Abstract
Proteins exposed to UV radiation are subject to irreversible photodamage through covalent modification of tryptophans (Trps) and other UV-absorbing amino acids. Crystallins, the major protein components of the vertebrate eye lens that maintain lens transparency, are exposed to ambient UV radiation throughout life. The duplicated β-sheet Greek key domains of β- and γ-crystallins in humans and all other vertebrates each have two conserved buried Trps. Experiments and computation showed that the fluorescence of these Trps in human γD-crystallin is very efficiently quenched in the native state by electrostatically enabled electron transfer to a backbone amide [Chen et al. (2006) Biochemistry 45, 11552−11563]. This dispersal of the excited state energy would be expected to minimize protein damage from covalent scission of the excited Trp ring. We report here both experiments and computation showing that the same fast electron transfer mechanism is operating in a different crystallin, human γS-crystallin. Examination of solved structures of other crystallins reveals that the Trp conformation, as well as favorably oriented bound waters, and the proximity of the backbone carbonyl oxygen of the n − 3 residues before the quenched Trps (residue n), are conserved in most crystallins. These results indicate that fast charge transfer quenching is an evolved property of this protein fold, probably protecting it from UV-induced photodamage. This UV resistance may have contributed to the selection of the Greek key fold as the major lens protein in all vertebrates.
Absorption of ultraviolet light is a major source of damage to DNA and other biological macromolecules. All prokaryotic, archaeal, and eukaryotic cells have extensive mechanisms to repair DNA damage (1,2). Proteins are also damaged by absorption of ultraviolet light, with one of the major pathways being scission of the indole ring of tryptophans (Trps),1 often described as photobleaching when observed in vitro (3,4). For plants and many prokaryotes and smaller eukaryotes exposed to UV radiation, UV photodamage is a serious problem. Pathways for the efficient repair of such covalently damaged proteins have not been reported. Damaged proteins are instead degraded and replaced by new synthesis (5).
A key feature of proteins determining susceptibility to such covalent photodamage is the lifetime of the Trp excited state and the efficiency of its energy loss through fluorescence emission or quenching of the excited state. A number of investigators have described a variety of mechanisms by which excited state Trp is quenched (6−10). The fluorescence intensity, lifetimes, and wavelength of Trp within proteins are highly sensitive to protein environment and have long been exploited to follow a plethora of protein transformations that affect the local Trp environment (6). The 30-fold range of fluorescence quantum yields and lifetimes has recently been quantitatively explained in terms of electrostatically tuned electron transfer (11,12). In general, the quenching of Trp fluorescence and associated electronic interactions have been viewed as an adventitious aspect of protein structure, not reflective of any fundamental biological properties.
Ocular tissues are directly exposed to ambient sunlight, including potentially damaging UV. Crystallins, the major protein class of the eye lens of many species, are important for maintaining the transparency and providing a proper refractive index gradient in the eye lens (13). Though the cornea absorbs a significant fraction of the incident UV radiation, the UV reaching the lens crystallins integrated over the lens lifetime is significant (14). The human retina is very sensitive to photodamage from UV, and therefore the lens must absorb the UV reaching it to protect the retina (15).
The Greek key domains of the major β- and γ-crystallins of vertebrate lenses contain four conserved Trp residues (Figure 1). Within the lens they presumably absorb the UV radiation reaching the lens and may thus play a crucial role in protecting the retina from UVB (290−320 nm) photodamage. One outcome of UV absorption by Trps is covalent scission of the indole ring, believed to be irreversible in proteins. Trp photodamage in the crystallins has long been believed to be one factor in the etiology of cataract formation (16). Among older adults, the World Health Organization estimates that one-fifth of world blindness from cataract may be due to the UV radiation exposure (17).
Figure 1.
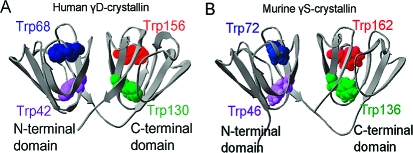
Homologous positions of four Trps in γ-crystallins. (A) The crystal structure of human γD-crystallin is shown as a ribbon diagram with four Trps in space-fill: Trp42 and Trp68 in the N-terminal domain and Trp130 and Trp156 in the C-terminal domain (PDB code 1HK0). (B) The NMR structure of murine γS-crystallin is depicted in ribbon representation showing the four Trps in space-fill: Trp46 and Trp72 in the N-terminal domain and Trp136 and Trp162 in the C-terminal domain (PDB code 1ZWM).
Among the human γ-crystallins, γC-, γD-, and γS-crystallins are the dominant species (13,18). The concentration of the crystallins in the lens reaches 200−400 mg/mL. γS-Crystallin represents an early evolutionary branch, which has been conserved and highly expressed throughout the vertebrates (19). Human γD-crystallin (HγD-Crys) is located in the center of the lens, the lens nucleus, and is synthesized early in lens differentiation. The terminally differentiated lens fiber cells lose their organelles so that there is neither new protein synthesis nor normal degradation within the lens nucleus. Human γS-crystallin (HγS-Crys) is especially abundant in the outer layer cortex, where protein turnover and regeneration occur in adulthood. These crystallins should not be confused with the enzymes which have been recruited as lens protein in birds and reptiles (13).
The other major crystallin family is the α-crystallins, members of the small heat shock family of chaperones, which function as passive chaperones suppressing aggregation from damaged or partially unfolded proteins (13). Failure of this process results in cataract, an aggregated insoluble form of crystallin which scatters visible light and degrades vision. Cataracts are the major cause of blindness in the world (16).
The crystallins are very stable and unusually long-lived. We have previously described the very rapid and efficient fluorescence quenching of the four conserved Trps in HγD-Crys in their native state (20,21). Recently developed methods (11,12,22−25) provide an understanding of Trp excited state quenching via local electrostatic promotion of electron transfer from the excited Trp to the surrounding. Examination of the conformation of the Trps within the tightly packed Greek key β-sheets reveals pathways for rapid loss of excitation energy, thereby reducing photodamage risk.
We have previously investigated the efficient quenching of Trp fluorescence in the native state of HγD-Crys (20,21). In HγD-Crys the homologous pair Trp68 and Trp156 is highly quenched in contrast to the homologous pair Trp42 and Trp130, which are moderately fluorescent. There is energy transfer from Trp42 to Trp68 in the N-terminal domain and from Trp130 to Trp156 in the C-terminal domain (20,21). Hybrid quantum mechanical−molecular mechanical (QM-MM) simulations indicated that electron transfer rates to the amide backbone of Trp68 and Trp156 are extremely fast relative to those for Trp42 and Trp130 (20). The charge transfer (CT) events of Trp68 and Trp156 are promoted by the net favorable location of several charged residues and nearby waters that electrostatically lower the energy of the resultant charge transfer state (20). This may be called an internal resonance Stark effect and is analogous to the external resonance Stark effect described by Treynor and Boxer (26).
