Abstract
The cool-water copepod Calanus finmarchicus is a key species in North Atlantic marine ecosystems since it represents an important food resource for the developmental stages of several fish of major economic value. Over the last 40 years, however, data from the Continuous Plankton Recorder survey have highlighted a 70 per cent reduction in C. finmarchicus biomass, coupled with a gradual northward shift in the species's distribution, which have both been linked with climate change. To determine the potential for C. finmarchicus to track changes in habitat availability and maintain stable effective population sizes, we have assessed levels of gene flow and dispersal in current populations, as well as using a coalescent approach together with palaeodistribution modelling to elucidate the historical population demography of the species over previous changes in Earth's climate. Our findings indicate high levels of dispersal and a constant effective population size over the period 359 000–566 000 BP and suggest that C. finmarchicus possesses the capacity to track changes in available habitat, a feature that may be of crucial importance to the species's ability to cope with the current period of global climate change.
Keywords: Calanus finmarchicus, climate change, dispersal, effective population size, North Atlantic
1. Introduction
One of the main predicted effects of global climate change on the Earth's biota is a shift in species' distribution ranges. Although correlations between temperature and species' ranges have been observed for many years (Hutchins 1947), range shifts in response to global warming are only now becoming apparent (Parmesan & Yohe 2003; Root et al. 2003). This concept of ‘habitat tracking’ was originally suggested by Darwin (1859) to describe the migration of species in response to climatic fluctuations during the Pleistocene ice ages and interglacials, but for organisms whose potential for dispersal is limited, their fate under scenarios of present and future phases of rapid climate change may be extinction (Thomas et al. 2004; Parmesan 2006; Foden et al. 2007).
Range shifts associated with climate change have also been observed in marine species, including several commercially important fish species (Perry et al. 2005). The full effects of global warming on marine communities may still not be fully apparent, however, since they may have differing impacts on species at a range of trophic levels. The cool-water copepod Calanus finmarchicus is an important food resource for the developmental stages of several fishes of major economic value and thus is a key species in North Atlantic marine ecosystems (Beaugrand et al. 2003). Over the last 40 years, data from the Continuous Plankton Recorder survey have highlighted a 70 per cent reduction in C. finmarchicus biomass, coupled with a gradual northward shift in the species's distribution, which have also been linked with climate change (Heath et al. 2001; Beaugrand et al. 2002; Helaouët & Beaugrand 2007). Given predictions of future temperature change, these trends are likely to continue or even accelerate (Thomas et al. 2004; Kohler et al. 2007) and, consequently, knowledge of its population dynamics is central to understanding and predicting the response of the regional marine ecosystem to future climatic impacts.
In zooplanktonic species, the combination of biological and physical factors such as extremely large population sizes, high dispersal capability, particularly of larvae, and an ostensible lack of barriers to dispersal would suggest large-scale homogeneity of populations in the pelagic realm (Palumbi 1992, 1994; Norris 2000). Empirical studies, however, have shown the existence of cryptic species in many planktonic taxa, indicating allopatric or parapatric divergence (de Vargas et al. 1999; Lee 2000; Goetze 2003), as well as evidence of genetic substructuring below the species level, even within ocean basins (Zane et al. 1998; Papetti et al. 2005; Darling et al. 2007; Kirby et al. 2007). Such patterns of differentiation are indicative of limited capacity for dispersal, contrary to expectations. Modelling (Bryant et al. 1998; Heath et al. 2001; de Young et al. 2004; Speirs et al. 2006) and population genetic studies (Bucklin & Kocher 1996; Bucklin et al. 1996, 2000; Kann & Wishner 1996) of C. finmarchicus have yielded somewhat conflicting results with respect to dispersal and connectivity across the species's range. Recent modelling studies have suggested that there may be extensive connectivity and transport across the entire range of C. finmarchicus (Speirs et al. 2006) but some genetic analyses report significant levels of genetic differentiation both between gyre systems and at basin-wide scales, suggesting limited dispersal (Bucklin & Kocher 1996; Bucklin et al. 2000). These early molecular studies relied largely on uniparentally inherited markers that exhibit limited levels of polymorphism and often commented on the need for high-resolution genetic analysis to better elucidate patterns of gene flow across the region (e.g. Kann & Wishner 1996; Bucklin et al. 2000).
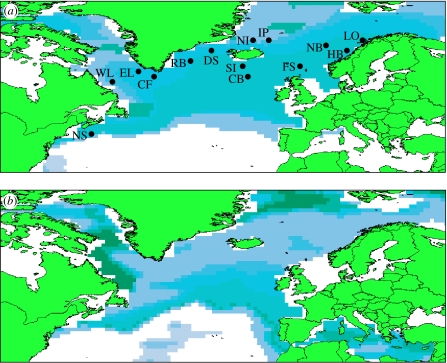
In this study, we have analysed samples spanning the entire North Atlantic range of C. finmarchicus (figure 1a; table 1) using recently developed microsatellite markers (Provan et al. 2007) as well as a portion of the mitochondrial cytochrome b (CYTB) gene to address three key issues concerning the distributional response of C. finmarchicus to the effects of climate change. First, we have estimated levels of gene flow between regions and across the entire range of the species to elucidate potential for dispersal. We particularly wanted to test whether the populations associated with the Irminger Sea and the Norwegian Sea gyre systems are genetically distinct, as previously suggested (Bucklin et al. 2000), since these represent the main centres of abundance for the species. If the levels of dispersal between the two areas are limited, decreases in population numbers in one are unlikely to be ameliorated by immigration from the other, for example, under a range-shift scenario induced by climate change. Second, we have used a coalescent approach together with palaeodistribution modelling to elucidate the historical population demography of C. finmarchicus and determine its response to previous fluctuations in Earth's climate during the Pleistocene glaciations. Knowledge of past population dynamics may provide valuable insights into how populations might respond to current and future climatic changes in terms of whether they possess the necessary potential for dispersal to track these changes and thus maintain large effective population sizes or whether these periods are characterized by alternating phases of extinction and recolonization. Finally, we have tested whether the recent decrease in census population numbers has been accompanied by a genetic bottleneck in any or all of the regions studied since such bottlenecks can compromise the future adaptive potential of impacted populations.
