Summary
We identified and attempted to disrupt the Cryptococcus neoformans homoserine and/or threonine biosynthetic genes encoding aspartate kinase (HOM3), homoserine kinase (THR1), and threonine synthase (THR4), however, each gene proved recalcitrant to disruption. By replacing the endogenous promoters of HOM3 and THR1 with the copper-repressible CTR4-1 promoter, we showed that HOM3 and THR1 were essential for the growth of C. neoformans in rich media, when ammonium was the nitrogen source, or when threonine was supplied as an amino acid instead of a dipeptide. Moreover, the severity of the growth defect associated with HOM3- or THR1-repression increased with increasing incubation temperature. This study comprises the first demonstration of threonine biosynthetic genes being essential in a fungus. The necessity of these genes for C. neoformans growth, particularly at physiologically relevant temperatures, makes threonine biosynthetic genes ideal anti-cryptococcal drug targets.
Introduction
Amino acid biosynthetic pathways provide attractive candidates for antifungal drug targets since many of these pathways are conserved throughout the fungi and are absent from humans. One such pathway of interest is the threonine biosynthetic pathway, in which threonine is produced from aspartate in five enzymatic steps via the intermediate homoserine, which is also required for methionine synthesis (Fig. 1, reviewed in (Jones & Fink, 1982)). In the yeast Saccharomyces cerevisiae, this pathway is regulated at the level of transcription by General Control (Hinnebusch, 1992; Mountain et al., 1991), and of enzyme activity, particularly by threonine feedback inhibition of aspartate kinase (Hom3p) at the initial step of the pathway (Martin-Rendon et al., 1993; Ramos & Calderon, 1992). In addition to auxotrophy, a number of deleterious phenotypes have been attributed to threonine biosynthetic mutants (Arevalo-Rodriguez et al., 2004; Birrell et al., 2001; Birrell et al., 2002; Care et al., 2004; Deutschbauer et al., 2002; Dunn et al., 2006; Enyenihi & Saunders, 2003; Giaever et al., 2002; Roberg et al., 1997); some defects of which, such as temperature sensitivity, salt sensitivity and being petite-negative, could also influence fungal survival in vivo. Moreover, the threonine-biosynthetic intermediate homoserine is also required for biosynthesis of methionine, itself a central metabolite, and threonine is required for isoleucine biosynthesis. Significantly, we and others have shown that various fungal methionine (Met2p, Met3p and Met6p) and isoleucine (Ilv2p), as well as threonine (Hom3p), biosynthetic enzymes are required for fungal survival in vivo and/or virulence (Kingsbury et al., 2004a; Kingsbury et al., 2006; Nazi et al., 2007; Pascon et al., 2004; Yang et al., 2002).
Figure 1.
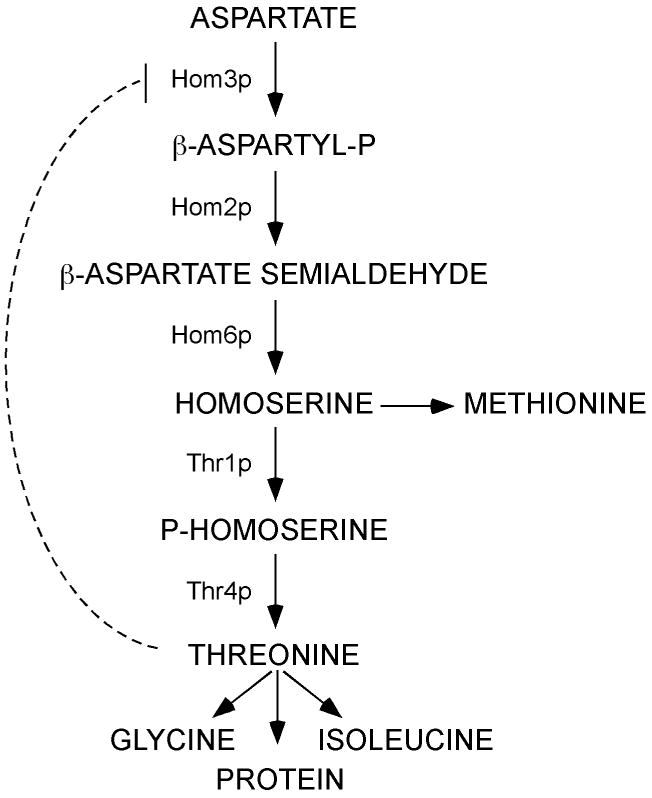
Fungal threonine biosynthetic pathway.