Fluorescence quenching mechanisms are very sensitive to small changes in Trp conformation, to the presence of the orientation of bound water molecules, and to the nearby side chain packing interactions. There is no a priori reason to believe that the detailed quenching mechanism would be conserved in different crystallins. In this paper, we report on the quantum yields of four Trps in HγS-Crys, which shows essentially parallel behavior to HγD-Crys. We also report from examination of crystallographic structures remarkable structural (but not sequence) homology supporting conserved fluorescence properties of the conserved Trps of many other β- and γ-crystallins. These results suggest that the efficient excited state quenching may be an evolved property of the protein fold that allows it to absorb ultraviolet light with minimal damage from associated photochemical covalent ring scission reactions.
To facilitate keeping track of the differences among the four conserved Trps, we will use italics to represent the two efficiently quenched Trp68 and Trp156 in HγD-Crys and Trp72 and Trp162 in HγS-Crys. The other moderately or highly fluorescent Trps will be in regular font.
Materials and Methods
Mutagenesis, Expression, and Purification of Recombinant HγS-Crys
The wild-type HγS-Crys gene was subcloned into the pQE-1 vector (Qiagen) as described before (27). Single, double, and triple Trp to Phe substitutions were constructed using site-directed mutagenesis. Mutant primers (IDT-DNA) were used to amplify a pQE.1 plasmid encoding HγS-Crys gene with an N-terminal 6-His tag. The single, double, and triple Trp to Phe substitutions (Trp46-only, Trp72-only, Trp136-only, and Trp162-only) were constructed similarly to the procedure described by Kosinski-Collins et al. (28). All of the mutations were confirmed by DNA sequencing (Massachusetts General Hospital). The wild-type and mutant HγS-Crys proteins were expressed by Escherichia coli M15 [pREP4] cells. All of the mutants accumulated as native-like soluble proteins. The proteins were purified to over 98% homogeneity by affinity chromatography with a Ni-NTA resin as previously described (28).
Fluorescence Spectroscopy and Quantum Yield Determinations
The emission spectra of native and denatured proteins and the quantum yields of different proteins were measured using a Hitachi F-4500 fluorescence spectrophotometer as described previously (20). The spectra of native proteins were recorded in S buffer (10 mM sodium phosphate, 5 mM DTT, and 1 mM EDTA at pH 7.0), and the denatured proteins were in S buffer plus 5.5 M guanidine hydrochloride (GuHCl). The band-pass was 10 nm for both excitation and emission. Trp fluorescence emission spectra of all the proteins were measured in the range of 310−420 nm with an excitation wavelength of 300 nm. The protein concentrations were 2.75 μM except for the quantum yields measurements.
Quantum yields of the Trps in HγS-Crys were measured using the proteins Trp46-only, Trp72-only, Trp136-only, Trp162-only, Trp46/Trp72, Trp136/Trp162, and wild type. An excitation wavelength of 300 nm was used in order to minimize tyrosine fluorescence. Because there were large portions of the blue edge of the emission spectra that could not be observed using a 300 nm excitation wavelength, the unobserved areas of the protein spectra were estimated by matching the longer wavelength areas of protein spectra with spectra of 3-methylindole (3MI) in different solvent systems (Figures 8S−21S in Supporting Information). The solvent system used to match 3MI spectra with the spectra of wild-type HγS-Crys, Trp72-only, and Trp162-only was cyclohexane−dioxane (66:34), and the spectra of 3MI in dioxane were used to match the spectra of Trp46-only and Trp46/Trp72. The spectra of 3MI in cyclohexane−dioxane (93:7) were used to match the spectra of Trp136-only, and the spectra of 3MI in cyclohexane−dioxane (75:25) were used to match the spectra of Trp136/Trp162.
QM-MM Simulations
The hybrid QM-MM method used in this work has been described in recent publications (11,12,22−25) for applications to Trp fluorescence quenching in proteins. The method grew from an earlier QM-MM procedure used to predict the fluorescence wavelengths of Trp in proteins (29). Briefly, the QM method is Zerner’s INDO/S-CIS method (30), modified to include the local electric fields and potentials at the atoms. The MM part is Charmm (version 31a) (31). Hydrogens were added to the crystal structure (PDB code 1HK0 and 1HA4), and the entire protein was solvated within a 30 or 35 Å radius sphere of TIP3 explicit water. The waters were held within the chosen radius with a quartic potential. The quantum mechanical part includes the selected Trp and the amide of the preceding residue, capped with hydrogens, i.e., N-formyltryptophanamide. The electric potential for each QM atom is determined by a simple Coulomb’s Law sum over all non-QM atoms in the protein and solvent with dielectric constant = 1 and is added to the diagonal elements of the QM Fock Hamiltonian in the atomic orbital basis.
Results
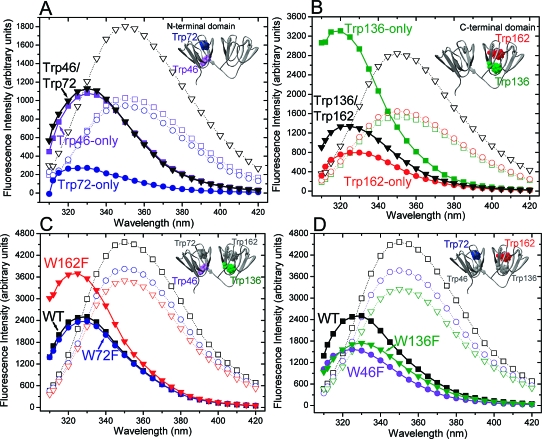
Figure 2 shows the fluorescence emission spectra of native and denatured wild-type HγS-Crys and the different Trp to Phe mutants. All of the spectra were taken with the same instrument settings and protein concentration, and therefore the areas under the spectra are in proportion to the quantum yields of the proteins. The fluorescence spectra of native and denatured double and triple Trp mutants, containing Trps only in the N-terminal domain (Trp46 and/or Trp72), are shown in Figure 2A. The spectra of Trps in the C-terminal domain are shown in Figure 2B (Trp136 and/or Trp162). Figure 2C displays comparison of the native and denatured wild type and single mutants of the weakly fluorescent Trps (W72F and W162F). Figure 2D shows the single substitutions of the moderately fluorescent Trp (W46F) and the strongly fluorescent Trp (W136F).
Figure 2.