Figure 1.
(a) Present and (b) LGM modelled distributions of C. finmarchicus based on current day and reconstructed sea surface temperature and bathymetry under the maximum entropy niche model. Labelled dots in (a) show sampling points (see table 1 for details).
Table 1.
Location of sample sites and sample numbers. (NNU: number of samples for nuclear microsatellite analysis. NMT: number of samples for mitochondrial CYTB analysis.)
| number | sample site | latitude | longitude | code | NNU | NMT |
|---|---|---|---|---|---|---|
| 1 | Nova Scotia | 42°51′N | 61°40′W | NS | 23 | 5 |
| 2 | West Labrador Sea | 55°06′N | 54°07′W | WL | 23 | 5 |
| 3 | East Labrador Sea | 60°23′N | 48°27′W | EL | 24 | 7 |
| 4 | Cape Farewell | 59°23′N | 44°00′W | CF | 24 | 5 |
| 5 | Irminger Basin | 62°50′N | 34°20′W | RB | 23 | 5 |
| 6 | Denmark Strait | 66°15′N | 27°42′W | DS | 22 | 5 |
| 7 | South Iceland | 63°00′N | 21°30′W | SI | 19 | 6 |
| 8 | Iceland Basin | 59°14′N | 19°35′W | CB | 24 | 7 |
| 9 | North Iceland | 68°00′N | 18°50′W | NI | 24 | 6 |
| 10 | Icelandic Plateau | 68°00′N | 12°40′W | IP | 19 | 6 |
| 11 | Faeroes/Shetland Channel | 61°28′N | 03°43′W | FS | 21 | 5 |
| 12 | Norwegian Basin | 68°15′N | 03°00′E | NB | 22 | 6 |
| 13 | Halten Bank | 65°15′N | 05°25′E | HB | 23 | 7 |
| 14 | Lofoten | 68°15′N | 13°00′E | LO | 22 | 7 |
| Total | 313 | 82 |
2. Material and methods
(a) Sample collection and DNA extraction
Samples of C. finmarchicus were collected from a total of 14 sites spanning the northern North Atlantic from off Nova Scotia to the Lofoten Islands (figure 1a). Details of the location of the sampling sites are given in table 1. The samples were collected from the surface 100 m layer from hauls of standard mesozooplankton nets or equivalent and stored in ethanol. Specimens of C. finmarchicus were sorted from each sample microscopically by N. Holliday (Marine Biological Association, Plymouth, UK). DNA was extracted from whole individual copepods using the DNeasy Tissue Kit (Qiagen). In total, 313 individuals from 14 populations were used for microsatellite genotyping and 82 were sequenced for the mitochondrial CYTB gene (table 1).
(b) Nuclear microsatellite genotyping
Samples were genotyped for the following six loci: EH666474, EH666870, EL585922, EL696609, EL773359 and EL773519. Primer sequences and details of polymerase chain reaction (PCR) and electrophoresis conditions are described in Provan et al. (2007).
(c) Mitochondrial CYTB sequencing
Previous analyses of mitochondrial variation in copepods have mostly used the cytochrome oxidase subunit 1 (COI) gene, which has been shown to evolve at an appropriate rate for intrapopulation studies in a range of animal species. Studies in C. finmarchicus, however, have suggested that there may be a nuclear pseudogene copy of the COI gene. Indeed, early papers by Bucklin et al. have reported variation in both the mitochondrial COI gene (Bucklin et al. 1999) and, subsequently, the nuclear pseudogene copy (Bucklin et al. 2000) using the same PCR primers. As other authors have suggested that they have studied a bona fide mitochondrial COI gene in other copepods within both the genera Calanus and Neocalanus (Papadopoulos et al. 2005; Kirby et al. 2007) and thus the situation still lacks clarity, we decided instead to use the CYTB region, which, while exhibiting less variation than COI, has still been shown to provide sufficient resolution for intraspecific and intrapopulation studies (Papadopoulos et al. 2005). A 307 bp fragment of the mitochondrial CYTB gene was amplified using the universal mollusc primers of Merritt et al. (1998). PCR was carried out on a MWG Primus thermal cycler using the following parameters: initial denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, extension at 72°C for 2 min and a final extension at 72°C for 5 min. PCR was carried out in a total volume of 20 μl containing 200 ng genomic DNA, 20 pmol of forward primer, 20 pmol of reverse primer, 1× PCR reaction buffer (7.5 mM Tris–HCl [pH9.0], 2.0 mM [NH4]2SO4, 5.0 mM KCl, 2.0 mM MgCl2) and 1.0 U BIOTOOLS DNA polymerase. Five microlitre PCR products were resolved on 1.5 per cent agarose gels and visualized by ethidium bromide staining and the remaining 15 μl sequenced commercially (Macrogen, Korea). Sequences were aligned using the ClustalW program in the BioEdit software package.
