006] Am. J. Hum. Genet. 79:792–806) in which either locus or haplotype scoring can be used to measure genetic distance. Although it allows various measures of genomic similarity and simultaneous analyses of multiple phenotypes, its power relative to other methods for case-control analyses is not well known. We compare the power of traditional methods with this new distance-based approach, for both locus-scoring and haplotype-scoring strategies. We discuss the relative power of these association methods with respect to five properties: (1) the marker informativity; (2) the number of markers; (3) the causal allele frequency; (4) the preponderance of the most common high-risk haplotype; (5) the correlation between the causal single-nucleotide polymorphism (SNP) and its flanking markers. We found that locus-based logistic regression and the global score test for haplotypes suffered from power loss when many markers were included in the analyses, due to many degrees of freedom. In contrast, the distance-based approach was not as vulnerable to more markers or more haplotypes. A genotype counting measure was more sensitive to the marker informativity and the correlation between the causal SNP and its flanking markers. After examining the impact of the five properties on power, we found that on average, the genomic distance-based regression that uses a matching measure for diplotypes was the most powerful and robust method among the seven methods we compared.
Keywords: canonical correlation, genomic distance, genotyping errors, linkage disequilibrium
INTRODUCTION
With the advent of high-throughput single-nucleotide polymorphism (SNP) data, using an appropriate statistical method to test the association between a large number of genetic markers and disease is of major interest. For case-control data, a multilocus association analysis is commonly used to evaluate the simultaneous association of several genetic markers with a trait. There are two broad categories of test statistics: locus-scoring methods that test only the main effects of markers, and haplotype-scoring methods that test haplotype effects. With binary traits for case-control data, logistic regression is widely used for modeling the association between markers and disease [Cordell and Clayton, 2002; Chapman et al., 2003]. Cordell and Clayton [2002] proposed a stepwise regression approach applicable to either case-control data or nuclear-family data, with case-control data modeled via unconditional and family data via conditional logistic regression. Chapman et al. [2003] derived a multilocus score test and showed that the power of association tests is determined by two coefficients of determination: that between markers and the unobserved causal locus and that between the causal locus and the trait. They also found that in most cases, the simplest method of scoring (locus coding), which does not require haplotype phase of the markers, is generally more powerful than scoring analyses that include haplotype information. The locus-scoring analysis has the merits of lower complexity and possibly fewer degrees of freedom for either the likelihood ratio test or the score test, potentially, providing greater power.
For a haplotype-scoring approach, Schaid et al. [2002] developed a score statistic for generalized linear models that tests the association between haplotypes and a wide variety of traits, including binary, ordinal, and quantitative traits. This haplotype-scoring approach can provide critical information regarding the function of a gene and can increase power when several mutations within a single gene interact to create a “super allele” that has a large effect on a disease. However, the phase ambiguity and the larger number of degrees of freedom usually compromise the potential gain in power. A similar haplotype approach, proposed by Zaykin et al. [2002], uses a likelihood ratio test to detect the association of haplotypes with discrete and continuous traits in samples of unrelated individuals. Schaid et al. [2002] and Zaykin et al. [2002] utilized the expectation-maximization (EM) algorithm to estimate the posterior distribution of pairs of haplotypes for each subject, yet power loss can occur due to haplotype phase uncertainty [Zaykin et al., 2002]. To design studies to test associations of haplotypes with traits, Schaid [2006] provided guidance to determine the sample size needed to achieve the desired power for a study. His derivations covered both phase-known and phase-unknown haplotypes, allowing evaluation of the loss of efficiency due to unknown phase.
Although haplotype-scoring approaches can be more powerful in some situations, Clayton et al. [2004] found that locus scoring of multiple tag-SNPs, not based on haplotypes, can be more powerful than analyses based on haplotypes. To investigate the ability of detecting association by locus-scoring and haplotype-scoring analyses, North et al. [2006] simulated data based on the patterns of linkage disequilibrium (LD) from the HapMap project [The International HapMap Consortium, 2003]. They used one to four markers surrounding the putative susceptibility locus as the marker sets and found that there was little difference in the performance of locus-scoring and haplotype-scoring methods.
In addition to the above methods, another branch of multilocus association methods uses genotype similarity for pairs of subjects, where genotype can span unphased markers as well as haplotype information. Here, the analyzed unit is a pair of subjects instead of a single subject. Several researchers have proposed statistics based on the excess in similarities among haplotypes in affected individuals [van der Meulen and Te Meerman, 1997; Cheung and Nelson, 1998; Grant et al., 1999; Devlin et al., 2000; Bourgain et al., 2001]. Tzeng et al. [2003a,b] and Schaid et al. [2005] proposed statistics to detect association by contrasting within-group similarities of cases and controls. Simulation studies have shown that neither Pearson's χ2 statistic nor the similarity-based method is uniformly more powerful than the other [Tzeng et al., 2003a]. The power of these two methods can be very different, depending on the population evolutionary history [Yuan et al., 2006]. The contrast of within-group similarities relies on a test statistic, T = δ/√Var(δ), where the numerator contrasts the within-group similarities, δ = p′Πsp − q′Πsq, and where p and q are vectors of the haplotype frequencies (or genotype frequencies in Schaid et al. [2005]) for the case and control samples, respectively; ΠS is a matrix of similarity measures among haplotypes or genotypes. How to determine an optimal genotype similarity was addressed by Schaid et al. [2005]. With one degree of freedom, the global U-statistic provided by Schaid et al. [2005], which combines information from multiple markers, does not appear to suffer from weak power, which plagues other methods with a large number of degrees of freedom.
A limitation of contrasting within-group similarities [Tzeng et al., 2003a,b; Schaid et al., 2005], however, is that there can exist large differences between p and q, but δ = p′Πsp − q′Πsq can be zero, a “blind spot.” To see how this can occur, suppose there are two SNPs constituting four possible haplotypes, and the haplotype frequencies are p for the cases and q for the controls. The match similarity measure (indicator of equivalent haplotypes) results in ΠS as the identity matrix, in which case δ = p′p − q′q, so δ = 0 whenever the sums of squared haplotype frequencies are the same for cases and controls, yet does not require p = q. The counting measure (counts of alleles alike in state along a pair of haplotypes) results in ΠS having 2′s on the down-sloping diagonal, 0′s on the up-sloping diagonal, and 1′s elsewhere, so that p′Πsp = 2′p′p + 2(p1 + p4)(p2 + p3). This illustrates that δ = 0 whenever p′p = q′q and (p1 + p4)(p2 + p3) = (q1 + q4)(q2 + q3). An extreme example that illustrates these points is p′ = (0.1, 0.1, 0.1, 0.7) and q′ = (0.7, 0.1, 0.1, 0.1). In contrast to using δ = p′Πsp − q′Πsq, Yuan et al. [2006] proposed a weighted two-sample U-statistic to eliminate this type of blind area, to enhance the power of detecting associations.
Recognizing the blind area of the existing similarity-based test [Tzeng et al., 2003a,b], Sha et al. [2007] provided a new association test using haplotype similarity, to eliminate the potential blind spot. They added the average within-group similarity of cases to that of controls, and compared this total within-group similarity with the average between-group similarity for cases and controls. Their statistic is S = U1/√Var(U1), where U = p′Πsp + q′Πsq − 2p′Πsq = (p − q). Their simulation results showed that, in most situations, their new haplotype similarity method was more powerful than both Pearson's χ2 test and the previous haplotype similarity method [Tzeng et al., 2003a,b].
In addition to the above similarity-based tests, a genomic distance-based regression for multilocus association analysis was recently proposed [Wessel and Schork, 2006], in which either locus or haplotype scoring can be applied. Wessel and Schork [2006] provided seven measures of genomic similarity, some based on genotypes and some based on haplotypes. The term “distance,” or “dissimilarity,” is dual with “similarity,” and so similarity can be transformed into genomic distance. A regression-based analysis was proposed to test the association of phenotype similarity with genotype similarity. This method allows various measures of genomic similarity and simultaneous analyses of multiple phenotypes. It can be used for gene expression data and can accommodate population stratification by simply including relevant covariates in the regression. Despite these merits and flexibility, the relative power of this approach for case-control data, versus some standard methods [e.g., logistic regression, Cordell and Clayton, 2002; Chapman et al., 2003] or haplotype score tests [Schaid et al., 2002]) is unknown. Surely the power performance of this new method will vary with the choice of similarity measure, and it will be helpful to have some priori knowledge about the optimal selection of similarity measure.