Fluorescence emission spectra of native and denatured wild type and various Trp mutants of HγS-Crys. (A) Fluorescence emission spectra of native Trp46-only (◼), Trp72-only (●), and Trp46/Trp72 (▼) and denatured Trp46-only (◻), Trp72-only (○), and Trp46/Trp72 (▽). (B) Fluorescence emission spectra of native Trp136-only (◼), Trp162-only (●), and Trp136/Trp162 (▼) and denatured Trp136-only (◻), Trp162-only (○), and Trp136/Trp162 (▽). (C) Fluorescence emission spectra of native wild type (WT) (◼), W72F (●), and W162F (▼) and denatured wild type (◻), W72F (○), and W162F (▽). (D) Fluorescence emission spectra of native wild type (◼), W46F (●), and W136F (▼) and denatured wild type (◻), W46F (○), and W136F (▽). The solid lines represent the emission spectra of native proteins, and the dotted lines represent the unfolded proteins. Native proteins were incubated in S buffer, and unfolded proteins were incubated in S buffer plus 5.5 M GuHCl for 6 h at 37 °C. The excitation wavelength was at 300 nm, and the protein concentration was 2.75 μM. The buffer signal was subtracted from all spectra. Because the crystal structure of full-length HγS-Crys is not available, the ribbon structure of murine γS-crystallin with Trps in space-fill is shown here instead at the upper right corner.
The fluorescence from Trps 72 and 162 is weaker than that of Trps 46 and 136 (Figure 2A,B). The difference is more significant for the Trps in the C-terminal domain than the ones in the N-terminal domain. The areas under the curves of the unfolded proteins are approximately in proportion to the number of Trps contained in the proteins. This suggests that the environments of the four Trps are approximately the same in the denatured state. Quantitative determination of the quantum yields for wild type and the triple mutants are summarized in Table 1S in Supporting Information. The average quantum yield for the triple Trp mutants was 0.090, about two times higher than that of the wild type.
Quenching by Electron Transfer
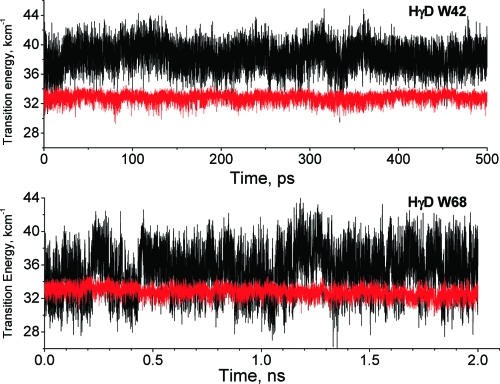
Figure 3 displays QM-MM trajectories that show INDO/S-computed transition energies for the fluorescing state (red) and the CT state (black) for Trps 42 and 68 of HγD-Crys. The relative gap between the states is the main determinant of electron transfer based fluorescence quenching and is seen to be considerably larger for Trp42 than for Trp68. These trajectories are representative of Trps 130 and 156 of HγD-Crys as well as Trps 136 and 162 of HγS-Crys. Electronic coupling of the fluorescing state to the CT state is similar in all cases. Trajectories were typically 150 ps in length. The longer trajectory shown for Trp68 in Figure 3 was prompted by the greater fluctuations noted for Trp68 during the first 150 ps. A longer trajectory for Trp42 was also done.
Figure 3.
QM-MM trajectories showing transition energies for the fluorescing state (red) and the CT state (black) for Trps 42 and 68 of HγD-Crys. The relative gap between the states is the main determinant of electron transfer based fluorescence quenching. The energy scale is in kcm−1 (8 kcm−1 = 1 eV = 96.5 kJ/mol). Fluctuations on the gap are on the order of 1 eV.
Table 1 and Figures 1S and 2S in Supporting Information compare calculated electron transfer (ET) rates and fluorescence quantum yields for HγD-Crys and HγS-Crys Trps using two recently published procedures, procedure I (11) and procedure II (12). Common to both procedures is the same QM-MM trajectory of 1La and CT transition energies, with the all-important energy gap between the fluorescing and CT states being ∼5000 cm−1 (0.6 eV) greater for the fluorescent Trps than for the nonfluorescent Trps. Both procedures are seen to predict the correct order of magnitude for the quantum yields. The low intrinsic electron affinity of amides means that a CT state involving electron transfer from the excited Trp ring to an amide is energetically accessible only if the protein/solvent environment lowers the relative energy by making the electric potential at the amide carbonyl carbon effectively the same as on the ring, i.e., bringing the 1La and CT states into resonance so that energy is conserved during the electron transfer. Figures 3S and 4S in Supporting Information show that the lower CT energy for Trps 68 and 156 comes from net average stabilization from both protein and water compared to Trps 42 and 130. The same pattern is found for the corresponding Trps of HγS-Crys (Figure 5S in Supporting Information).
Table 1. Comparison of Predicted and Experimental Quantum Yields.
| residue | trajectory length (ps) | CT-1La gap (kcm−1) | std dev (kcm−1) | ET rate constant (109 s−1)a | predicted quantum yieldsa | ET rate constant (109 s−1)b | predicted quantum yields and std devb | exptl quantum yields |
|---|---|---|---|---|---|---|---|---|
| HγSTrp136 | 50 | 7.93 | 1.85 | 0.053 | 0.218 | 0.034 | 0.24 | 0.25 |
| HγSTrp162 | 50 | 0.42 | 2.64 | 11.2 | 0.004 | 270 | 0.0001 | 0.03 |
| HγDTrp42 | 500 | 5.6 | 1.7 | 0.51 | 0.067 | 0.52 ± 0.17 | 0.07 ± 0.02 | 0.13c |
| HγDTrp68 | 2000 | 2.6 | 2.9 | 6.3 | 0.006 | 136 ± 68 | 0.0003 ± 0.0002 | 0.008 |
| HγDTrp130 | 150 | 6.0 | 1.6 | 0.33 | 0.087 | 1.2 ± 0.8 | 0.09 ± 0.09 | 0.17 |
| HγDTrp156 | 150 | 1.0 | 2.8 | 10.0 | 0.004 | 173 ± 100 | 0.0005 | 0.01 |
Computed by procedure I (11).
Computed by procedure II (12). Quantum yields and deviations are based on 50 ps segments of the trajectory.
Data in italics have been reported previously (20).
About 10% errors on the quantum yields are estimated due to the systematic errors (extrapolation, instrument wavelength-dependent sensitivity, polarization effects, etc.). Although it is unlikely that an error in the quantum yield due to the estimation technique employed would exceed 10−20%, the computational method is expected to have much larger uncertainty. The uncertainty depends on the degree of quenching. Several fold errors are possible at the low quantum yield end, but when quenching is slight, errors in the electron transfer rate have much less effect. This stems in part from the difficulty in being sure that the somewhat short simulations adequately represent the average environment effects to which the electron transfer rate is so sensitive.