(d) Palaeodistribution modelling
Ecological niche modelling (ENM) was carried out to determine suitable habitats for C. finmarchicus at the last glacial maximum (LGM, ca 18 KYA) using the maximum entropy approach implemented in the Maxent software package (v. 3.2.1; Phillips et al. 2006). Present-day species occurrence data for C. finmarchicus were downloaded from the Global Biodiversity Information Facility data portal (www.gbif.org). A distribution model based on current sea surface temperature (SST) and bathymetry grids at one degree resolution (Bigg et al. 2008) was generated using Maxent with the default parameters for convergence threshold (10−5) and number of iterations (500) and projected onto reconstructed LGM data (Bigg et al. 2008) for the same parameters to identify potential refugial areas. Duplicate records from the same locality were removed to reduce the effects of spatial autocorrelation. The performance of the model was tested using 25 per cent of the occurrence data points to determine the area under the receiver operating characteristic (ROC) curve.
(e) Data analysis
Nuclear microsatellite allele sizes were scored using a 10 bp ladder and were checked by comparison with previously sized control samples. To give an indication of levels of dispersal, population pairwise estimates of gene flow (Nm) based on nuclear microsatellites were calculated using the private alleles method (Slatkin 1985; Barton & Slatkin 1986) as implemented in the Genepop software package (v. 3.4; Raymond & Rousset 1995). Population pairwise ΦST values, which give an analogue of FST (Weir & Cockerham 1984) calculated within the analysis of molecular variance (AMOVA) framework (Excoffier et al. 1992), were also calculated for both the nuclear microsatellite and mitochondrial CYTB data using the Arlequin software package (v. 3.01; Excoffier et al. 2005). Significance of population differentiation was estimated by permuting genotypes between populations. Overall levels of genetic differentiation between populations and between regions were also estimated from allele and haplotype frequencies using Arlequin. In addition, a test for isolation by distance (IBD; Rousset 1997) was carried out using a Mantel test to assess the relationship between genetic distance, measured as FST/(1− FST), and geographical distance in Genepop. To identify possible population structuring, the software package Baps (v. 3.2; Corander et al. 2003) was used to identify clusters of genetically similar populations using a Bayesian approach. Ten replicates were run for all possible values of the maximum number of clusters (K) up to K=14, the number of populations sampled in the study.
As our results suggested genetic homogeneity, simulations were carried out using the Powsim software package (v. 4.0; Ryman & Palm 2006) to eliminate the possibility of a type II error (false negative for population differentiation) based on the microsatellite data. Simulations were carried out for effective population sizes Ne =1000 and Ne =10 000 to yield FST values of 0.005, 0.01 and 0.02. Although C. finmarchicus undoubtedly has a much larger effective population size, this is not relevant to the analysis, since Ne only determines the time necessary to reach the target FST. Thus, the use of larger values of Ne is unjustified as the difference between, say, Ne=10 000 and 100 000 (and higher) is not important at values of FST as small as those identified in our study (Nils Ryman, personal communication). The tests were carried out for both the global (all 14 populations) and two groups (Irminger Gyre v. Norwegian Sea Gyre) scenarios. In all cases, 1000 replicates were run and the power of the analysis was indicated by the proportion of tests that were significant at p<0.05 using the respective allele frequencies at the six loci studied.
Long-term population demography was assessed using a generalized skyline plot implemented in the Genie software package (v. 3.0; Pybus & Rambaut 2002). An unweighted pair-group method with arithmetic averages (UPGMA) ultrametric tree was generated from the mitochondrial CYTB data in PAUP* (v. 4.0.b10; Swofford 2002) and this was used to generate the skyline plot with the smoothing parameter (ε) estimated using the ‘maximize optimization’ option. A mutation rate for the CYTB gene in Calanus was estimated based on the average divergence between Atlantic, Adriatic and Black Sea populations of C. helgolandicus and C. euxinus (Papadopoulos et al. 2005). The average divergence between CYTB sequences was 0.371 per cent, compared with 0.467 per cent for COI. Thus, given that the COI mutation rate is 1.4–2.2 per cent/Myr (Knowlton & Weigt 1998), the equivalent rate for the CYTB gene is 1.11–1.75 per cent/Myr. To test for the occurrence of a more recent genetic bottleneck in any of the populations, the Wilcoxon test for heterozygote excess in the microsatellite data was performed under the infinite alleles model, the stepwise mutation model and a two-phase model incorporating 90 per cent single-stepwise mutations using the program Bottleneck (v. 1.2; Piry et al. 1999). The Wilcoxon test was used as it is recommended for a relatively low number of loci.