In this work, we first show that the genomic distance-based regression reduces to a diplotype dissimilarity test when a specific similarity measure is applied to analyses of case-control data, linking this method with the class of haplotype similarity/dissimilarity tests [Tzeng et al., 2003a,b; Yuan et al., 2006; Sha et al., 2007; Klei and Roeder, 2007]. Although there are some power studies, previous work focused on comparing the haplotype similarity tests with frequency-based statistics, such as Pearson's χ2 test. The relative power of the class of haplotype similarity tests with some standard association methods is not clear. Here, we compare the power of the genomic distance-based regression with that of logistic regression and score tests for haplotypes [Schaid et al., 2002] under the scenario of one causal locus simulated on genotypes from the HapMap data [The International HapMap Consortium, 2005]. From simulations, we explore the limitations and the relative power of a variety of multilocus association methods.
METHODS
LIKELIHOOD RATIO TEST FOR LOCUS-SCORING LOGISTIC REGRESSION: GENO-LRT
Suppose that there are L diallelic markers, and xL×1 is a vector of length L that codes the number of copies of an allele at each locus, with possible values of 0, 1, or 2. Let Y be the disease status, with 1 for affected and 0 for unaffected. The logistic regression model is
| (1) |
To test whether any of the L markers are associated with the disease, we test the hypothesis H0 : β = 0 versus H1 : β ≠ 0. The log-likelihood for Y is
| (2) |
where N is the number of subjects. We can test H0 versus H1 using the difference of the deviance statistics [Dobson, 2002],
| (3) |
where D0 is the deviance for the null model, D1 is the deviance for the alternative model, and β̂ and β̃ are the MLEs under H1 and H0, respectively. Asymptotically, ΔD has a χ2 distribution with degrees of freedom equal to L. Rejection of the null hypothesis suggests that at least one of the L markers is associated with the disease. This method is denoted “geno-LRT.” Model (1) does not directly capture the correlation structure of SNPs as haplotypes would. Although interactions between SNPs can be included, this generally reduces power to detect an association [Chapman et al., 2003; Balding, 2006]. Thus, we only model the main effects of markers in the method “geno-LRT.”
GENOMIC DISTANCE-BASED REGRESSION
Similarity measure for genotypes: geno-sim
Let and denote the genotypes of the lth locus for the ith and jth subjects, respectively. A similarity measure of genotypes is the average of identity-by-state (IBS) for the L loci, i.e.,
| (4) |
where is the IBS of the ith and jth subjects for the lth locus. Since the possible values for are 0, 1, or 2, ranges from 0 to 1. We call the method based on equation (4) “geno-sim.”
Similarity measure for diplotypes—counting measure: haplo-sim
Let hiu = (hiu1/hiu2) be the uth possible diplotype (i.e., the pair of haplotypes a subject possesses) of the ith subject, where u = 1, …, nhi, and where nhi is the number of possible diplotypes for the ith subject. P(hiu|gi) is the posterior probability that the ith subject has the uth possible diplotype, given the unphased genotypes. An intuitive haplotype-similarity measure proposed by Wessel and Schork [2006] that summarizes similarity contributed by all L loci on the haplotypes is
| (5) |
where refers to the allele at position l on one of the two chromosomes (c = 1 or 2) for the uth possible diplotype of the ith subject. The score equals 1 if the alleles at the lth locus match for subjects i and j, for the first haplotype, given a particular diplotype, otherwise this score equals 0. Equation (5) is the expected haplotype-similarity over the posterior distribution of pairs of haplotypes, given the observed unphased genotypes. In this measure, max{} ensures that the similarity will not depend on the order of the two haplotypes in each possible haplotype pair.
Similarity measure for diplotypes—matching measure: haplo-match
Another measure that treats haplotypes as “super alleles” is
| (6) |
where s(hiu1, hjv1) equals 1 only when all alleles on the first chromosome for subject i are the same as those on the first chromosome for subject j, otherwise s(hiu1, hjv1) equals 0. With the appropriate standardization in the denominator, and range from 0 to 1. To implement the haplotype-scoring analysis based on the distance-based regression, we first used the function “haplo.em” in the package haplo.stats [Schaid et al., 2002] to infer haplotype phase by the EM algorithm. Then, the haplotype similarity between any two subjects can be calculated by equations (5) or (6). We call the method based on equation (5) “haplosim” and that based on equation (6) “haplo-match.” Equation (5) corresponds to the “counting measure” in Tzeng et al. [2003a], while equation (6) corresponds to their “matching measure”.
Tzeng et al. [2003a] has shown that the numerator of their haplotype similarity test statistic for the counting measure can be computed directly from unphased genotype data [see also Schaid, 2004a for an intuitive explanation]. To simplify notation, we assume there is no phase ambiguity. When measuring the similarity of the ith and jth subjects, each with two haplotypes, the counting measure provided by Tzeng et al. [2003a] is
This measure is the total count of allele matches between two subjects, which does not depend on the linkage phase and so can be computed directly from unphased genotype data. However, for “haplo-sim,” the numerator of is
which depends on the linkage phase.
A conceptual multivariate regression
If we have N subjects, a similarity matrix, S = [Sij]N×N , can be constructed by the above similarity measures for genotypes or diplotypes, with the (i,j)th element defined by expressions (4), (5), or (6). Note that S is different from ΠS of Tzeng et al. [2003a] that is described in our introduction. S is an N-by-N matrix of similarities between subjects, while ΠS is an nH-by-nH matrix of similarities between distinct haplotypes, where nH is the number of haplotype categories. A recently proposed regression-based procedure can be used to evaluate the correlation between genetic similarity and phenotype similarity [Wessel and Schork, 2006], which originated in work by Gower [1966] and McArdle and Anderson [2001]. We apply this approach to case-control data. The procedure can be summarized as follows:
Genetic part:
(1) Calculate a distance matrix for all pairs of N subjects by D = [Dij]N×N = [1 − Sij]N×N, with elements Dij ranging from 0 to 1.
(2) Compute .
(3) Center A according to , where I is the identity matrix and 1 is a vector with all elements 1 [see Wessel and Schork, 2006 for more details].
Phenotype part:
(4) Denote the N-vector y with elements “−1” for controls and “1” for cases.
(5) Compute the projection matrix HN×N = y(y′y)−1y′. Note that H is a similarity matrix for the phenotype.
The pseudo-F statistic:
- (6) Compute the pseudo-F statistic by
(7)
where tr() denotes the trace of a matrix. This statistic follows the conceptual framework of multivariate analysis of variance (MANOVA) [Anderson, 2001]. Because the distribution of F is unknown, permutations of traits are used to obtain empirical p values. Note that coding 0 for controls is not appropriate for step 4 because that would obscure the phenotype similarity with the multiplication in step 5. With similarity measured between any two subjects and p values obtained by permutations, the computational burden for this method is heavier than other approaches. However, this method is attractive because it flexibly allows various similarity measures, as well as simultaneous analysis of multiple phenotypes, either binary or continuous traits, by use of the projection matrix in step 5. This approach differs from those proposed by Tzeng et al. [2003a,b] and Schaid et al. [2005], which measure the difference of within-group similarities among haplotypes or genotypes between cases and controls. Because Wessel and Schork's [2006] approach uses both the between-group distance and the within-group distances, there is no blind area.
RELATIONSHIP BETWEEN GENOMIC DISTANCE-BASED REGRESSION AND HAPLOTYPE SIMILARITY METHODS
When applying the genomic distance-based regression to a balanced case-control study (equal numbers of cases and controls), the pseudo-F statistic can be simplified to
| (8) |
where p and q are vectors of genotype/diplotype frequencies for the case and control samples, respectively; ΠD2 is a distance matrix among distinct genotypes/ diplotypes (Appendix A). The elements of ΠD2 are squared distances, i.e., , where Dij is the distance between the genotypes (or diplotypes) of the ith and jth subjects. The term p′ΠD2q is the between-group distance, while p′ΠD2p and q′ΠD2q are the within-group distances for cases and controls, respectively. From equation (8), we can see that the genomic distance-based regression uses both the between-group distance and the within-group distances.