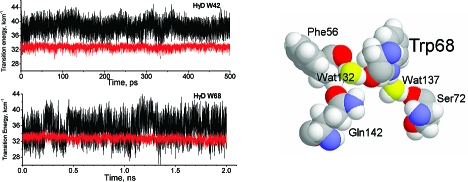
Examination of the electrostatic contributions from nearby waters shows that the nonfluorescent Trps 68 and 156 of HγD-Crys and their counterparts in HγS-Crys have two tightly bound waters that contribute strongly to stabilizing the CT state. These two waters are prominent in the crystal structures of 10 γ- and β-crystallins from a variety of species (Figure 4A and Table 2) (32−37). One is usually an H-bond donor to the O of the Trp amide and stabilizes the electron transfer because the protons are near the amide C atom. The other is usually an H-bond acceptor from the HE1 on the N of the indole ring. This stabilizes because of the proximity of the negative water O atom to the ring, which is positive in the CT state.
Figure 4.
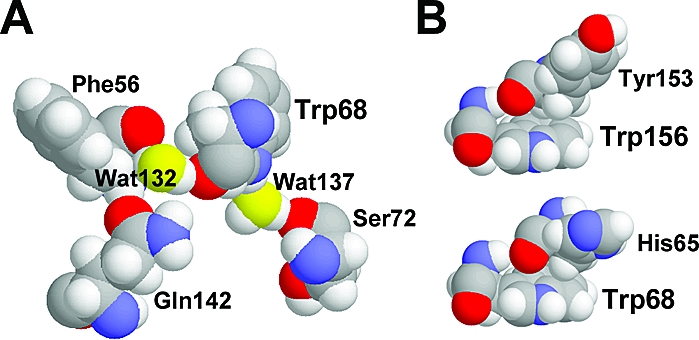
Representative conserved nearby waters and aromatic residues of quenched Trps in HγD-Crys. (A) The two crystallographic water molecules (yellow oxygens) that most stabilize the electron transfer from the indole ring to the amide backbone of Trp68 in the HγD-Crys (PDB code 1HK0). These sites do not appear to be accessible, but Wat132 is trapped by an additional H-bond to Gln142 and/or Phe56, and Wat137 is usually also H-bonded to Ser72. (B) Representative conserved proximity of the backbone carbonyl bond of n − 3 residues before the nonfluorescent Trps (residue n) in the HγD-Crys.
Table 2. Conformation of Trps at Homologous Position (Trps 68 and 157) in β- and γ-Crystallinsa.
| crystallin | quenching of Trp b | PDB code | dihedral angles of Trps and distance between Trp and its close-by waters and n − 3 residue |
|---|---|---|---|
| human γD-crystallin | yesc | 1HK0 | Trp68: ω = 167.7°, ψ = −108.66°, φ = 18.77°, χ1 = 60.5, χ2 = 79.1; H2O137 (2.9 Å from NE1), H2O132 (2.8 Å from O=C); His65 O is ∼3.5 Å from CG |
| Trp157:d ω = 171.83°, ψ = −101.27°, φ = 12.10°, χ1 = 58.4, χ2 = 86.3; H2O270 (2.9 Å from NE1), H2O263 (2.7 Å from O=C); Tyr154 O is ∼3.5 Å from CG | |||
| human γS-crystallin | yes | 1HA4 (C-td) | Trp157: ω = 176.55°, ψ = −111.57°, φ = 23.87°, χ1 = 61.0, χ2 = 74.4; H2O37 (2.9 Å from NE1), H2O36 (2.7 Å from O=C); Pro154 O is ∼3.5 Å from CG |
| human βB1-crystallin | yes | 1OKI | Trp126: ω = 170.08°, ψ = −100.10°, φ = −22.53°, χ1 = 67.7, χ2 = 65.8; H2O110 (2.8 Å from NE1); Trp123 O is ∼3.5 Å from CG |
| Trp218: ω = 170.49°, ψ = −103.52°, φ = 13.71°, χ1 = 60.4, χ2 = 81.9; H2O190 (3.1 Å from NE1), H2O172 (2.9 Å from O=C); Trp215 O is ∼3.5 Å from CG | |||
| bovine γB-crystallin | −,e predicted | 1AMM | Trp68: ω = 173.90°, ψ = −112.42°, φ = 15.92°, χ1 = 59.4, χ2 = 83.4; H2O229 (2.9 Å from NE1), H2O237 (2.8 Å from O=C); Tyr65 O is 3.5 Å from CG |
| Trp157: ω = 175.83°, ψ = −101.18°, φ = 8.82°, χ1 = 57.9, χ2 = 87.8; H2O210 (2.9 Å from NE1), H2O226 (2.8 Å from O=C); Tyr154 O is 3.5 Å from CG | |||
| rat γE-crystallin | − | 1A5D | Trp68: ω = 176.00°, ψ = −113.21°, φ = 17.53°, χ1 = 64.7, χ2 = 76.3; H2O218 (3.3 Å from NE1), H2O286 (2.8 Å from O=C); Tyr65 O is 3.3 Å from CG |
| Trp157: ω = 179.52°, ψ = −107.75°, φ = 21.00°, χ1 = 59.0, χ2 = 78.0; H2O220 (3.0 Å from NE1), H2O246 (3.1 Å from O=C); Tyr154 O is 3.6 Å from CG | |||
| ciona β,γ-crystallin | −, predicted | 2BV2 | Trp70: ω = 168.47°, ψ = −105.76°, φ = 5.63°, χ1 = 67.9, χ2 = 82.0; H2O50 (3.1 Å from NE1), H2O37 (3.1 Å from O=C), H2O45 (2.6 Å from O=C); Pro67 O is 3.5 Å from CG |
Only the conformation of Trps at homologous position (Trps68 and 157) in chain A of representative β- and γ-crystallins is listed above, and that of bovine γS-crystallin, bovine γD-crystallin, bovine γE-crystallin, and bovine γF-crystallin is listed in Table 3S in Supporting Information.
Quenching of Trp fluorescence emission refers to the lower fluorescence intensity in the native state protein with respect to the denatured state protein.
The experimental characterizations of the fluorescence emission spectrum of various crystallins can be found in refs (20),(28), and (40).
The amino acid sequence number is consistent with the protein database.
−: experimental data of the fluorescence emission spectrum of native and unfolded state protein is unavailable.
Together, the two waters stabilize the CT state by ∼0.5 eV on average. In the case of HγD-Crys, these water hydrogen bonds to the Trp are transiently broken multiple times during the 150 ps trajectories, but never for more than about 100 fs (Figure 6S in Supporting Information). When the Wat132 H-bond is broken, it is H-bonded to the backbone O of Phe56 or the side chain HE of Gln142 and trapped nearby. A similar behavior is found with Wat137, which is kept nearby by an H-bond donated to the backbone O of Ser72. This pattern is widely repeated. For Trp156 of HγD-Crys, Wat263 is trapped near the carbonyl by HN of Leu144 and O of Gln54, and Wat270 is held by Asn160. For Trp157 of HγS-Crys, Arg142 holds a water near the carbonyl and Ser161 holds a water near the HN of the ring.