Amino acid auxotrophy has been shown to be particularly deleterious in the human pathogenic fungus Cryptococcus neoformans. Compared with S. cerevisiae, various auxotrophies are less well supplemented by the amino acids for which they are lacking, particularly in the presence of ammonium, suggesting fewer or less active permeases, or a greater proportion subject to nitrogen repression (Kingsbury et al., 2004a; Kingsbury et al., 2004b; Nazi et al., 2007; Pascon et al., 2004). In addition, auxotrophs show defects in known Cryptococcal virulence traits such as the ability to proliferate at 37 °C, and melanin and capsule production (Kingsbury et al., 2004a; Kingsbury et al., 2004b; Pascon et al., 2004; Yang et al., 2002). We were therefore interested in evaluating the potential of threonine biosynthetic enzymes as anti-cryptococcal targets in C. neoformans. Several attempts to disrupt the homoserine and threonine biosynthetic gene HOM3 (encoding aspartate kinase, EC: 2.7.2.4), and the threonine biosynthetic genes THR1 (encoding homoserine kinase, EC: 2.7.1.39) and THR4 (encoding threonine synthase, EC: 4.2.3.1) were unsuccessful. We demonstrate that this is because HOM3 and THR1, and likely other threonine biosynthetic enzymes, are essential for C. neoformans growth in most conditions.
Methods
Strains, media and growth conditions
All S. cerevisiae strains used in this study were isogenic with S288c and C. neoformans strains were isogenic with H99 (Serotype A Mat α, (Perfect et al., 1993)), and listed in Table 1. One Shot Top10 Chemically Competent Escherichia coli (Invitrogen) was used for plasmid propagation. Standard yeast and bacterial media were prepared as described previously (Sambrook et al., 1989; Sherman et al., 1974). Where specified, media was supplemented with nourseothricin (Nat; 100 μg ml-1, Hans Knöll Institute für Naturstoff-Forschung, Jena, Germany), geneticin (200 μg ml-1; Life Technologies), proline (1 g L-1), sorbitol (1 M), bathocuproinedisulfonic acid (BCS; 200 μM), cupric sulfate (CuSO4, 25 μM), ascorbic acid (1 mM), threonine (2.5 mM), homoserine (2.5 mM), methionine (0.13 mM), Ala-Thr (2.5 mM), and Met-Leu (0.13 mM).
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| C. neoformans | ||
| H99 | Mat α | (Perfect et al., 1993) |
| H99-73 | NAT1-PCTR4-1-THR1 | This study |
| H99-76 | NAT1-PCTR4-1-HOM3 | This study |
| S. cerevisiae | ||
| S157 | ura3Δ | (Yang et al., 2002) |
| S318 | ura3Δ thr4Δ∷natMX4 | This study |
| YJK1358 | ura3Δ thr1Δ∷kan MX4 | This study |
| YJK2416 | ura3Δ hom3Δ∷natMX4 | This study |
Identification of C. neoformans serotype A HOM3, THR1 and THR4
The predicted C. neoformans HOM3, THR1 and THR4 genes were initially identified from a C. neoformans serotype D strain JEC21 database that had been annotated by a genome-wide BLAST search (Loftus et al., 2005). NCBI accession numbers for the predicted Hom3p, Thr1p and Thr4p included XP_572658, XP_572893 and XP_568789, respectively. Sequences were then BLASTed against the Cryptococcus neoformans Serotype A strain H99 sequence. Serotype A HOM3, THR1 and THR4 occurred in sequence with the NCBI accession numbers AACO02000077.1, AACO02000074.1 and AACO02000068.1, respectively.