When computing genotype distances (ignoring phase) for L diallelic markers, p and q are the 3L-length vectors of the joint-genotype frequencies of the markers for the case and control samples, respectively. The 3L × 3L matrix ΠD2 with elements contains the squared distances between any two genotype categories.
When computing distances between diplotypes (pairs of haplotypes), suppose that there are nH distinct haplotypes. Then, p and q have lengths nH(nH þ 1)/2 for the case and control samples, respectively. The matrix ΠD2 contains the squared distances between any two categories of diplotypes, so the numbers of rows and columns are both nH(nH þ 1)/2.
Computing the pseudo-F statistic with equation (8) can be time consuming when many markers or haplotypes are involved, because the size of the matrix ΠD2 can be quite large. While the computational intensity of equation (8) increases with the number of markers or the number of distinct haplotypes, that of equation (7) increases with the number of subjects. From equation (8), the pseudo-F statistic is determined by the ratio of the between-group distance to the total within-group distance. Under the null hypothesis, H0 : p = q, so F equals zero. When p ≠ q, F gets larger as the discrepancy increases, and hence a more significant result. When diplotypes are used to construct distances, the genomic distance-based regression can be linked to the new association test using haplotype similarity [Sha et al., 2007]. They both compare the similarity/distance within cases and within controls to the similarity/distance between cases and controls. However, there are two basic differences between these two methods: (1) while Sha et al. [2007] test the association between disease and the haplotypes, the genomic distance-based regression that uses equations (5) or (6) as the measure of similarity tests the association between disease and the diplotypes; (2) Sha et al. [2007] use similarity (dual with distance) among haplotypes in their statistic. The genomic distance-based regression uses the squared distances among diplotypes (ΠD2) in the statistic. The pseudo-F statistic of equation (8) can also be expressed as
| (9) |
with the numerator illustrating the contrast of between-group distance with the total within-group distance. The statistic proposed by Sha et al. [2007] can be expressed as
| (10) |
which is similar to the numerator of the pseudo-F statistic.
CANONICAL CORRELATION: CAN-COR
Canonical correlation can be used to measure the correlation between genotypes and multiple phenotypes. We applied it to case-control data because of its simplicity and ability to analyze multiple phenotypes. Conceptually, canonical correlation creates two new variables for two sets of variables such that the correlation of the two new variables is maximized. Let the genetic set, x = (x1, …, xL), contain the counts for copies of some prespecified allele of the L markers, and let the phenotype set, y, be an N-vector with element “0” for controls and “1” for cases; y can be a matrix with several columns if more phenotypes are measured. In our situation, the canonical correlation reduces to a multiple correlation, which measures the correlation between one phenotype and several marker genotypes. Let Σ be the variance-covariance matrix of y and x,
| (11) |
where contains the sample covariances of y with x1, …, xL, and Σxx is the sample covariance matrix of the x's. The squared multiple correlation between y and x's can be computed from the partitioned covariance matrix,
| (12) |
where is the sample variance of y. The multiple correlation R can be defined alternatively as the maximum correlation between y and a linear combination of the x's [Rencher, 2002], and R2 is the coefficient of determination (or proportion of variance explained) in multiple linear regression. It is equivalent to the score statistic derived from a logistic regression (Appendix B), so the coefficient of determination is asymptotically equivalent to the likelihood ratio test statistic for logistic regression. When multiple phenotypes are measured, the canonical correlation is
| (13) |
where det() denotes the determinant of a matrix. This provides a simultaneous test of association for more than one phenotype, and it is implemented by the function “cancor” in the R package. A permutation p value is used to test whether the L markers are associated with the phenotype. An advantage of canonical correlation is that the computational speed is much faster than that for the genomic distance-based regression because comparisons between all pairs of subjects are not required. This method is denoted “can-cor.”
SIMULATION STUDY
Our simulation compared the five methods described in the previous section (denoted “geno-LRT,” “geno-sim,” “haplo-sim,” “haplo-match,” and “can-cor”), as well as “haplo-score,” a global score test for haplotypes, and “haplo-max,” the maximum score statistic over all haplotype scores. The last two tests were proposed by Schaid et al. [2002] and provided by their package haplo.stats. When computing the haplotype-scoring distance-based statistics (“haplo-sim” and “haplo-match”), we first inferred haplotype phases by the EM algorithm. All the haplotypes with frequencies less than 0.01, a cutoff value suggested by Sha et al. [2007], were considered to be rare haplotypes and were merged with their most similar common haplotype. When more than one common haplotype had the same counting similarity with a rare haplotype, the rare haplotype was merged with the common haplotype with the highest frequency. When computing the score statistics for haplotypes (“haplo-score” and “haplo-max”), only haplotypes with frequencies larger than 0.01 were scored. We evaluated these seven methods in the scenario of one causal locus simulated on genotypes from the HapMap data. Power comparisons were based on 1,000 repetitions; in each repetition, p values were calculated by 1,000 permutations.
To study association methods under real human LD structure, we downloaded SNP genotype data on a chromosomal region from the HapMap website (www.hapmap.org). We used data from CEU - CEPH (Utah residents with ancestry from northern and western Europe), and kept the genotypes of 60 unrelated subjects—parents in the 30 trios comprising the CEPH data set. A total of 25 SNPs on chromosome 17 were selected, chosen to have minor allele frequency >5% and without missing genotypes, spanning 68.2 kb (40,248,321–40,316,535), yielding an average (median) distance between SNPs of 2.84 kb (1.65 kb). These 25 SNPs were highly correlated, with an average D′ of 0.97 for all adjacent loci. Following North et al. [2006], to model the effects of the susceptibility locus for a complex disease, we assigned the probabilities of being affected, conditional on possessing 0, 1, or 2 copies of the causal allele, as 0.029, 0.076, and 0.214, respectively. These penetrances mimic Alzheimer's disease for the APOE-4 genotype [Kuusisto et al., 1994]. The odds ratios for genotypes Aa and AA, relative to genotype aa, were 2.75 and 9.12, respectively. To simulate one causal locus, we let the rarer allele be the causal allele and we let each SNP locus be the disease susceptibility locus, with the remaining 24 SNPs serving as markers. The causal SNP was assumed not to be genotyped and so was not contained in the analysis. According to the genotype frequencies for each SNP, the prevalence of the disease in the population would vary from 3.84 to 7.87%, with an average of 5.35%. The total sample size was set at 100 subjects, of which half were cases and half were controls. In each repetition, 50 cases and 50 controls were sampled with replacement from the CEPH data composed of 60 unrelated subjects, and the disease status was generated according to the genotypes of the causal SNP and the disease model.
We further extended the above simulation scenario to fewer markers, with more HapMap data on different chromosomes. For chromosome 17, we selected eight SNPs from 25 SNPs, by the clustering method [Tzeng et al., 2003a]. With the clustering method, six common haplotypes were retained, and the other five rare haplotypes were clustered into one of the six categories through a one-step or two-step mutation. The retained haplotypes were constructed by eight SNPs. In addition, we randomly selected regions along chromosomes, to collect more genotype data. The background information of these chromosomal regions is listed in Table I. Every studied chromosomal region spanned within 100 kb, and we list the average pairwise LD for all loci within regions in Table I. We did not study the power under the situation of weak LD between markers, because multilocus association analyses would not be preferred in that situation. For diplotype data, haplotypes are treated as partially missing when haplotype frequencies are estimated among unrelated subjects. This ambiguity can increase the variance of the estimated haplotype frequencies, reducing the statistical efficiency especially when LD is weak [Schaid, 2002]. Furthermore, with weak LD, there are many more distinct haplotypes, leading to weak power of haplotype methods.
TABLE I.