Förster Resonance Energy Transfer
The fluorescence emission spectra of single, double, and triple Trp mutants (Figure 2) strongly suggest that there is intradomain Förster resonance energy transfer (FRET) in both the C-terminal domain and N-terminal domain. In the C-terminal domain, the highly fluorescent Trp136 acts as the energy donor, and the weakly fluorescent Trp162 functions as the energy acceptor. In Figure 2C, when Trp162 was substituted with Phe, the integrated fluorescence intensity of W162F was 34% higher than wild type. In contrast, as shown in Figure 2D, the integrated fluorescence intensity of W136F decreased 25% compared to wild type. The spectra provide convincing evidence that Trp136 is the energy donor. The maximal emission wavelength for Trp136 is blue shifted about 10 nm compared to Trp162, consistent with a less polar environment for Trp136. A large portion of the excitation energy from Trp136 is not emitted due to the energy transfer to the weakly emitting Trp162. As shown in Figure 2D, because of the positive contribution of Trp136 to the overall fluorescence intensity of wild-type HγS-Crys, a decrease in fluorescence intensity was observed for its Phe substitution (W136F).
The results of the fluorescence emission spectra of double Trp mutants (Trp136/Trp162) further confirmed resonance energy transfer in the C-terminal domain. Fluorescence intensity of the double mutant Trp136/Trp162 was not equal to the simple addition of Trp136-only and Trp162-only fluorescence. It was 1.6 times higher than the intensity of Trp162-only and 51% lower than the intensity of Trp136-only (Figure 2B).
Resonance energy transfer was also observed in the N-terminal domain. Moderately fluorescent Trp46 functions as the energy donor, and weakly fluorescent Trp72 is the energy acceptor. Because of the energy transfer from Trp46 to Trp72 in the N-terminal domain, the intensity of the double Trp mutant, Trp46/Trp72, is essentially equal to that of Trp46-only (Figure 2A), whereas it would be equal to the addition of the intensity of both Trp46-only and Trp72-only if there is no energy transfer. The energy donor, Trp46, makes the dominant contribution to the overall fluorescence intensity of wild-type HγS-Crys, and therefore substitution of Trp46 (W46F) caused a decrease in fluorescence intensity compared to wild type (Figure 2D). In Figure 2C, when the energy acceptor Trp72 was substituted with Phe, the fluorescence intensity of W72F was approximately equal to that of wild type. Because the intensity of the donor Trp46 is somewhat low, even though the Förster energy transfer is efficient in the N-terminal domain, the intensity of W72F was not higher than that of wild type. In summary, because there is little uncertainty in the absolute value of the fluorescence intensity, and because of the somewhat low quantum yield of Trp46, the differences between various Trp spectra in the N-terminal domain were not as large as those in the C-terminal domain, but the energy transfers are quite efficient in both domains.
There is no significant interdomain energy transfer from the highly fluorescent Trp136 in the C-terminal domain to the weakly fluorescent Trp72 in the N-terminal domain based on the spectra of the double Trp mutant Trp136/Trp162 in Figure 2B and the single Trp mutant W46F (containing Trps 136, 162, and 72) in Figure 2D. In the case of the interdomain energy transfer from Trp136 to Trp72, the fluorescence intensity of the double Trp mutant Trp136/Trp162 (containing Trp136 and Trp162 but not potential energy acceptor Trp72) should be higher than the single Trp mutant W46F (containing Trps 136 and 162 and potential energy acceptor Trp72). Because of the similar fluorescence intensities of both Trp136/Trp162 and W46F, there is probably no interdomain energy transfer from Trp136 to Trp72. Similarly, the comparison of the double Trp mutant Trp46/Trp72 (containing Trps 46 and 72 but not potential energy acceptor Trp162) in Figure 2A and single Trp mutant W136F (containing Trps 46 and 72 and potential energy acceptor Trp162) in Figure 2D suggests that there is little or no energy transfer from moderately fluorescent Trp46 in the N-terminal domain to the weakly fluorescent Trp162 in the C-terminal domain.
Discussion
Conservation of Fluorescence Quenching by Charge Transfer in β- and γ-Crystallins
The mechanism of fluorescence quenching by charge transfer appears to be conserved in β- and γ-crystallins and probably represents a physiological function of the crystallins selected for at some stage in lens evolution. Based on the available fluorescence data, fluorescence quenching phenomena of native state crystallins have been confirmed for human γC-, γD-, and γS-Crys, mouse γN-crystallin, and human βA1-, βA3-, βA4-, and βB1-crystallin (19,20,27,28,38−40). Thus far, the crystal structures of nine different γ-crystallins have been resolved. It is striking that in these γ-crystallins all of the Trps at homologous positions (as Trps68/156 in HγD-Crys) have similar dihedral angles and two water molecules within 3.1 Å (Table 2 and Table 3S in Supporting Information). The highly quenched Trps (Trps68/156 in HγD-Crys and Trps72/162 in HγS-Crys) are buried in the hydrophobic core and forming H-bonds with two water molecules nearby. The two water molecules stabilize the charge transfer events by forming H-bonds with the Trp indole ring and backbone, respectively (Figure 4A). Unlike γ-crystallin, β-crystallins contain not only buried Trps but also solvent-exposed Trps. In human βB1-crystallin, buried Trp218 also displays similar dihedral angles and H-bond formations with two water molecules.
A second strikingly conserved structural feature is the presence of an aromatic residue or proline occurring at sequence number n − 3, where n is the position of a conserved weakly fluorescent Trp. This may be relevant to increasing the quenching of these Trps because the backbone carbonyl from the n − 3 residue is always pressed into the face of the Trp ring, with the n − 3 O atom always close to 3.5 Å from the CG of the Trp (Figure 4B, Table 2, and Table 3S in Supporting Information). This arrangement of amide C=O and Trp ring has recently been implicated in ultrafast quenching of Trp in myoglobin by Zhong and co-workers (10). A preliminary simulation shows that ∼50% of the time the lowest CT state has the electron transferred to the n − 3 amide instead of the Trp amide. In Table 2 it is seen that for γB-, γD-, and γE-crystallins the n − 3 residue is Tyr, with the exception of Trp68 of HγD-Crys, for which it is His. For the γS-crystallins, it is Pro, and for the β-crystallins, it is Trp. Förster transfer is expected to be rapid and complete to the nearby low-fluorescent Trp so that the additional Trps in the β-crystallins should not fluoresce well. The type of arrangement of n − 3 residue relative to the conserved weakly fluorescent Trp may contribute to the high incidence of aromatic−aromatic interactions in proteins reported by Burley and Petsko (41).