We also attempted to isolate the C. neoformans HOM3, THR1 and THR4 cDNAs by complementation of the methionine and/or threonine auxotrophies of S. cerevisiae hom3Δ, thr1Δ, and thr4Δ strains, using a C. neoformans cDNA library. Specifically, S. cerevisiae strains YJK2416 (hom3Δ ura3Δ), YJK1358 (thr1Δ ura3Δ), and S318 (thr4Δ ura3Δ), were transformed by lithium acetate-mediated transformation (Gietz et al., 1995), with a library that contained C. neoformans H99 cDNAs under the control of the S. cerevisiae GAL1 promoter in the pYES2.0 vector (Invitrogen) (Suvarna et al., 2000). Ura+ transformants were screened for the acquisition of methionine and/or threonine prototrophy in the presence of galactose, but not dextrose, as a carbon source. Plasmids that conferred prototrophy were isolated, propagated in E. coli DH10B, then analyzed by restriction analysis, and sequenced by the Duke University Cancer Center Sequencing Facility. Plasmids included pJO373 (pYES2.0 + C. neoformans THR1 cDNA, NCBI accession number EU623435), and pJO378 (pYES2.0 + C. neoformans THR4 cDNA, NCBI accession number EU635873).
Plasmid and strain construction
In vitro C. neoformans thr4∷NAT1, thr4∷NEO, thr1∷NAT1, and hom3∷NAT1 targeting cassettes were constructed using a modified PCR fusion technique (Davidson et al., 2002). To construct the thr4∷NAT1 and thr4∷NEO targeting cassettes, the first rounds of PCR amplified 5′ and 3′ THR4 sequence from H99 genomic DNA (primer pairs ZY125 + JO257, and ZY126 + JO255, respectively), and the NAT1 cassette from pGMC200 (McDade & Cox, 2001) or the NEO cassette from pJAF1 (Fraser et al., 2003) (primers JO254 + JO256). The gel-purified products were used as a template in the final fusion PCR reaction with primers ZY125 + ZY126. To construct the thr1∷NAT1 targeting construct, the first round of PCR consisted of amplification of 5′ and 3′ THR1 sequence from H99 genomic DNA (primer pairs JO298 + JO303, and JO302 + JO300, respectively), and the NAT1 cassette from pGMC200 (primers JO301 + JO304), then products were combined for the fusion PCR using primers JO298 + JO300. Construction of the hom3∷NAT1 cassette consisted of amplification of 5′ and 3′ HOM3 sequence from H99 genomic DNA (primer pairs JO318 + JO316, and JO320 + JO315, respectively), and NAT1 from pGMC200 (primers JO314 + JO317), followed by fusion of products in the final PCR reaction with primers JO318 + JO320. All gel-purified constructs were cloned into pCR2.1-TOPO (Invitrogen) according to manufacturer's instructions. Plasmids consisted of pJO193 (thr4∷NAT1), pJO221 (thr4∷NEO), pJO249 (thr1∷NAT1), and pJO260 (hom3∷NAT1).
The NAT1-PCTR4-1 cassette-containing plasmid pJO306 was also created by fusion PCR. The first round of PCR consisted of amplification of NAT1 from pGMC200 (primers JO357 + JO412) and PCTR4.1 from template pCTR4.2 (Ory et al., 2004) (primers JO408 + JO409). The purified products were used as a template in the final fusion PCR reaction using primers JO357 + JO409, and the gel-purified final product was cloned into pCR2.1-TOPO.
In vitro targeting cassettes were constructed to place HOM3 and THR1 under control of the CTR4-1 promoter. To create the NAT1- PCTR4-1 -HOM3 cassette, HOM3 upstream and 5′ gene sequence were PCR-amplified from H99 genomic DNA (primer pairs JO414 + JO415, and JO416 + JO413), and NAT1- PCTR4-1 was amplified from pJO306 (primers JO357 + JO409). The products were combined in the fusion PCR reaction (primers JO413 + JO414), and the resulting product was gel-purified and cloned into pCR2.1-TOPO (pJO310). The first round of PCR for construction of the NAT1- PCTR4-1 -THR1 cassette consisted of amplification of THR1 upstream and 5′ gene sequence (primer pairs JO359 + JO360, and JO410 + JO362), and NAT1- PCTR4-1 from pJO306 (primers JO357 + JO409). The products were used as a template for the final fusion PCR reaction using primers JO359 + JO362, and the resulting product was cloned into pCR2.1-TOPO (pJO308). All plasmid constructions were confirmed by restriction digestion and PCR analyses.