3. Results
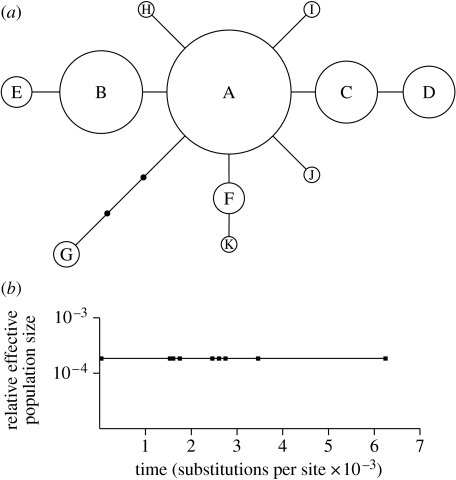
The six microsatellites analysed were moderately to highly polymorphic, between five and 20 alleles (mean 9.83) detected at each locus. A total of 12 substitutions were detected in the 307 bp alignment of the mitochondrial CYTB sequences, giving 11 haplotypes (figure 2a; electronic supplementary material, table S1). No significant genetic differentiation was observed at the inter-population level or across the species's range in either the nuclear or mitochondrial datasets. Only three of 91 pairwise ΦST values based on nuclear microsatellites were significantly different from zero (NS v. CB, EL v. NB and CB v. LO) and the only significant pairwise ΦST value based on the mitochondrial CYTB data was between the EL and NB samples (data not shown). None of these was significant after sequential Bonferroni correction for multiple tests. On a range-wide scale, no significant genetic differentiation was found either between all populations across the species's range or between populations from the Irminger Gyre and those in the Norwegian Sea Gyre using both nuclear and mitochondrial markers (table 2) and there was no evidence of IBD. The Baps analysis likewise found no evidence of any genetic structuring and assigned all 14 populations to a single cluster. The lack of population differentiation suggested by these analyses was reflected in high values of Nm between populations, which ranged from 1.87 (CB v. IP) to 7.37 (FS v. NB) and a range-wide value across all populations of 6.02 (electronic supplementary material, table S2). All values were above 1.00, which represents the theoretical threshold for population differentiation due to genetic drift (Wright 1931). The simulation studies suggested that a type II error due to low power of the markers used was unlikely. For both the global and two-groups scenarios, the microsatellite data were able to detect FST values of as low as 0.01 at least 95 per cent of the time (electronic supplementary material, table S3).
Figure 2.
(a) Mitochondrial CYTB haplotype network. Circle sizes are approximately proportional to haplotype frequency. Haplotype codes correspond to those in electronic supplementary material, table S1. (b) Generalized skyline plot for C. finmarchicus generated from the CYTB haplotype network. Based on the calculated mutation rate of 1.11–1.75 per cent/Myr, the final coalescent (i.e. rightmost) point corresponds to a time of ca 359 000–566 000 BP.
Table 2.
Levels of genetic differentiation between populations/groups.
| per cent of genetic differentiation | ||||
|---|---|---|---|---|
| partition | nuclear | mitochondrial | ||
| between all populations | 0.88% (ΦST=0.0088) | p=0.391 | 2.01% (ΦST=0.0201) | p=0.271 |
| Irminger Gyre (RB/SI/CB) v. Norwegian Sea Gyre (NB/HB/LO) | 0.13% (ΦST=0.0013) | p=0.345 | −0.19% (ΦST=−0.0190) | p=0.509 |
For the ecological niche model generated by Maxent under current SST and bathymetry, the area under the ROC curve value of 0.791 indicated a better than random prediction. A binomial test of omission confirmed that the model predictions were significantly better than random (p<10−8) for all typical threshold values. The current model (figure 1a) was a good representation of the actual distribution of C. finmarchicus, while the palaeodistribution model (figure 1b) indicated potentially suitable habitats in three main areas, the Labrador Sea, the eastern North Atlantic and the Greenland Sea, that could have acted as refugia for C. finmarchicus during the LGM. Long-term persistence of the species in a refugium in one of these areas is supported by the generalized skyline plot, which suggested that the effective population size of C. finmarchicus has remained constant over the coalescent time of the sampled genealogy (figure 2b). Based upon the calculated mutation rate for the CYTB region studied (1.11–1.75%/Myr), this corresponds to a time of ca 359 000–566 000 BP. Finally, the Wilcoxon test for heterozygote excess did not indicate the recent genetic bottleneck in any of the populations under any of the three mutation models.
4. Discussion
The findings of this study suggest that high levels of dispersal in the indicator copepod species C. finmarchicus have maintained a stable effective population size throughout previous episodic fluctuations in Earth's climate. Two main theories exist concerning the distributional responses of organisms to climate change. The hypothesis of habitat tracking implies that, given sufficient capacity for dispersal, species may shift their distributions to use available habitat and thus maintain stable community structures (Eldredge 1989; Coope 1994). Under the alternative scenario, species with limited dispersal capabilities would become extinct in large parts of their ranges due to lack of suitable habitats, and subsequently recolonize from greatly reduced populations in refugial areas as the climate ameliorated (Bennett et al. 1991; Dalén et al. 2007; Provan & Bennett 2008).