Selection for UV Radiation Resistance May Have Contributed to the Recruitment of the Crystallin Fold as a Lens Protein
The efficient quenching by charge transfer depends critically on the precise orientation of the Trp side chain, the presence of the oriented waters, and the presence of nearby favorably charged groups. Though functions of bound waters have been shown to assist catalytic activities in a variety of active sites and also to stabilize the collagen fold through H-bond to hydroxyprolines (42,43), the results reported here suggest an additional role for bound waters within protein structures. The tightly packed buried core of the Greek key provides an appropriate environment for maintaining such relationships and also excludes solvent. The lack of motion of the Trp ring is clear not only from the high-resolution crystal structure (32) but from fluorescence anisotropy measurements of the protein in solution (21). Thus this fast charge transfer quenching pathway is intimately tied to the detailed fold of the crystallins.
The image-forming lens is associated with the origin of the vertebrates and is a more recent event than the emergence of the Greek key β-sheet fold in proteins. Bird lens proteins include molecules that have clearly been recruited from other tissues because of some features of their solubility or stability or regulation (13). Of considerable evolutionary interest is the question of the ancestry of the crystallin family. The earliest homologue is a monomeric crystallin from the sea squirt Ciona intestinalis, believed to be related to early vertebrate progenitors (36). The sea squirt lacks a lens or image-forming eye. However, its crystallin has Trps in the homologous position as the human γ-crystallins (Table 2).
Single cell and small metazoans are more exposed to radiation than large organisms. The conformations of the crystallins conferring UV resistance presumably evolved hundreds of millions of years ago, long before the emergence of the vertebrates. Perhaps one factor that led to the recruitment of the crystallin family for the vertebrate lens was that this fold already exhibited efficient UV absorption, with minimal photodamage, due to efficient quenching. The lack of nuclei, ribosomes, proteasomes, and other cell organelles in the mature lens limits the possibility for replacing damaged crystallins through the degradative pathways used by other cell types. Though the damaged proteins are probably bound by α-crystallin, they remain within the lens, often as constituents of cataracts (44).
A great variety of terrestrial vertebrates, as well as Homo sapiens through most of human evolution, spent much of their lifetime exposed to sunlight. Selection for the vertebrate lens to remain transparent and flexible over a reproductive lifetime has probably been significant.
Have Crystallin Folds Evolved To Protect Trps from UV Irradiation Damage by Photoinduced Electron Transfer?
Though the crystallins may be resistant to UV-induced photodamage, over a lifetime we would expect accumulation of some level of covalently damaged proteins. Bachem (45) reported that the action spectrum for in vivo cataract formation in guinea pigs and rabbits rose sharply from zero at 293 nm to a maximum near 300 nm and fell to near zero at 313 nm. Kurzel et al. (46) demonstrated that this action spectrum is closely duplicated by the product of the transmission spectrum of the cornea and the Trp phosphorescence action spectrum of intact human lenses at 77 K, thereby strongly implicating excitation of Trp as a primary cause of cataracts.
In humans cataract is rare through the first four decades of life but increases sharply with age over 50 (47). We suspect that the low incidence in younger adults is not due to lack of crystallin damage but to the protective function of the α-crystallins. These oligomeric complexes which bind damaged crystallins, but do not refold them, may become saturated in later life. There is considerable epidemiologic evidence showing a direct association between cumulative exposure to UVB radiation and prevalence of cortical (outer one-third of lens) cataract in humans (16).
The presence of the conserved Trps, whether for essential structure or for UV protective purposes, poses a challenge because of the enormous cumulative photooxidative stress from ambient UV light absorbed by Trps that do not turn over. This raises the question of why the Trps have been conserved. We suspect that the conservation of the four Trps in modern mammals may reflect its function as a UV filter to protect the retina. The lens also contains small molecule UV absorbers which are believed to function as UV filters (48).
A 6-fold repeated form of the crystallin domain has recently been described in an epidermal protein absent in melanoma 1 (AIM1) (49). As described in its name, the protein was identified because it is missing in melanoma tumors. It may be that the normal function of the AIM protein is as a UV filter and that in its absence epidermal cells are sensitive to UV-induced mutation and carcinogenesis.
Alternate models of Trp quenching, such as electron transfer to metals or protonated cysteines or other amino acids (11), might also be expected to protect the Trps from photodamage. Why has efficient quenching by fast electron transfer to the amide bond been so conserved? The most attractive hypothesis is that the fast electron transfer rapidly returns the excited Trp to the ground state, in a manner that bypasses photochemical pathways known to be accessible from the excited state, most likely the lowest triplet state. Previous time-resolved fluorescent measurements indeed show that the highly quenched Trp68 and Trp156 have very short lifetimes, τ ∼ 0.1 ns. In contrast, the moderately fluorescent Trp42 and Trp130 have longer lifetimes, τ ∼ 3 ns (21).
Strong support for this hypothesis comes from the observation by Tallmadge and Borkman (50) that irradiation of bovine γB-crystallin (γB-Crys) in vitro with 295 nm UV light photodegrades Trps 42 and 131 three times more efficiently than Trps68 and 157. Our QM-MM based mechanism powerfully links structure to experimental quenching results, allowing, through Table 2, the highly plausible supposition that Trps68 and 157 will exhibit a fast electron transfer deactivation of the excited state. Further protection by FRET from the photoreactive Trps 42 and 131 to the less reactive Trps 68 and 157 is also inferred by this scheme.
If, however, photolysis were not possible following electron transfer, the ratio of photolysis yields should equal the ratio of fluorescence quantum yields. This follows from the reasonable notion that the photolysis rate is much slower than the fluorescence rate, so that both quantum yields would be reduced in proportion by a competing process. We must therefore conclude that either the ratio of fluorescence quantum yields in the bovine γB-Crys is only about 3:1 or that some photolysis is possible from the CT state. The fluorescence intensity ratio for Trps 42, 131 to Trps 68, 157 is unknown for bovine γB-Crys. Assuming it is similar to what we have measured for the HγD-Crys and HγS-Crys, the ratio of photolysis yields should be ∼10:1 if no photolysis were possible from the CT state. A reasonable possibility is that there is some singlet to triplet conversion from the CT state. It is known that this could be fast because the singlet and triplet CT states are very close in energy, making conversion much easier. The excited triplet state is the most likely precursor of permanent photodamage because of its many orders of magnitude longer decay time.
Conclusion
We conclude that the conservation of electrostatically enabled excited state quenching of conserved Trps, in the absence of precise sequence homology, is an evolved property of the crystallin fold. This permits the vertebrate lens to contain Trps, which are essential for structural integrity and UV protection of the retina, with minimal photodegradation over a lifetime of UVB exposure.
Acknowledgments
We thank Prof. Ludwig Brand and Dr. Dmitri Toptygin at Johns Hopkins University for helpful discussions.