Information of the HapMap chromosomal regions studied and hypothetical disease-causing SNP used for simulations.
| Chr. | Spanning (base pair) | No. of SNPs |
LD of adjacent loci mean (range) |
LD of all pairs of loci mean (range) |
Average prevalencea (%) |
Causal allele frequency |
||
|---|---|---|---|---|---|---|---|---|
| D′ | r2 | D′ | r2 | |||||
| 1 | 85.0 kb (75,996,416–76,081,443) | 8 | 0.99 (0.90–1.00) | 0.25 (0.04–0.76) | 0.91 (0.11–1.00) | 0.27 (0.002–0.86) | 6.30 (3.68–8.17) | 0.28 (0.08–0.38) |
| 2 | 74.7 kb (165,005,914–165,080,660) | 4 | 1.00 (1.00–1.00) | 0.31 (0.02–0.88) | 0.77 (0.31–1.00) | 0.19 (0.02–0.88) | 4.31 (4.00–4.62) | 0.14 (0.12–0.17) |
| 3 | 70.0 kb (48,181,106–48,251,081) | 4 | 1.00 (1.00–1.00) | 0.08 (0.02–0.19) | 1.00 (1.00–1.00) | 0.22 (0.02–0.86) | 5.67 (3.45–7.02) | 0.25 (0.06–0.34) |
| 4 | 97.8 kb (95,656,914–95,754,677) | 4 | 0.82 (0.45–1.00) | 0.39 (0.09–0.95) | 0.82 (0.45–1.00) | 0.29 (0.07–0.95) | 6.12 (4.85–7.79) | 0.26 (0.18–0.38) |
| 5 | 67.8 kb (86,285,103–86,352,855) | 8 | 0.99 (0.91–1.00) | 0.23 (0.02–0.65) | 0.97 (0.49–1.00) | 0.25 (0.02–0.94) | 5.37 (3.91–8.79) | 0.21 (0.09–0.43) |
| 6 | 24.5 kb (85,437,983–85,462,480) | 4 | 1.00 (1.00–1.00) | 0.32 (0.02–0.90) | 1.00 (1.00–1.00) | 0.17 (0.01–0.90) | 4.17 (3.68–5.23) | 0.11 (0.08–0.18) |
| 7 | 75.9 kb (79,276,107–79,352,038) | 4 | 1.00 (1.00–1.00) | 0.67 (0.48–0.90) | 0.93 (0.68–1.00) | 0.55 (0.31–0.90) | 4.09 (3.68–4.70) | 0.12 (0.08–0.18) |
| 8 | 24.2 kb (73,087,988–73,112,209) | 4 | 1.00 (1.00–1.00) | 0.04 (0.02–0.09) | 0.99 (0.92–1.00) | 0.13 (0.02–0.54) | 4.70 (3.99–5.79) | 0.17 (0.10–0.28) |
| 9 | 42.1 kb (69,172,556–69,214,705) | 4 | 0.98 (0.94–1.00) | 0.40 (0.01–0.69) | 0.99 (0.94–1.00) | 0.28 (0.01–0.69) | 4.65 (3.53–5.85) | 0.15 (0.07–0.23) |
| 10 | 32.9 kb (67,662,628–67,695,506) | 4 | 0.97 (0.90–1.00) | 0.10 (0.04–0.21) | 0.93 (0.73–1.00) | 0.28 (0.04–0.88) | 5.81 (4.54–8.55) | 0.23 (0.14–0.39) |
| 17 | 68.2 kb (40,248,321–40,316,535) | 25, 8 | 0.97 (0.23–1.00) | 0.22 (0.003–1.00) | 0.94 (0.17–1.00) | 0.20 (0.002–1.00) | 5.35 (3.84–7.87) | 0.22 (0.10–0.39) |
| 22 | 70.5 kb (33,606,466–33,676,951) | 8 | 1.00 (1.00–1.00) | 0.04 (0.004–0.15) | 0.84 (0.12–1.00) | 0.08 (0.001–0.67) | 4.86 (3.37–9.10) | 0.17 (0.05–0.45) |
| The total number of simulation scenarios | 89 | |||||||
The prevalence was calculated under a disease model representing Alzheimer's disease. In the parentheses, we list the minimum and the maximum prevalence when each SNP in turn was considered the causal locus. In real data (population of Kuopio, eastern Finland, 980 people aged 69–78), the prevalence of Alzheimer's disease was 4.7% [Kuusisto et al., 1994]. SNP, single-nucleotide polymorphism, LD, linkage disequilibrium.
SIMULATION RESULTS
TYPE-I ERROR RATES
We evaluated the Type-I error rate of each method under the 89 simulation scenarios described above, but now generated case-control status independent of genotype. That is, we allowed the SNP allele frequencies to vary, as well as the LD structure, according to that in the CEPH data. The total sample size was set at 100 subjects, of which half were cases and half were controls. Simulation results were based on 1,000 repetitions; in each repetition, p values were calculated by 1,000 permutations. We also allowed a reasonable range of genotyping errors in the data, described later in the “Power Comparisons in the Presence of Genotyping Errors.” The Type-I error rates of each method under three nominal significance levels are presented in Table II. All seven methods were quite conservative with our sampling from the small CEPH diplotype pool. This conservativeness is likely because the CEPH is a small pool to sample from, causing many duplicates in the case-control samples. These duplicates cause ties in the resulting test statistics, leading to conservative test results. Indeed, when we simulated from a larger haplotype pool using the coalescent-based program ms [Hudson, 2002], in most situations, the over-conservativeness faded away and the Type-I error rates were close to the nominal significance levels.
TABLE II.
Type–I error rates, averaged over the 89 simulation scenarios (sample size = 100)
| Nominal significance level | Geno-sim | Haplo-sim | Geno-LRT | Can-cor | Haplo-score | Haplo-match | Haplo-max |
|---|---|---|---|---|---|---|---|
| 0.05 | 0.00864 | 0.00855 | 0.01721 | 0.01433 | 0.01516 | 0.01120 | 0.01545 |
| 0.01 | 0.00083 | 0.00081 | 0.00259 | 0.00186 | 0.00191 | 0.00109 | 0.00173 |
| 0.005 | 0.00033 | 0.00024 | 0.00117 | 0.00092 | 0.00074 | 0.00042 | 0.00060 |
POWER COMPARISONS–STRATIFIED ANALYSIS
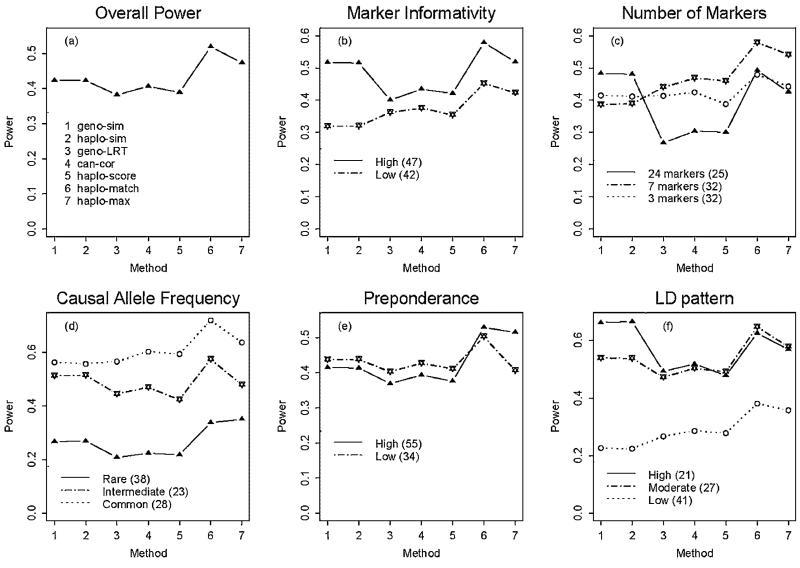
Figure 1(a) presents the overall power performance of the seven methods, showing that the best method was “haplo-match.” The methods “geno-sim” and “haplo-sim” have similar power, because the diplotype similarity based on the counting measure largely depends on genotypes. These two methods can be viewed as a group. Another group includes the methods “geno-LRT” and “can-cor.” They are asymptotically equivalent, and are expected to have similar power under large sample sizes. We discuss the relative power of the seven association methods with respect to five properties: (1) the marker informativity; (2) the number of markers; (3) the causal allele frequency; 4) the preponderance of the most common high-risk haplotype; (5) the LD pattern between the causal SNP and its flanking markers.
Fig. 1.
Power according to five properties. The x-axis gives a number for each method and the y-axis is the average power under the significance level 0.05. (a) The overall power performance—the average power over all 89 scenarios. (b) The power performance stratified by marker informativity. There were 47 simulation scenarios with high marker informativity and 42 scenarios with low marker informativity (in the parentheses are the numbers of scenarios and the power is the average power over these scenarios). (c) The power performance stratified by the number of markers. (d) The power performance stratified by the causal allele frequency. (e) The power performance stratified by the preponderance of the most common high-risk haplotype. (f) The power performance stratified by the linkage disequilibrium (LD) pattern between the causal single-nucleotide polymorphism (SNP) and its flanking markers.