The coalescent analysis carried out in this study shows a constant effective population size in C. finmarchicus over the period ca 359 000–566 000 BP, a period that includes several glacial/interglacial cycles of major climate change. Although the accuracy of molecular clocks is a much debated subject, we would have to invoke an unfeasibly high mutation rate of greater than 20 per cent/Myr if this population stability were only post-LGM. The long-term stability, combined with high levels of dispersal indicated by the lack of genetic structuring in extant populations across the species's range, suggests that the ‘habitat tracking’ scenario best explains the response of C. finmarchicus to climatic fluctuations. The alternative ‘extinction–recolonization’ hypothesis would be reflected in decreases in effective population size, which are not apparent. Evidence of habitat tracking has also been reported previously for the planktonic foraminifera Globorotalia truncatulinoides over the last 140 000 years (Renaud & Schmidt 2003). Although the palaeodistribution model for C. finmarchicus highlighted multiple suitable areas of habitat, it is extremely likely that the species persisted through the LGM as a single, continuous population, since vicariance in multiple refugia would have led to the formation of distinct evolutionary lineages (Provan & Bennett 2008). The presence of year-round sea ice at the LGM in the Labrador and Greenland Seas would appear to rule out these areas as potentially suitable habitats due to the lack of phytoplankton prey and observations of limited development at low temperatures (Hirche & Kosobokova 2007), leaving the area in the eastern North Atlantic to the southwest of the British Isles as the most likely area for persistence of C. finmarchicus during the LGM.
It has been suggested that ocean current systems may represent hydrographic barriers to dispersal in planktonic species, including C. finmarchicus (Bucklin et al. 2000; Goetze 2005; Papetti et al. 2005). If this is the case, scope for habitat tracking may be limited due to sequestration of large parts of the population within discrete gyre systems. Our finding of a lack of significant genetic variation between the Irminger Gyre and the Norwegian Sea Gyre populations does not support this idea. Bryant et al. (1998) used a Lagrangian particle-tracking model to demonstrate the potential for the two sub-gyres of the Norwegian Sea Gyre to retain substantial proportions of populations of C. finmarchicus for up to a year but that finding does not necessarily imply that populations from the main gyre systems are hydrographically isolated. Exchange of individuals between the Norwegian Sea Gyre and the Irminger/Labrador Gyres may occur during the reproductive season as a result of lateral advective and mixing processes in the surface layers (Ivers 1975). This process is assisted by an apparent tendency for the C. finmarchicus adult stages to be associated with the margins of the two gyres where mixing between populations is more likely (Heath et al. 2008). Such potential range-wide connectivity of C. finmarchicus populations implied by our study would appear to confirm the findings of a recent modelling study by Speirs et al. (2006). Using more sophisticated techniques than employed previously, they demonstrated that inoculation of a single spatial cell of the model in either the Labrador Sea in the west or the Norwegian Sea in the east resulted in range-wide dispersal of the populations throughout the northern North Atlantic within a 6-year period, suggesting the high levels of connectivity across the region that would be necessary for successful habitat tracking.
The response of calanoid copepods to climate change is of crucial importance to the entire North Atlantic ecosystem in general, and to fisheries in particular. C. finmarchicus is central to the ‘bottom–up’ maintenance of healthy fish stocks in the region, and the survival of larval cod is partly dependent on an abundance of planktonic prey (Beaugrand et al. 2003; Alheit et al. 2005). It is believed that replacement of C. finmarchicus by congeneric species as a result of climate change may lead to a mismatch between the timings of development of juvenile cod and their calanoid prey (Beaugrand et al. 2003; Hirche & Kosobokova 2007). A recent ENM study, however, has suggested that cod survived the LGM in North Atlantic refugia (Bigg et al. 2008) and this may also indicate the concurrent survival of calanoid prey.
Despite the recorded decrease in census population numbers (Helaouët & Beaugrand 2007), the inferred long-term persistence of C. finmarchicus and the apparent absence of a significant recent genetic bottleneck in the population suggest that any detrimental effects of climate-induced range shifts have not yet become manifest. The apparent discrepancy between maintaining stable and/or large effective population sizes despite observed decreases in census population size is not unknown (e.g. Dutton et al. 1999; Ribeiro et al. 2008) and, given that effective population size is strongly correlated with evolutionary potential (Frankham et al. 2003), this is an extremely positive sign in the extant C. finmarchicus population. Nevertheless, although our results suggest that the dispersal capacity of C. finmarchicus may be sufficient to ameliorate the genetic effects of climate-induced changes in habitat availability, they do not preclude the need for future monitoring, both in terms of genetics and biomass.
Acknowledgments
We are particularly grateful to our colleagues in Canada (Professor E. Head, Bedford Institute of Oceanography, Dartmouth, Nova Scotia), Iceland (Professor A. Gislason, Marine Research Institute, Reykjavik) and Norway (Professor W. Melle, Institute for Marine Research, Bergen), and also to our colleagues in the UK Marine Productivity Programme, especially Mr S. Hays, for their very willing assistance in the collection of the samples. We are extremely grateful to Grant Bigg, who supplied the current and LGM reconstructed model layers, Nils Ryman for advice on the Powsim software, Rob Paxton, Pierre Helaouët and Phil Williamson for helpful comments on an earlier version of the manuscript, Martin Thiel and two anonymous referees for further helpful comments, and Grant Bigg and Dougie Speirs for advice on modelling. We are also grateful to Nicola McAreavey for laboratory assistance. This research was funded under the NERC Marine Productivity programme (grant NER/T/S/2001/0015).