Supporting Information Available
Figures 1S and 2S showing the comparison of experimental and calculated fluorescence quantum yields for Trps in HγD-Crys or HγS-Crys; Figures 3S−5S showing computed electrostatic stabilization of the charge transfer state due to protein and water or protein for Trps of HγD-Crys or HγS-Crys; Figure 6S showing Wat132 H-bond distance to Trp68 in HγD-Crys; Figure 7S showing the amino acid sequence alignment of γ- and β-crystallin; Table 1S showing the quantum yields of wild type and Trp mutants of HγS-Crys; Table 2S showing the dihedral angles of Trp72 and Trp162 of murine γS-crystallin; Table 3S showing the fluorescence quenching phenomena, conformation of Trps at homologous position (Trp68 and Trp156), and the nearby waters and n − 3 residue of these Trps in crystallins; Figures 8S−21S showing the fluorescence emission spectra of 3MI and the spectra of Trp mutants (Trp46-only, Trp72-only, Trp46/Trp72, Trp136-only, Trp162-only, and Trp136/Trp162) and wild type for the extrapolation of the quantum yields of Trps in HγS-Crys on the blue side. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: Trps, tryptophans; HγD-Crys, human γD-crystallin; HγS-Crys, human γS-crystallin; QM-MM, hybrid quantum mechanics−molecular mechanics; GuHCl, guanidine hydrochloride; 3MI, 3-methylindole; CT, charge transfer; ET, electron transfer; FRET, Förster resonance energy transfer; AIM1, absent in melanoma 1; γB-Crys, γB-crystallin.
Supplementary Material
References
- Jun S. H.; Kim T. G.; Ban C. (2006) DNA mismatch repair system. Classical and fresh roles. FEBS J. 273, 1609–1619. [DOI] [PubMed] [Google Scholar]
- White M. F. (2003) Archaeal DNA repair: paradigms and puzzles. Biochem. Soc. Trans. 31, 690–693. [DOI] [PubMed] [Google Scholar]
- Davies M. J.; Truscott R. J. (2001) Photo-oxidation of proteins and its role in cataractogenesis. J. Photochem. Photobiol. B 63, 114–125. [DOI] [PubMed] [Google Scholar]
- Grossweiner L. I. (1984) Photochemistry of proteins: a review. Curr. Eye Res. 3, 137–144. [DOI] [PubMed] [Google Scholar]
- Barber J.; Andersson B. (1992) Too much of a good thing: light can be bad for photosynthesis. Trends Biochem. Sci. 17, 61–66. [DOI] [PubMed] [Google Scholar]
- Beechem J. M.; Brand L. (1985) Time-resolved fluorescence of proteins. Annu. Rev. Biochem. 54, 43–71. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Barkley M. D. (1998) Toward understanding tryptophan fluorescence in proteins. Biochemistry 37, 9976–9982. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Liu B.; Yu H.-T.; Barkley M. D. (1996) The peptide bond quenches indole fluorescence. J. Am. Chem. Soc. 118, 9271–9278. [Google Scholar]
- Petrich J. W.; Chang M. C.; Mcdonald D. B.; Fleming G. R. (1983) On the origin of non-exponential fluorescence decay in tryptophan and its derivatives. J. Am. Chem. Soc. 105, 3824–3832. [Google Scholar]
- Qiu W.; Li T.; Zhang L.; Yang Y.; Kao Y.-T.; Wang L.; Zhong D. (2008) Ultrafast quenching of tryptophan fluorescence in proteins: interresidue and intrahelical electron transfer. Chem. Phys. 350, 154–164. [Google Scholar]
- Callis P.; Liu T. (2004) Quantitative predictions of fluorescence quantum yields for tryptophan in proteins. J. Phys. Chem. B 108, 4248–4259. [Google Scholar]
- Callis P. R.; Petrenko A.; Muino P. L.; Tusell J. R. (2007) Ab initio prediction of tryptophan fluorescence quenching by protein electric field enabled electron transfer. J. Phys. Chem. B 111, 10335–10339. [DOI] [PubMed] [Google Scholar]
- Bloemendal H.; de Jong W.; Jaenicke R.; Lubsen N. H.; Slingsby C.; Tardieu A. (2004) Ageing and vision: structure, stability and function of lens crystallins. Prog. Biophys. Mol. Biol. 86, 407–485. [DOI] [PubMed] [Google Scholar]
- Lerman S. (1980) Radiant energy and the eye. Chapter 3: Biological and chemical effects of ultraviolet radiation, Macmillan Publishing, New York. [Google Scholar]
- Li Z. L.; Tso M. O.; Jampol L. M.; Miller S. A.; Waxler M. (1990) Retinal injury induced by near-ultraviolet radiation in aphakic and pseudophakic monkey eyes. A preliminary report. Retina 10, 301–314. [DOI] [PubMed] [Google Scholar]
- Robman L.; Taylor H. (2005) External factors in the development of cataract. Eye 19, 1074–1082. [DOI] [PubMed] [Google Scholar]
- WHO (2006) Global disease burden from solar ultraviolet radiation, WHO, WHO sites, Media centre, Fact sheets
- Lampi K. J.; Ma Z.; Shih M.; Shearer T. R.; Smith J. B.; Smith D. L.; David L. L. (1997) Sequence analysis of betaA3, betaB3, and betaA4 crystallins completes the identification of the major proteins in young human lens. J. Biol. Chem. 272, 2268–2275. [DOI] [PubMed] [Google Scholar]
- Wistow G.; Wyatt K.; David L.; Gao C.; Bateman O.; Bernstein S.; Tomarev S.; Segovia L.; Slingsby C.; Vihtelic T. (2005) gammaN-crystallin and the evolution of the betagamma-crystallin superfamily in vertebrates. FEBS J. 272, 2276–2291. [DOI] [PubMed] [Google Scholar]
- Chen J.; Flaugh S. L.; Callis P. R.; King J. (2006) Mechanism of the highly efficient quenching of tryptophan fluorescence in human gammaD-crystallin. Biochemistry 45, 11552–11563. [DOI] [PubMed] [Google Scholar]
- Chen J.; Toptygin D.; Brand L.; King J. (2008) Mechanism of the efficient tryptophan fluorescence quenching in human gammaD-crystallin studied by time-resolved fluorescence. Biochemistry 47, 10705–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis P.; Vivian J. (2003) Understanding the variable fluorescence quantum yield of tryptophan in proteins using QM-MM simulations. Quenching by charge transfer to the peptide backbone. Chem. Phys. Lett. 369, 409–414. [Google Scholar]
- Kurz L. C.; Fite B.; Jean J.; Park J.; Erpelding T.; Callis P. (2005) Photophysics of tryptophan fluorescence: link with the catalytic strategy of the citrate synthase from Thermoplasma acidophilum. Biochemistry 44, 1394–1413. [DOI] [PubMed] [Google Scholar]