Marker informativity
Figure 1(b) presents the power performance stratified by the marker informativity, showing notable power loss of “geno-sim” and “haplo-sim” due to low marker informativity. Here we used the average of minor allele frequencies of markers as an index for marker informativity, and a threshold 0.215 was set to classify the marker informativity into two categories. Note that the threshold was not absolute to categorize high or low marker informativity—we chose it for convenience of explanation. Power of “geno-sim” and “haplo-sim” relies on higher marker informativity. To obtain better power of these two methods, intermediate marker allele frequency (close to 0.5, representing higher marker informativity) is required—a similar result reported by Klei and Roeder [2007]. The intuition is that subjects possessing common alleles are not easily distinguished, and because we use markers to detect the unobserved causal SNP, high marker informativity will help to distinguish subjects. In contrast, “haplo-match” does not suffer from such a great loss in power due to low marker informativity. The haplotypes constructed by SNPs can serve as many alleles on a highly informative marker, and “haplo-match” is like “geno-sim’ working on an informative marker. Thus, “haplo-match’ is more powerful than other distance-based approaches, especially when the marker informativity is not high.
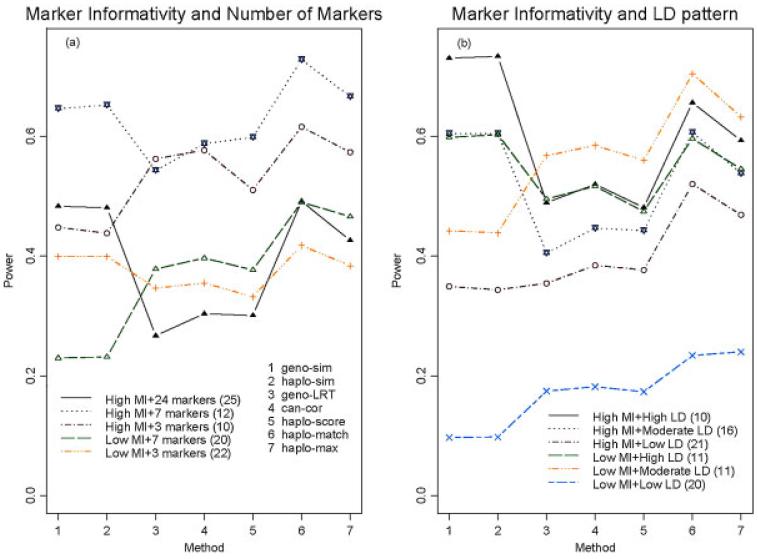
Number of markers
The number of markers that should be considered simultaneously in multilocus association analyses remains a difficult balance between degrees of freedom and power. As illustrated in Figure 1(c), no general power trend can be deciphered for this factor. Because of the dependence between the marker informativity and the number of markers, we examined the power performance stratified by the marker informativity and the number of markers. Due to coincidence, all the 25 scenarios using 24 markers were classified into the “High” group for the marker informativity. Figure 2(a) shows that locus-based logistic regression (i.e., “geno-LRT” and “can-cor”) was relatively less powerful when many markers were included in the analyses, because more markers diluted the association signal and this method does not directly capture the haplotype LD structure of the SNPs.
Fig. 2.
Power according to marker informativity, number of markers, and the linkage disequilibrium (LD) pattern. The x-axis gives a number for each method and the y-axis is the average power under the significance level 0.05. (a) The power performance stratified by the marker informativity and the number of markers. The first stratum “High MI+24 markers (25)” means that there were 25 simulation scenarios under high marker informativity and using 24 markers, and the power is the average power over these 25 scenarios. (b) The power performance stratified by the marker informativity and the LD pattern between the causal SNP and its flanking markers. The first stratum “High MI+High LD (10)” means that there were 10 simulation scenarios under high marker informativity and high linkage disequilibrium (LD), and the power is the average power over these ten scenarios.
Causal allele frequency
The causal allele frequency and the preponderance of the most common high-risk haplotype are unknown when conducting association analyses, so studying their impact on the power performance might be limiting. Nonetheless, it is worthwhile to know the relative power of different methods. Figure 1(d) presents the power performance for different levels of the causal allele frequencies, which have been classified into three groups: Rare [0.05, 0.15); Intermediate [0.15, 0.25); Common [0.25, 0.5). For all levels of causal allele frequencies, “haplo-match” was the most powerful method. When the causal allele was rare, “haplo-max” had comparable power with “haplo-match.” Because lower causal allele frequency was confounded with fewer high-risk haplotypes (when the causal allele was rarer, it tended to occur on one or few haplotypes), “haplo-max” was also powerful when the causal allele was rare. In general, the causal allele frequency did not seem to make a crucial difference to the relative power performances of these seven methods.
The preponderance of the most common high-risk haplotype
To aid the interpretation of our simulations, we created a haplotype preponderance index. Suppose that there are H high-risk haplotypes, h1; h2, …, hH (from the most common to the least common), and the haplotype frequencies for them are fh1, fh2, …, fhH (fh1 ≥ fh2 ≥ ⋯ ≥ fhH), respectively. We define an index for the preponderance of the most common high-risk haplotype as , which ranges from 0 to 1. We estimated this index with 500 cases and 500 controls when each SNP in turn was considered the causal locus. When there is only one high-risk haplotype, Ipre = 1; when there are several high-risk haplotypes, Ipre<1. There are two extreme cases for several high-risk haplotypes: (1) there is one major high-risk haplotype (with relatively high haplotype frequency) and several minor high-risk haplotypes (with relatively low haplotype frequencies), Ipre ≈ 1; 2) several high-risk haplotypes have similar frequencies, in which case, Ipre is much lower than 1. We set a threshold 0.8 to classify the preponderance of the most common high-risk haplotype into two categories. This threshold was set to uncover the general trend of power. From Figure 1(e), except for “haplo-max,” the preponderance of the most common high-risk haplotype did not influence the power of the association methods. The method “haplo-max” is more powerful than the other methods if a single risk haplotype is much more frequent than all other risk haplotypes.
LD pattern of causal SNP and flanking markers
The correlation between the causal SNP and its neighboring markers plays an important role in multilocus association studies. We expect power loss due to low LD between the unobserved causal SNP and the neighboring markers, because low LD implies the lack of information from markers to make correct inference. To investigate this, we categorized the squared correlation coefficient (r2) between the casual SNP and its adjacent markers into three groups. The LD of a simulation scenario was labeled as “High” if the causal SNP was in high correlation with at least one adjacent marker (r2>0.6); “Moderate” if 0.15<r2 ≤ 0.6; “Low” if r2 0.15. Figure 1(f) shows the power performance stratified by this pattern. When the causal SNP had high LD with at least one of its flanking markers, the three methods derived from the genomic distance-based regression attained higher power than the conventional methods. However, when LD was not high, the power loss was substantial for “geno-sim” and “haplo-sim” (i.e., the genomic distance-based regression with the counting measures). In contrast, the conventional methods and the genomic distance-based regression with the matching measure did not suffer from such a dramatic loss in power, though there was some power loss due to reduced information from markers.
Our results for the influence of LD pattern on power of the genomic distance-based regression that used the counting measures were similar to those for a new regression-based multimarker test that uses haplotype similarity [Tzeng et al., 2007]. Tzeng et al. [2007] proposed a gene-trait similarity regression analytically united with the variance-component approaches [Tzeng and Zhang, 2007]; see similar derivations by Goeman et al. [2004]. Although Tzeng et al. [2007] reversed the roles of genetic similarity and trait similarity compared with the regression system of Wessel and Schork's, the two methods should have similar power because both methods measure correlations between these two measures of similarity. Tzeng et al. [2007] observed that their gene-trait similarity regression with the counting measure can be more powerful than the standard regression [Schaid et al., 2002] when the causal SNP was tagged (with r2>0.7 defining tagged) by at least one nearby marker, but suffered from a greater loss in power when the causal SNP was not tagged (r2 ≤ 0.7). In our work, we found that using the matching measure in the similarity regression can reduce the loss in power when the causal SNP was not tagged, probably because the whole haplotype provided greater information for the unobserved causal SNP.