Supplementary Material
Mitochondrial CYTB haplotype frequencies
Population pairwise estimates of gene flow (Nm) calculated from nuclear microsatellite data using the private alleles method of <ce:cross-ref refid="bib2">Barton & Slatkin (1986)</ce:cross-ref>
Statistical power for detecting varying levels of population differentiation (FST) based on loci, allele frequencies and population sizes used in the present study. Power is expressed as proportion of significant FST values (p<0.05)
References
- Alheit J., Möllmann C., Dutz J., Kornilovs G., Loewe P., Morholz V., Wasmund N. Synchronous ecological regime shifts in the central Baltic and North Sea in the late 1980s. ICES J. Mar. Sci. 2005;62:1205–1215. doi:10.1016/j.icesjms.2005.04.024 [Google Scholar]
- Barton N.H., Slatkin M. A quasi-equlibrium theory of the distribution of rare alleles in a subdivided population. Heredity. 1986;56:409–415. doi: 10.1038/hdy.1986.63. doi:10.1038/hdy.1986.63 [DOI] [PubMed] [Google Scholar]
- Beaugrand G., Reid P.C., Ibañez F., Lindley J.A., Edwards M. Reorganization of North Atlantic marine copepod biodiversity and climate. Science. 2002;296:1692–1694. doi: 10.1126/science.1071329. doi:10.1126/science.1071329 [DOI] [PubMed] [Google Scholar]
- Beaugrand G., Brander K.M., Lindley J.A., Souissi S., Reid P.C. Plankton effect on cod recruitment in the North Sea. Nature. 2003;426:661–664. doi: 10.1038/nature02164. doi:10.1038/nature02164 [DOI] [PubMed] [Google Scholar]
- Bennett K.D., Tzedakis P.C., Willis K.J. Quaternary refugia of north European trees. J. Biogeogr. 1991;18:103–115. doi:10.2307/2845248 [Google Scholar]
- Bigg G.R., Cunningham C.W., Ottersen G., Pogson G.H., Wadley M.R., Williamson P. Ice-age survival of Atlantic cod: agreement between palaeoecology models and genetics. Proc. R. Soc. B. 2008;275:163–172. doi: 10.1098/rspb.2007.1153. doi:10.1098/rspb.2007.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant A.D., Hainbucher D., Heath M. Basin-scale advection and population persistence of Calanus finmarchicus. Fish. Oceanogr. 1998;7:235–244. doi:10.1046/j.1365-2419.1998.00074.x [Google Scholar]
- Bucklin A., Kocher T.D. Source regions for recruitment of Calanus finmarchicus to Georges bank: evidence from molecular population genetic analysis of mtDNA. Deep Sea Res. II. 1996;43:1665–1881. doi:10.1016/S0967-0645(96)00059-8 [Google Scholar]
- Bucklin A., Sundt R.C., Dahle G. The population genetics of Calanus finmarchicus in the North Atlantic. Ophelia. 1996;44:29–45. [Google Scholar]
- Bucklin A., Guarnieri M., Hill R.S., Bentley A.M., Kaartvedt S. Taxonomic and systematic assessment of planktonic copepods using mitochondrial COI sequence variation and competitive, species-specific PCR. Hydrobiologia. 1999;401:239–254. doi:10.1023/A:1003790411424 [Google Scholar]
- Bucklin A., Astthorsson O.S., Gislason A., Allen L.D., Smolenak S.B., Wiebe P.H. Population genetic variation of Calanus finmarchicus in Icelandic waters: preliminary evidence of genetic differences between Atlantic and Arctic populations. ICES J. Mar. Sci. 2000;57:1592–1604. doi:10.1006/jmsc.2000.0957 [Google Scholar]
- Coope G.R. The response of insect faunas to glacial-interglacial climatic fluctuations. Phil. Trans. R. Soc. B. 1994;344:19–26. doi:10.1098/rstb.1994.0046 [Google Scholar]
- Corander J., Waldmann P., Sillanpää M.J. Bayesian analysis of genetic differentiation between populations. Genetics. 2003;163:367–374. doi: 10.1093/genetics/163.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalén L., Nyström V., Valdiosera C., Germonpré M., Sablin M., Turner E., Angerbjörn A., Arsuaga J.L., Götherström A. Ancient DNA reveals lack of postglacial habitat tracking in the arctic fox. Proc. Natl Acad. Sci. USA. 2007;104:6726–6729. doi: 10.1073/pnas.0701341104. doi:10.1073/pnas.0701341104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling K.F., Kucera M., Wade C.M. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proc. Natl Acad. Sci. USA. 2007;104:5002–5007. doi: 10.1073/pnas.0700520104. doi:10.1073/pnas.0700520104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C.R. John Murray; London: 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. [PMC free article] [PubMed] [Google Scholar]
- de Vargas C., Norris R., Zaninetti L., Gibb S.W., Pawlowski J. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proc. Natl Acad. Sci. USA. 1999;96:2864–2868. doi: 10.1073/pnas.96.6.2864. doi:10.1073/pnas.96.6.2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Young B., Heath M., Werner F., Chai F., Megrey B., Monfray P. Challenges of modeling ocean basin ecosystems. Science. 2004;304:1463–1466. doi: 10.1126/science.1094858. doi:10.1126/science.1094858 [DOI] [PubMed] [Google Scholar]