The targeting cassettes were PCR-amplified from their respective plasmids, and introduced into strain H99 by biolistic transformation (Toffaletti et al., 1993). For transformation with the thr4∷NAT1, thr4∷NEO, thr1∷NAT1, and hom3∷NAT1 constructs, transformation was performed on YPD + sorbitol, and after a 4 h incubation, cells were scraped off plates and spread on YPD + NAT or G418 plates to select for transformants. Transformants were purified and plated on SD to screen for acquisition of auxotrophy. For transformation with the NAT1- PCTR4-1 -HOM3 and NAT1- PCTR4-1 –THR1 constructs, cells were plated on YPD + sorbitol + BCS and incubated for 2-3 h prior to transformation to allow for expression from PCTR4. Following transformation, plates were incubated for 4 h and cells were replated on YPD + BCS + NAT. Purified transformants were screened from acquisition of auxotrophy on SD + CuSO4 + ascorbic acid plates (PCTR4-1-repressing conditions). The NAT1-PCTR4-1-THR1 genotype in strain H99-73 and NAT1-PCTR4-1-HOM3 genotype in strain H99-76 was confirmed by PCR (primer pairs JO281 + JO300, and JO506 + JO280 for H99-73, and JO281 + JO320, and JO505 + JO280 for H99-76), and Southern hybridization analysis (Figure 2).
Figure 2.
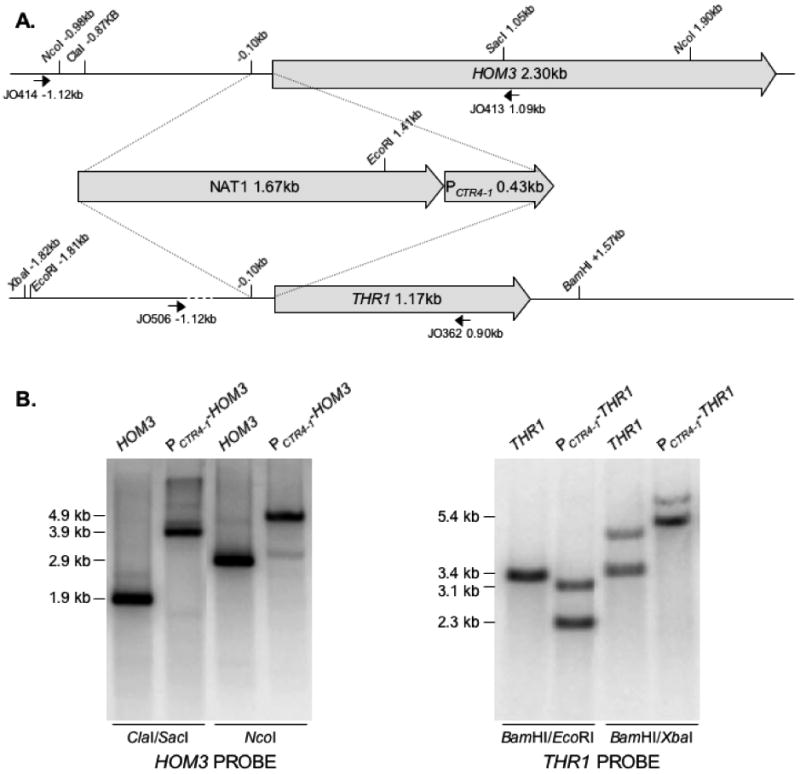
(A) The HOM3 and THR1 genes were placed under the control of the PCTR4-1 promoter by inserting the NAT1-PCTR4-1 construct immediately upstream of the predicted ORF, as shown in the diagram. (B) Southern hybridization analysis confirming correct positioning of the NAT1-PCTR4-1 construct in strains H99-73 (PCTR4-1-THR1) and H99-76 (PCTR4-1-HOM3). Genomic DNA from strains H99 (wildtype), H99-73 and H99-76 was digested with the restriction enzymes indicated, and blots were hybridized as indicated with a HOM3 or THR1 DNA probe, amplified using primer pairs JO414+JO413 and JO506+JO362, respectively.