For the properties of marker informativity and LD pattern, we observed that “geno-sim” and “haplo-sim” performed well when the marker informativity and the correlation between the causal SNP and markers were high. We further examined the power performance stratified by the marker informativity and the LD pattern. Figure 2(b) shows that both factors have a significant influence on the power of association methods, especially for “geno-sim” and “haplo-sim.”
Overall, our results suggest that “haplo-match” was a better method because it performed well under a variety of situations. The methods “geno-sim” and “haplo-sim” can be adopted if high marker informativity and a strong correlation between causal SNP and markers can be assured. The locus-based logistic regression, “geno-LRT” and “can-cor,” had relatively low power when many unassociated SNPs were involved in the analyses. The power of “haplo-max” was comparable to that of “haplo-match,” when the preponderance of the most common high-risk haplotype was high.
POWER COMPARISONS—REGRESSION ANALYSIS
To evaluate which factors had the strongest independent effects on the power of each method, we regressed the simulated power of each method on six variables, and used a stringent level of significance, 0.01. Note that some factors might still influence power (i.e., p value >0.01), but we wanted to screen for the most influential factors. The parameter estimates of significant variables are listed in Table III, with p values listed in parentheses. We found that the causal allele frequency and the LD pattern between the causal SNP and its flanking markers played key roles in predicting the power of all seven methods. The small p values suggested their significant influences, and the positive parameter estimates suggested positive associations. These results are similar to those formed in the stratified analyses. Higher causal allele frequency and stronger correlation between causal SNP and markers increase power of all methods. In addition to “geno-sim” and “haplo-sim,” “geno-LRT,” “can-cor,” and “haplo-score” can also be improved by enhancing the marker informativity. On the other hand, most of the haplotype-scoring methods (“haplo-match” and “haplo-max”) are less influenced by low per SNP marker informativity, because haplotypes constructed by SNPs can be viewed as many alleles on a highly informative marker.
TABLE III.
Regression of simulated power on six properties for each association statistic, with most significant factors listed (p value <0.01)
| Properties | Geno-sim | Haplo-sim | Geno-LRT | Can-cor | Haplo-score | Haplo-match | Haplo-max |
|---|---|---|---|---|---|---|---|
| Marker informativity | 1.72 (0.0063) | 1.73 (0.0063) | 1.23 (0.0026) | 1.28 (0.0021) | 1.15 (0.0059) | ||
| 3 Markers | 0.27 (<.0001) | 0.25 (<.0001) | 0.21 (<.0001) | ||||
| 7 Markers | 0.24 (<.0001) | 0.23 (<.0001) | 0.22 (<.0001) | 0.18 (0.0019) | |||
| Causal allele frequency | 1.04 (0.0002) | 1.00 (0.0004) | 1.52 (<.0001) | 1.59 (<.0001) | 1.63 (<.0001) | 1.60 (<.0001) | 1.43 (<.0001) |
| Preponderance Index | 0.39 (0.0006) | ||||||
| LD pattern | 0.45 (<.0001) | 0.46 (<.0001) | 0.21 (0.0002) | 0.21 (0.0001) | 0.18 (0.0014) | 0.23 (0.0012) | 0.21 (0.0015) |
The simulated power of each method was regressed on the six properties (“3 markers” and “7 markers” are indicator variables, while others are continuous variables). The parameter estimates of significant variables are listed, with p values given in parentheses. LD, linkage disequilibrium.
Because of many degrees of freedom, the locus-based logistic regression (“geno-LRT” and “can-cor”) and the global score test (“haplo-score”) suffered from power loss when 24 markers were included in the analyses. As a strategy to reduce the degrees of freedom, the similarity-based methods (“geno-sim,” “haplo-sim,” and “haplo-match”) were less vulnerable to more markers or more haplotypes. Finally, when one high-risk haplotype was most frequent, the maximum score statistic over all haplotype scores (“haplo-max”) had good power.
POWER COMPARISONS IN THE PRESENCE OF GENOTYPING ERRORS
As discussed by Tzeng et al. [2003a] and Sha et al. [2007], the matching measure is not robust to genotyping errors, missing data, and recent marker mutation. We further compared the power of the seven methods in the presence of genotyping errors. The error model considered in our simulation was the asymmetric allele dropout model [Morris and Kaplan, 2004], where heterozygotes were misclassified twice as frequently as homozygotes. We considered error rates γ0→1 = 0.025, γ0→2 = 0, γ1→0 = 0.05, γ1→2 = 0.05, γ2→0 = 0, γ2→1 = 0.025, where γi→j was the conditional probability that true genotype gi was identified as genotype gj, and where i, j = 0, 1, 2. Based on the frequencies of heterozygotes, the expected error rates for each marker would vary from 0.025 to 0.1. According to previous studies [Abecasis et al., 2001; Tintle et al., 2005; Cheng and Lin, 2007], error rates less than 0.05 are moderate and a 0.08 error rate is the maximum genotyping error rate when the missing genotypes are included in the calculation of the genotyping error rate. Thus, the error rates considered in our simulation were in a reasonable range.
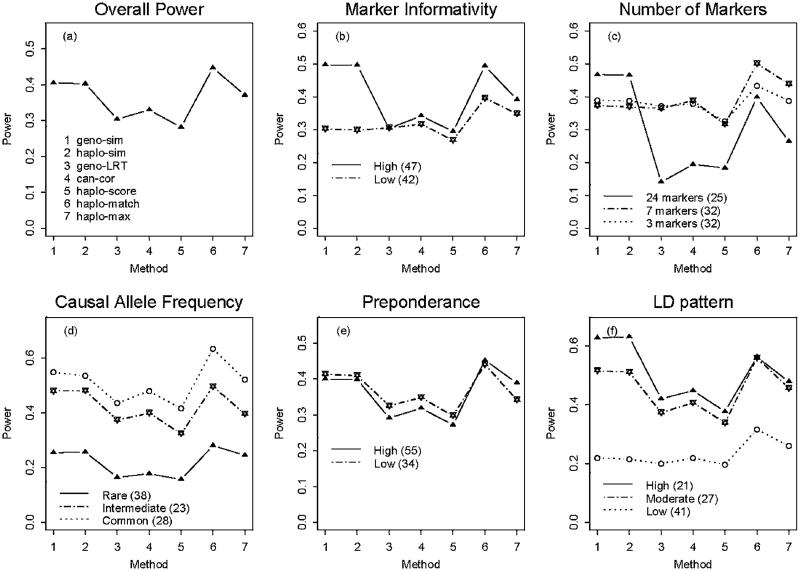
Similar to previous presentations, the power results in the presence of genotyping errors are shown in Table IV, Figures 3 and 4. The genotyping errors did not plague the method “haplo-match” very much, which remained a better method in the sense that it was more robust to the varying marker informativity and the low LD between the causal SNP and its neighboring markers. This suggested that the matching measure was still a desirable similarity measure regarding a reasonable range of genotyping errors. Among the seven methods, although the distance-based approach that used the counting measure (“geno-sim” and “haplo-sim”) seemed to be less vulnerable to genotyping errors, it suffered from a large drop in power when the marker informativity or the LD between the causal SNP and its neighboring markers was low.
TABLE IV.
Regression of simulated power on six properties for each association statistic (in the presence of genotyping errors); p value < 0.01
| Properties | Geno-sim | Haplo-sim | Geno-LRT | Can-cor | Haplo-score | Haplo-match | Haplo-max |
|---|---|---|---|---|---|---|---|
| Marker informativity | 1.57 (0.0075) | 1.62 (0.0069) | 1.32 (0.0007) | 1.31 (0.0008) | 1.01 (0.0035) | ||
| 3 Markers | 0.34 (<.0001) | 0.30 (<.0001) | 0.24 (<.0001) | 0.22 (<.0001) | |||
| 7 Markers | 0.28 (<0001) | 0.25 (<.0001) | 0.18 (<.0001) | 0.17 (0.0022) | 0.23 (<.0001) | ||
| Causal allele frequency | 1.07 (<.0001) | 0.99 (0.0002) | 1.13 (<.0001) | 1.25 (<.0001) | 1.13 (<.0001) | 1.50 (<.0001) | 1.30 (<.0001) |
| Preponderance Index | 0.22 (0.0176)a | ||||||
| LD pattern | 0.42 (<.0001) | 0.44 (<.0001) | 0.20 (0.0001) | 0.21 (<.0001) | 0.16 (0.0007) | 0.23 (0.0004) | 0.21 (0.0003) |
Although this p value was greater than 0.01, we kept it for comparison with Table III.