- Dutton P.H., Bowen B.W., Owens D.W., Barragan A., Davis S.K. Global phylogeography of the leatherback turtle (Dermochelys coriaceae) J. Zool. 1999;248:397–409. doi:10.1111/j.1469-7998.1999.tb01038.x [Google Scholar]
- Eldredge N. McGraw-Hill; New York: 1989. Macroevolutionary dynamics: species, niches and adaptive peaks. [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Laval L.G., Schneider S. Arlequin, v. 3.0: An integrated software package for population genetic data analysis. Evol. Bioinf. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Foden W., et al. A changing climate is eroding the geographical range of the Namib desert tree aloe through population declines and dispersal lags. Divers. Distrib. 2007;13:645–653. [Google Scholar]
- Frankham R., Ballou J.D., Briscoe D.A. Cambridge University Press; Cambridge, UK: 2003. Introduction to conservation genetics. [Google Scholar]
- Goetze E. Cryptic species on the high seas: global phylogenetics of the copepod family Eucalanidae. Proc. R. Soc. B. 2003;270:2321–2331. doi: 10.1098/rspb.2003.2505. doi:10.1098/rspb.2003.2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze E. Global population genetic structure and biogeography of the oceanic copepods Eucalanus hyalinus and E. spinifer. Evolution. 2005;59:2378–2398. doi:10.1111/j.0014-3820.2005.tb00948.x [PubMed] [Google Scholar]
- Heath M., Carlotti F., de Young B., Fiksen Ø., Werner C. Secondary production in the oceans and the response to climate change. ICES/GLOBEC Newsl. 2001;8:4–6. [Google Scholar]
- Heath M.R., et al. Spatial demography of Calanus finmarchicus in the Irminger Sea. Prog. Oceanog. 2008;76:39–88. doi:10.1016/j.pocean.2007.10.001 [Google Scholar]
- Helaouët P., Beaugrand G. Macroecology of Calanus finmarchicus and C. helgolandicus in the North Atlantic Ocean and adjacent seas. Mar. Ecol. Prog. Ser. 2007;345:147–165. doi:10.3354/meps06775 [Google Scholar]
- Hirche H.-J., Kosobokova K. Distribution of Calanus finmarchicus in the northern North Atlantic and Arctic Ocean—expatriation and potential colonization. Deep Sea Res. II. 2007;54:2729–2747. doi:10.1016/j.dsr2.2007.08.006 [Google Scholar]
- Hutchins L.W. The bases for temperature zonation in geographical distribution. Ecol. Monogr. 1947;17:323–335. doi:10.2307/1948663 [Google Scholar]
- Ivers, W. D. 1975 The deep circulation in the northern North Atlantic, with especial reference to the Labrador Sea. PhD thesis, University of California, San Diego.
- Kann L.M., Wishner K. Genetic population structure of the copepod Calanus finmarchicus in the Gulf of Maine: allozyme and amplified mitochondrial DNA variation. Mar. Biol. 1996;125:65–75. doi:10.1007/BF00350761 [Google Scholar]
- Kirby R.R., Lindley J.A., Batten S.D. Spatial heterogeneity and genetic variation in the copepod Neocalanus cristatus along two transects in the North Pacific sampled by the Continuous Plankton Recorder. J. Plankt. Res. 2007;29:97–106. doi:10.1093/plankt/fbl074 [Google Scholar]
- Knowlton N., Weigt L.A. New dates and new rates for divergence across the Isthmus of Panama. Proc. R. Soc. Lond. B. 1998;265:2257–2263. doi:10.1098/rspb.1998.0568 [Google Scholar]
- Kohler J., James T.D., Murray T., Nuth C., Brandt O., Barrand N.E., Aas H.F. Acceleration in thinning rate on western Svalbard glaciers. Geophys. Res. Lett. 2007;34:L18502. doi:10.1029/2007GL030681 [Google Scholar]
- Lee C.E. Global phylogeography of a cryptic copepod species complex and reproductive isolation between genetically proximate ‘populations’. Evolution. 2000;54:2014–2027. doi: 10.1111/j.0014-3820.2000.tb01245.x. doi:10.1111/j.0014-3820.2000.tb01245.x [DOI] [PubMed] [Google Scholar]
- Merritt T.J.S., Shi L., Chase M.C., Rex M.A., Etter R.J., Quattro J.M. ‘Universal’ cytochrome b primers facilitate intraspecific studies in molluscan taxa. Molec. Mar. Biol. Biotechnol. 1998;7:7–11. [PubMed] [Google Scholar]
- Norris R.D. Pelagic species diversity, biogeography and evolution. Paleobiology. 2000;26:236–258. doi:10.1666/0094-8373(2000)26[236:PSDBAE]2.0.CO;2 [Google Scholar]
- Palumbi S.R. Marine speciation on a small planet. Trends Ecol. Evol. 1992;7:114–118. doi: 10.1016/0169-5347(92)90144-Z. doi:10.1016/0169-5347(92)90144-Z [DOI] [PubMed] [Google Scholar]
- Palumbi S.R. Genetic divergence, reproductive isolation and marine speciation. Ann. Rev. Ecol. Syst. 1994;25:547–572. doi:10.1146/annurev.es.25.110194.002555 [Google Scholar]
- Papadopoulos L.N., Peijnenburg K.T.C.A., Luttikhuizen P.C. Phylogeography of the calanoid copepods Calanus helgolandicus and C. euxinus suggests Pleistocene divergences between Atlantic, Mediterranean, and Black Sea populations. Mar. Biol. 2005;147:1353–1365. doi:10.1007/s00227-005-0038-x [Google Scholar]