The HOM3, THR1 and THR4 genes were replaced in the S. cerevisiae S157 strain by the natMX4 or kanMX4 cassettes, using PCR-mediated gene disruption (Goldstein & McCusker, 1999; Wach et al., 1994). Gene deletions were confirmed by PCR, and by acquisition of methionine and/or threonine auxotrophy.
Manipulation of nucleic acids
Plasmid DNA from E. coli was extracted using the QIAprep Spin Miniprep kit (Qiagen), according to the manufacturer's instructions. Extraction of plasmid DNA from S. cerevisiae, and genomic DNA from C. neoformans for PCR analysis, was performed as described previously (Hoffman & Winston, 1987). Genomic DNA from C. neoformans for Southern hybridization analysis was isolated as described previously (Yang et al., 2002), 2 μg of which was digested with various restriction enzymes, separated by electrophoresis on a 0.75 % (w/v) agarose gel, denatured and transferred to a nylon membrane (Roche), as described previously (Sambrook et al., 1989).
RNA for Northern analyses was prepared from cells that had first been grown to a density of approximately 2×108 cells ml-1, in 50 ml YPD + BCS. Cells were harvested, washed twice in sterile water, then split four ways and incubated with shaking in 50 ml YPD + BCS or YPD + CuSO4 + Ascorbic acid, at 25 °C or 37 °C. Following incubation for 5 h, RNA was isolated as described previously (Yang et al., 2002). Each sample was prepared in duplicate, and 10 μg duplicates of each preparation were separated in a 1 % (w/v) agarose-formaldehyde gel, and transferred to a nylon membrane.
Probes for Southern and Northern hybridizations were prepared from gel-purified PCR products. Specifically, probes for Southern hybridizations were amplified using primer pairs JO413 + JO414 (HOM3), and JO362 + JO506 (THR1). Primer pairs for amplification of Northern hybridization probes included JO770 + JO772 (HOM3), JO298 + JO362 (THR1), JO223 + JO225 (GPD), and JO765 + JO766 (CTR4). Probes were labeled with [α-32P]dCTP (Perkin-Elmer) using the RediprimeII Random Prime Labeling System (Amersham Biosciences), according to the manufacturer's instructions. Blots were prehybridized and hybridized in ULTRAhyb buffer (Ambion), and washed according to manufacturer's instructions. Membrane signal was visualized using a Typhoon 9200 Variable Mode Imager (Molecular Dynamics), and band signal intensity was quantified using ImageQuaNT 5.2 software (Molecular Dynamics).
All primers used in this study are listed in Table S1 (Supplemental Information).
Results and Discussion
Identification of HOM3, THR1 and THR4 in C. neoformans
Given the absence of the threonine biosynthetic pathway in humans (Payne & Loomis, 2006) and the avirulence or inability to survive in vivo of various amino acid auxotrophs, we were interested in assessing the potential of threonine biosynthetic enzymes as antifungal drug targets in the human pathogenic fungus C. neoformans. In particular, we focused on first, the aspartate kinase (encoded by HOM3), the initial and key feedback regulatory enzyme of the pathway. The avirulence of other Cryptococcal methionine auxotrophs (Nazi et al., 2007; Pascon et al., 2004; Yang et al., 2002) indicates that C. neoformans is unable to supplement this auxotrophy in the in vivo environment, thus we reasoned that the combined threonine and methionine auxotrophies of hom3 mutants should be even more detrimental to survival in vivo and/or virulence. Moreover, HOM3 is required for the in vivo survival of S. cerevisiae (Kingsbury et al., 2006). We were also interested in the final two steps of threonine biosynthesis, catalyzed by homoserine kinase (encoded by THR1) and threonine synthase (encoded by THR4). Mutation of these genes in S. cerevisiae results in a plethora of deleterious phenotypes in addition to auxotrophy (Birrell et al., 2001; Birrell et al., 2002; Care et al., 2004; Deutschbauer et al., 2002; Dunn et al., 2006; Enyenihi & Saunders, 2003; Giaever et al., 2002; Roberg et al., 1997), which may also influence in vivo survival and/or virulence.