The simulated power of each method was regressed on the six properties (“3 markers” and “7 markers” are indicator variables, while others are continuous variables). The parameter estimates of significant variables are listed, with p values given in parentheses. LD, linkage disequilibrium.
Fig. 3.
Power according to five properties (in the presence of genotyping errors).
Fig. 4.
Power according to marker informativity, number of markers, and the linkage disequilibrium (LD) pattern (in the presence of genotyping errors).
DISCUSSION
Genetic epidemiologists have struggled to identify genetic polymorphisms involved in the development of diseases. Many have collected data on large numbers of markers, but the most powerful analysis methods are not always obvious [Heidema et al., 2006]. For multilocus association studies, it is important to choose powerful and appropriate statistical methods that are designed to relate genotype or haplotype information to phenotypes of interest. The most powerful method for multilocus association studies, however, changes under different genetic architectures, such as the patterns of LD, and the number of causal loci located in a chromosomal region. There has been a considerable debate about whether one should use a locus-scoring approach or a haplotype-scoring approach [Chapman et al., 2003; Clayton et al., 2004; North et al., 2006; Humphreys and Iles, 2005; Bardel et al., 2006]. A locus-scoring approach is appealing because it does not require haplotype phase resolution. A haplotype-scoring approach can be more powerful in some cases, such as when several mutations within a single gene interact to create a “super allele” that has a large effect on a disease [Schaid et al., 2002]. The HapMap project [The International HapMap Consortium, 2005] characterizes patterns of LD in the human genome, and ideas like haplotype blocks [Cardon and Abecasis, 2003] might prove useful to map complex human trait loci. If all SNPs within a haplotype block are highly correlated among themselves, then any SNP within the block should capture sufficient information to fully interrogate a particular region of the genome, allowing one to economically detect an association, with large samples, possibly also allowing one to rule out the role of a region if no significant association is detected [Schaid 2004a,b]. Because haplotypes are difficult to directly measure over long stretches of DNA in diploid organisms, ambiguous phase can lead to a loss of statistical efficiency [Douglas et al., 2001; Schaid, 2002].
To evaluate previously proposed association methods under various genetic situations, we compared the power for three locus-scoring approaches (“geno-sim,” “geno-LRT,” “can-cor”) and four haplotype-scoring approaches (“haplo-sim,” “haplo-score,” “haplo-match,” “haplo-max”). The method “geno-LRT” is a likelihood ratio test for logistic regression that models the main effects of loci, while “can-cor” is equivalent to a score test for logistic regression. These two tests are asymptotically equivalent. The methods “geno-sim,” “haplo-sim,” and “haplo-match” are based on the distance-based regression [Wessel and Schork, 2006], for locus coding and two ways of haplotype coding, respectively. They are derived from an approach that involves similarity for pairs of subjects based on their diploid genotypes at multiple loci in the region of interest, and relates variation in a measure of genotype similarity to variation in a measure of trait similarity. The method “haplo-score” is a global score test that includes phase resolution by the EM algorithm, while “haplo-max” is the maximum score statistic among all haplotype-specific scores [Schaid et al., 2002].
We have shown that the distance-based regression can be included in the class of haplotype similarity tests [Yuan et al., 2006; Sha et al., 2007] when a specific similarity measure is used. In fact, Wessel and Schork's method is more general because it allows various similarity measures and multiple phenotypes. We evaluated the power of seven association methods under the scenario of one causal locus simulated on genotypes from the HapMap data [The International HapMap Consortium, 2005]. Based on our simulation results, the distance-based regression that uses the matching measure of diplotypes had better power under a variety of situations. The maximum score statistic over all haplotype scores can have comparable power, however, it suffered from power loss when there were several high-risk haplotypes of equal frequency.
With similarity measured between any two subjects and the need for permutation p values, the distance-based regression is computationally intensive. Because of this, it was difficult to study more similarity measures. Nonetheless, because this procedure depends critically on the choice of similarity measure, it is expected to have different power performance for other genetic architectures. Different from testing the association between disease and the haplotypes [Sha et al., 2007], the distance-based regression, using equations (5) or (6) as the similarity measure, tests the association between disease and the diplotypes. We have shown that the distance-based regression, using the similarity measure in equation (6), performs well under a variety of situations.
Following Sha et al. [2007], we merged rare haplotypes with similar common haplotypes, which may provide the advantage of robustness to genotyping errors. According to previous studies [Tzeng et al., 2003a; Sha et al., 2007], the matching measure is not robust to genotyping errors, missing data, and recent marker mutation. We also evaluated the power of the seven methods under these situations. Our results showed that genotyping errors did compromise the power of the method “haplo-match,” but not much. Although “geno-sim” and “haplo-sim” were more robust to genotyping errors, they were less desirable than “haplo-match,” when the marker informativity or the correlation between the causal SNP and markers was low. Generally speaking, the power trends were quite similar to those for the situation of no genotyping errors.
The statistic of the distance-based regression is similar to that proposed by Sha et al. [2007]. The simulation results of Sha et al. [2007] showed that on average their tests are more powerful than the χ2 test and the tests proposed by Tzeng et al. [2003a], and which test and similarity measure to use depends upon the nature of the markers. Our study further discussed the impact of marker properties on power of several prevailing association methods. Results of Sha et al. [2007] also showed that the matching measure is better than other measures when there is only one high-risk haplotype. On the other hand, when there are several high-risk haplotypes, the counting measure is better. However, the average performances of varying similarity measures do not have big differences. Our results showed that the key points to evaluate the counting measure and the matching measure would be the marker informativity and the correlation between the causal SNP and markers. The preponderance of the most common high-risk haplotype did not influence the power of the two similarity measures very much.
Finally, although canonical correlation can also measure the correlation between genotypes and multiple phenotypes, we did not compare the power between it and the distance-based regression for analyses of multiple phenotypes. Here, we focused on a single phenotype, disease status, for case-control studies. Exploring powerful association approaches for multiple phenotypes deserves further research.
ACKNOWLEDGMENTS
We are grateful for the constructive comments from the anonymous reviewers that improved this work. We also thank the investigators and participants in the International HapMap Project for making the data available to the scientific community. This research was supported by the US Public Health Service, National Institutes of Health, contract grant number GM065450 (D.J.S.), and the Graduate Students Visiting Abroad Scholarship awarded by the National Science Council of Taiwan (W.-Y.L).
Contract grant sponsor: VS Public Health Service; Contract grant sponsor: National Institutes of Health; Contract grant number: GM065450; Contract grant sponsor: National Science Council of Taiwan.
APPENDIX A. SIMPLIFICATION OF THE PSEUDO-F STATISTIC IN EQUATION (7)
If there are N subjects, of which half are controls and half are cases, the numerator of the pseudo-F statistic is
where A is the association matrix from step 2, A11 is the partitioned matrix containing the associations between controls and controls, A22 is the partitioned matrix containing the associations between cases and cases, A12 is the partitioned matrix containing the associations between controls and cases, 1 is a vector with all elements 1, ΣA11 is the sum of elements in A11, p and q are vectors of the genotype (or diplotype) frequencies for the case and control samples, respectively, and ΠA is the matrix of associations among genotypes (or diplotypes). Note that ΠA is different from A. The former contains associations between distinct genotypes/diplotypes, while the latter contains associations between subjects. The denominator of the pseudo-F statistic is
where O is a zero matrix with dimension (N/2 × N/2). The pseudo-F statistic is thus
where the (i,j)th element in ΠA is , because of step 2
APPENDIX B. DERIVATION OF THE EQUIVALENCE OF THE SCORE STATISTIC FOR LOGISTIC REGRESSION AND THE CANONICAL CORRELATION IN EQUATION (12)
Following the logistic regression model in equation (1) and the log-likelihood shown in equation (2), the score vector for β is
| (B1) |
and its variance is
| (B2) |
The score statistic is then
| (B3) |
where and are evaluated under the null hypothesis. Ṽ−1 is the generalized inverse of Ṽ. The covariance terms Σyx, Σxx, and are defined in the subsection of canonical correlation. The statistic T has an approximate χ2 distribution with degrees of freedom equal to the rank of the matrix Ṽ, which is usually the number of markers. The statistic T can be expressed as
| (B4) |
Thus, the score statistic derived from logistic regression is equivalent to the canonical correlation in equation (12), and is asymptotically equivalent to the likelihood ratio test statistic for logistic regression.