- Papetti C., Zane L., Bortolotto E., Bucklin A., Patarnello T. Genetic differentiation and local temporal stability of population structure in the euphausiid Meganyctiphanes norvegica. Mar. Ecol. Prog. Ser. 2005;289:225–235. doi:10.3354/meps289225 [Google Scholar]
- Parmesan C. Ecological and evolutionary response to recent climate change. Ann. Rev. Ecol. Evol. Syst. 2006;37:637–669. doi:10.1146/annurev.ecolsys.37.091305.110100 [Google Scholar]
- Parmesan C., Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. doi:10.1038/nature01286 [DOI] [PubMed] [Google Scholar]
- Perry A.L., Low P.J., Ellis J.R., Reynolds J.D. Climate change and distribution shifts in marine fishes. Science. 2005;308:1912–1915. doi: 10.1126/science.1111322. doi:10.1126/science.1111322 [DOI] [PubMed] [Google Scholar]
- Phillips S.J., Anderson R.P., Schapire R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006;190:231–259. doi:10.1016/j.ecolmodel.2005.03.026 [Google Scholar]
- Piry A., Luikart G., Cornuet J.M. Bottleneck: a computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999;90:502–503. doi:10.1093/jhered/90.4.502 [Google Scholar]
- Provan J., Beatty G.E., Maggs C.A., Savidge G. Expressed sequence tagged-derived microsatellites for the cool-water marine copepod Calanus finmarchicus. Mol. Ecol. Notes. 2007;7:1369–1371. doi:10.1111/j.1471-8286.2007.01889.x [Google Scholar]
- Provan J., Bennett K.D. Phylogeographic insights into cryptic glacial refugia. Trends. Ecol. Evol. 2008 doi: 10.1016/j.tree.2008.06.010. doi:10.1016/j.tree.2008.06.010 [DOI] [PubMed] [Google Scholar]
- Pybus O.G., Rambaut A. Genie: estimating demographic history from molecular phylogenies. Bioinformatics. 2002;18:1404–1405. doi: 10.1093/bioinformatics/18.10.1404. doi:10.1093/bioinformatics/18.10.1404 [DOI] [PubMed] [Google Scholar]
- Raymond M., Rousset F. Genepop (v. 1.2): population genetic software for exact tests and ecumenicism. J. Hered. 1995;86:248–249. [Google Scholar]
- Renaud S., Schmidt D.N. Habitat tracking as a response of the planktonic foraminifer Globorotalia truncatulinoides to environmental fluctuations during the last 140 kyr. Mar. Micropalaeontol. 2003;49:97–122. doi:10.1016/S0377-8398(03)00031-8 [Google Scholar]
- Ribeiro A., Moran P., Caballero A. Genetic diversity and effective size of the Atlantic salmon Salmo salar L. inhabiting the River Eo (Spain) following a stock collapse. J. Fish Biol. 2008;72:1933–1944. doi:10.1111/j.1095-8649.2007.01769.x [Google Scholar]
- Root T.L., Price J.T., Hall K.R., Schneider S.H., Rosenzweig C., Pounds J.A. Fingerprints of global warming on wild animals and plants. Nature. 2003;421:57–60. doi: 10.1038/nature01333. doi:10.1038/nature01333 [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman N., Palm S. Powsim: a computer program for assessing statistical power when testing for genetic differentiation. Mol. Ecol. Notes. 2006;6:600–602. doi:10.1111/j.1471-8286.2006.01378.x [Google Scholar]
- Slatkin M. Rare alleles as indicators of gene flow. Evolution. 1985;39:53–65. doi: 10.1111/j.1558-5646.1985.tb04079.x. doi:10.2307/2408516 [DOI] [PubMed] [Google Scholar]
- Speirs D.C., Gurney W.S.C., Heath M.R., Horbelt W., Wood S.N., de Cuevas B.A. Ocean-scale modeling of the distribution, abundance and seasonal dynamics of the copepod Calanus finmarchicus. Mar. Ecol. Prog. Ser. 2006;313:173–192. doi:10.3354/meps313173 [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. Sunderland, Massachesetts:Sinauer & Associates.
- Thomas C.D., et al. Extinction risk from climate change. Nature. 2004;427:145–148. doi: 10.1038/nature02121. doi:10.1038/nature02121 [DOI] [PubMed] [Google Scholar]
- Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi:10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zane L., Ostellari L., Maccatrozzo L., Bargelloni L., Battaglia B., Patarnello T. Molecular evidence for genetic subdivision of Antarctic krill (Euphausia superba Dana) populations. Proc. R. Soc. B. 1998;265:2387–2391. doi: 10.1098/rspb.1998.0588. doi:10.1098/rspb.1998.0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mitochondrial CYTB haplotype frequencies
Population pairwise estimates of gene flow (Nm) calculated from nuclear microsatellite data using the private alleles method of <ce:cross-ref refid="bib2">Barton & Slatkin (1986)</ce:cross-ref>
Statistical power for detecting varying levels of population differentiation (FST) based on loci, allele frequencies and population sizes used in the present study. Power is expressed as proportion of significant FST values (p<0.05)