- Liu T.; Callis P. R.; Hesp B. H.; de Groot M.; Buma W. J.; Broos J. (2005) Ionization potentials of fluoroindoles and the origin of nonexponential tryptophan fluorescence decay in proteins. J. Am. Chem. Soc. 127, 4104–4113. [DOI] [PubMed] [Google Scholar]
- Xu J.; Toptygin D.; Graver K. J.; Albertini R. A.; Savtchenko R. S.; Meadow N. D.; Roseman S.; Callis P. R.; Brand L.; Knutson J. R. (2006) Ultrafast fluorescence dynamics of tryptophan in the proteins monellin and IIA(Glc). J. Am. Chem. Soc. 128, 1214–1221. [DOI] [PubMed] [Google Scholar]
- Treynor T. P.; Boxer S. G. (2004) Probing excited-state electron transfer by resonance stark spectroscopy: 3. Theoretical foundations and practical applications. J. Phys. Chem. B 108, 13513–13522. [Google Scholar]
- Mills I. A.; Flaugh S. L.; Kosinski-Collins M. S.; King J. A. (2007) Folding and stability of the isolated Greek key domains of the long-lived human lens proteins gammaD-crystallin and gammaS-crystallin. Protein Sci. 16, 2427–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski-Collins M. S.; Flaugh S. L.; King J. (2004) Probing folding and fluorescence quenching in human gammaD crystallin Greek key domains using triple tryptophan mutant proteins. Protein Sci. 13, 2223–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian J. T.; Callis P. R. (2001) Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 80, 2093–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley J.; Zerner M. (1973) An intermediate neglect of differential overlap technique for spectroscopy: pyrrole and the azines. Theor. Chim. Acta (Berlin) 32, 111–134. [Google Scholar]
- MacKerell A. D.; Bashford D.; Bellott M.; Dunbrack R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; Joseph-McCarthy D.; Kuchnir L.; Kuczera K.; Lau F. T. K.; Mattos C.; Michnick S.; Ngo T.; Nguyen D. T.; Prodhom B.; Reiher W. E.; Roux B.; Schlenkrich M.; Smith J. C.; Stote R.; Straub J.; Watanabe M.; Wiórkiewicz-Kuczera J.; Yin D.; Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Basak A.; Bateman O.; Slingsby C.; Pande A.; Asherie N.; Ogun O.; Benedek G. B.; Pande J. (2003) High-resolution X-ray crystal structures of human gammaD crystallin (1.25 Å) and the R58H mutant (1.15 Å) associated with aculeiform cataract. J. Mol. Biol. 328, 1137–1147. [DOI] [PubMed] [Google Scholar]
- Kumaraswamy V. S.; Lindley P. F.; Slingsby C.; Glover I. D. (1996) An eye lens protein-water structure: 1.2 Å resolution structure of gammaB-crystallin at 150 K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 52, 611–622. [DOI] [PubMed] [Google Scholar]
- Norledge B. V.; Hay R. E.; Bateman O. A.; Slingsby C.; Driessen H. P. (1997) Towards a molecular understanding of phase separation in the lens: a comparison of the X-ray structures of two high Tc gamma-crystallins, gammaE and gammaF, with two low Tc gamma-crystallins, gammaB and gammaD. Exp. Eye Res. 65, 609–630. [DOI] [PubMed] [Google Scholar]
- Purkiss A. G.; Bateman O. A.; Goodfellow J. M.; Lubsen N. H.; Slingsby C. (2002) The X-ray crystal structure of human gamma S-crystallin C-terminal domain. J. Biol. Chem. 277, 4199–4205. [DOI] [PubMed] [Google Scholar]
- Shimeld S. M.; Purkiss A. G.; Dirks R. P.; Bateman O. A.; Slingsby C.; Lubsen N. H. (2005) Urochordate betagamma-crystallin and the evolutionary origin of the vertebrate eye lens. Curr. Biol. 15, 1684–1689. [DOI] [PubMed] [Google Scholar]
- Van Montfort R. L.; Bateman O. A.; Lubsen N. H.; Slingsby C. (2003) Crystal structure of truncated human betaB1-crystallin. Protein Sci. 12, 2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman O. A.; Sarra R.; van Genesen S. T.; Kappe G.; Lubsen N. H.; Slingsby C. (2003) The stability of human acidic beta-crystallin oligomers and hetero-oligomers. Exp. Eye Res. 77, 409–422. [DOI] [PubMed] [Google Scholar]
- Liang J. J. (2004) Interactions and chaperone function of alphaA-crystallin with T5P gammaC-crystallin mutant. Protein Sci. 13, 2476–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk M.; Herbst R.; Hoeger D.; Kretschmar M.; Lubsen N. H.; Jaenicke R. (2000) Gamma S-crystallin of bovine and human eye lens: solution structure, stability and folding of the intact two-domain protein and its separate domains. Biophys. Chem. 86, 95–108. [DOI] [PubMed] [Google Scholar]
- Burley S. K.; Petsko G. A. (1985) Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229, 23–28. [DOI] [PubMed] [Google Scholar]
- Bella J.; Brodsky B.; Berman H. M. (1995) Hydration structure of a collagen peptide. Structure 3, 893–906. [DOI] [PubMed] [Google Scholar]
- Seibert C. M.; Raushel F. M. (2005) Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44, 6383–6391. [DOI] [PubMed] [Google Scholar]
- Harrington V.; Srivastava O. P.; Kirk M. (2007) Proteomic analysis of water insoluble proteins from normal and cataractous human lenses. Mol. Vision 13, 1680–1694. [PubMed] [Google Scholar]
- Bachem A. (1956) Ophthalmic ultraviolet action spectra. Am. J. Ophthalmol. 41, 969–975. [DOI] [PubMed] [Google Scholar]
- Kurzel R. B.; Wolbarsht M.; Yamanashi B. S.; Staton G. W.; Borkman R. F. (1973) Tryptophan excited states and cataracts in the human lens. Nature (London) 241, 132–133. [DOI] [PubMed] [Google Scholar]
- (2008) Vision problems in the U.S. Prevalence of adult vision impairment and age-related eye disease in America, pp 22−36, National Eye Institute, Bethesda, MD
- Hains P. G.; Mizdrak J.; Streete I. M.; Jamie J. F.; Truscott R. J. (2006) Identification of the new UV filter compound cysteine-l-3-hydroxykynurenine O-beta-d-glucoside in human lenses. FEBS Lett. 580, 5071–5076. [DOI] [PubMed] [Google Scholar]
- Aravind P.; Wistow G.; Sharma Y.; Sankaranarayanan R. (2008) Exploring the limits of sequence and structure in a variant betagamma-crystallin domain of the protein absent in melanoma-1 (AIM1). J. Mol. Biol. 381, 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallmadge D. H.; Borkman R. F. (1990) The rates of photolysis of the four individual tryptophan residues in UV exposed calf gamma-II crystallin. Photochem. Photobiol. 51, 363–368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.