Footnotes
ELECTRONIC-DATABASE INFORMATION
URLs for data in this article are as follows
R package, http://www.r-project.org/
Package haplo.stats, http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm
The HapMap website, http://www.hapmap.org
The R code for simulations is available by sending an email to W-YL, d92842006@ntu.edu.tw
REFERENCES
- Abecasis GR, Cherny SS, Cardon LR. The impact of genotyping error on family-based analysis of quantitative traits. Eur J Hum Genet. 2001;9:130–134. doi: 10.1038/sj.ejhg.5200594. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Aust Ecol. 2001;26:32–46. [Google Scholar]
- Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- Bardel C, Darlu P, Genin E. Clustering of haplotypes based on phylogeny: how good a strategy for association testing? Eur J Hum Genet. 2006;14:202–206. doi: 10.1038/sj.ejhg.5201501. [DOI] [PubMed] [Google Scholar]
- Bourgain C, Genin E, Holopainen P, Mustalahti K, Maki M, Partanen J, Clerget-Darpoux F. Use of closely related affected individuals for the genetic study of complex diseases in founder populations. Am J Hum Genet. 2001;68:154–159. doi: 10.1086/316933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Abecasis GR. Using haplotype blocks to map human complex trait loci. Trends Genet. 2003;19:135–140. doi: 10.1016/S0168-9525(03)00022-2. [DOI] [PubMed] [Google Scholar]
- Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: A class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- Cheng KF, Lin WJ. Simultaneously correcting for population stratification and for genotyping error in case-control association studies. Am J Hum Genet. 2007;81:726–743. doi: 10.1086/520962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Nelson SF. Genomic mismatch scanning identifies human genomic DNA shared identical by descent. Genomics. 1998;47:1–6. doi: 10.1006/geno.1997.5082. [DOI] [PubMed] [Google Scholar]
- Clayton DG, Chapman JM, Cooper JD. Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol. 2004;27:415–428. doi: 10.1002/gepi.20032. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet. 2002;70:124–141. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L. Genomic control for association studies: a semiparametric test to detect excess-haplotype sharing. Biostatistics. 2000;1:369–387. doi: 10.1093/biostatistics/1.4.369. [DOI] [PubMed] [Google Scholar]
- Dobson AJ. An Introduction to Generalized Linear Models. 2nd edition Chapman & Hall/CRC; New York: 2002. [Google Scholar]
- Douglas JA, Boehnke M, Gillanders E, Trent JM, Gruber SB. Experimentally derived haplotypes substantially increase the efficiency of linkage disequilibrium studies. Nat Genet. 2001;28:361–364. doi: 10.1038/ng582. [DOI] [PubMed] [Google Scholar]
- Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- Gower JC. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–338. [Google Scholar]
- Grant GR, Manduchi E, Cheung VG, Ewens WJ. Significance testing for direct identity-by-descent mapping. Ann Hum Genet. 1999;63:441–454. doi: 10.1046/j.1469-1809.1999.6350441.x. [DOI] [PubMed] [Google Scholar]
- Heidema AG, Boer JMA, Nagelkerke N, Mariman ECM, van der ADL, Feskens EJM. The challenge for genetic epidemiologists: how to analyze large numbers of SNPs in relation to complex diseases. BMC Genet. 2006;7:23–37. doi: 10.1186/1471-2156-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Humphreys K, Iles MM. Fine-scale mapping in case-control samples using locus scoring and haplotype-sharing methods. BMC Genet. 2005;6:S74. doi: 10.1186/1471-2156-6-S1-S74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klei L, Roeder K. Testing for association based on excess allele sharing in a sample of related cases and controls. Hum Genet. 2007;121:549–557. doi: 10.1007/s00439-007-0345-z. [DOI] [PubMed] [Google Scholar]
- Kuusisto J, Koivisto K, Kervinen K, Mykkanen L, Helkala E-L, Vanhanen M, Hanninen T, Pyorala K, Kesaniemi YA, Piekkinen P, Laasko M. Association of apolipoprotein E phenotypes with late onset Alzheimer's disease: population based study. Br Med J. 1994;309:636–638. doi: 10.1136/bmj.309.6955.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- Morris RW, Kaplan NL. Testing for association with a case-parents design in the presence of genotyping errors. Genet Epidemiol. 2004;26:142–154. doi: 10.1002/gepi.10297. [DOI] [PubMed] [Google Scholar]
- North BV, Sham PC, Knight J, Martin ER, Curtis D. Investigation of the ability of haplotype association and logistic regression to identify associated susceptibility loci. Ann Hum Genet. 2006;70:893–906. doi: 10.1111/j.1469-1809.2006.00301.x. [DOI] [PubMed] [Google Scholar]
- Rencher AC. Methods of Multivariate Analysis. 2nd edition Wiley; New York: 2002. [Google Scholar]
- Schaid DJ. Relative efficiency of ambiguous vs. directly measured haplotype frequencies. Genet Epidemiol. 2002;23:426–443. doi: 10.1002/gepi.10184. [DOI] [PubMed] [Google Scholar]
- Schaid DJ. Evaluating associations of haplotypes with traits. Genet Epidemiol. 2004a;27:348–364. doi: 10.1002/gepi.20037. [DOI] [PubMed] [Google Scholar]
- Schaid DJ. The complex genetic epidemiology of prostate cancer. Hum Mol Genet. 2004b;13:R103–R121. doi: 10.1093/hmg/ddh072. [DOI] [PubMed] [Google Scholar]
- Schaid DJ. Power and sample size for testing associations of haplotypes with complex traits. Ann Hum Genet. 2006;70:116–130. doi: 10.1111/j.1529-8817.2005.00215.x. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Hebbring SJ, Cunningham JM, Thibodeau SN. Nonparametric tests of association of multiple genes with human disease. Am J Hum Genet. 2005;76:780–793. doi: 10.1086/429838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Q, Chen H-S, Zhang S. A new association test using haplotype similarity. Genet Epidemiol. 2007;31:577–593. doi: 10.1002/gepi.20230. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintle NL, Ahn K, Mendell NR, Gordon D, Finch SJ. Characteristics of replicated single-nucleotide polymorphism genotypes from COGA: Affymetrix and Center for Inherited Disease Research. BMC Genet. 2005;6:S154. doi: 10.1186/1471-2156-6-S1-S154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J-Y, Zhang D. Haplotype-based association analysis via variance-components score test. Am J Hum Genet. 2007;81:927–938. doi: 10.1086/521558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J-Y, Devlin B, Wasserman L, Roeder K. On the identification of disease mutations by the analysis of haplotype similarity and goodness of fit. Am J Hum Genet. 2003a;72:891–902. doi: 10.1086/373881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J-Y, Byerley W, Devlin B, Roeder K, Wasserman L. Outlier detection and false discovery rates for whole-genome DNA matching. J Am Stat Assoc. 2003b;98:236–246. [Google Scholar]
- Tzeng J-Y, Chang S-M, Zhang D, Thomas DC, Davidian M. Regression-based multi-marker analysis for genome-wide association studies using haplotype similarity. 2007 submitted for publication. [Google Scholar]
- Van der Meulen MA, Te Meerman GJ. Haplotype sharing analysis in affected individuals from nuclear families with at least one affected offspring. Genet Epidemiol. 1997;14:915–920. doi: 10.1002/(SICI)1098-2272(1997)14:6<915::AID-GEPI59>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Wessel J, Schork NJ. Generalized genomic distance-based regression methodology for multilocus association analysis. Am J Hum Genet. 2006;79:792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Yue Q, Apprey V, Bonney G. Detecting disease gene in DNA haplotype sequences by nonparametric dissimilarity test. Hum Genet. 2006;120:253–261. doi: 10.1007/s00439-006-0216-z. [DOI] [PubMed] [Google Scholar]
- Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]