We identified the putative C. neoformans H99 HOM3, THR1 and THR4 genes from the Cryptococcus neoformans Serotype A strain H99 through sequence similarity with the respective predicted ORFs in Serotype D. Furthermore, cDNAs matching the predicted THR1 and THR4 genes were isolated from a C. neoformans H99 cDNA library based on the ability to confer threonine prototrophy to S. cerevisiae thr1Δ and thr4Δ strains respectively (Figure 3), thus verifying that the identified genes encoded the predicted enzyme activities. We were unable to isolate the HOM3 cDNA by complementation of a S. cerevisiae hom3Δ strain, however, likely due to under representation of the HOM3 cDNA in the library. Consistent with this, we were unable to PCR-amplify the HOM3 cDNA from the library DNA. The C. neoformans H99 HOM3, THR1 and THR4 genes were predicted to contain six, two, and five introns, respectively. The predicted C. neoformans Hom3p, Thr1p and Thr4p sequences were highly similar to the corresponding proteins in S. cerevisiae, with approximately 50 %, 56 % and 50 % amino acid identity, respectively.
Figure 3.
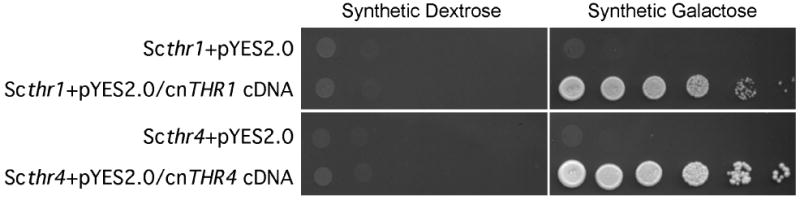
Functional complementation of S. cerevisiae thr1 (YJK1358) and thr4 (S318) strains by C. neoformans THR1 and THR4 cDNA. Ten-fold dilutions of YJK1358 transformed with pYES2.0 or pJO373 (pYES2.0/cnTHR1 cDNA), and S318 was transformed with pYES2.0 or pJO378 (pYES2.0/cnTHR4 cDNA), were plated on Synthetic Dextrose or Synthetic Galactose, and incubated at 30 °C for 3 days.
C. neoformans HOM3, THR1 and THR4 are recalcitrant to disruption
In order to study the phenotypes of C. neoformans hom3, thr1 and thr4 mutants, we attempted to disrupt HOM3, THR1 and THR4 in the Serotype A strain H99, using hom3∷NAT1, thr1∷NAT1, thr4∷NAT1 and thr4∷NEO targeting cassettes. However, no auxotrophic mutants were obtained after screening 151 transformants for HOM3 disruption, 89 transformants for THR1 disruption, and 469 transformants for THR4 disruption. An inability to disrupt these genes may be because HOM3, THR1 and THR4 are essential for C. neoformans growth.
C. neoformans HOM3 and THR1 are essential
To determine whether threonine biosynthetic genes are essential in C. neoformans, we replaced the endogenous promoters of HOM3 and THR1 with the PCTR4-1 promoter (Ory et al., 2004), thereby placing the genes under copper-repressible control (Figure 2). Growth of the wildtype (H99), PCTR4-1-HOM3 (H99-76) and PCTR4-1-THR1 (H99-73) strains were compared by plating 10-fold spot dilutions of strains that had been pre-grown in YPD + BCS, onto YPD + BCS (promoter-inducing conditions) and YPD + Cu + ascorbic acid (promoter-repressing conditions) (Figure 4). After incubation at 30 °C for three days in promoter-inducing conditions, PCTR4-1-HOM3 and PCTR4-1-THR1 strains grew considerably, although less well than the wildtype as judged by colony size, indicating that HOM3 and THR1 were expressed at different than normal levels in these strains. However, no colony formation was observed for the PCTR4-1-HOM3 and PCTR4-1-THR1 strains in promoter-repressing conditions. Since the Yeast Extract and Bacto Peptone in YPD contains significant levels of threonine and methionine for supplementation of Thr and Met auxotrophies (Difco Manual, 11th Edition), these results indicate that THR1 and HOM3 are essential in C. neoformans, even in the presence of abundant threonine and methionine.
Figure 4.
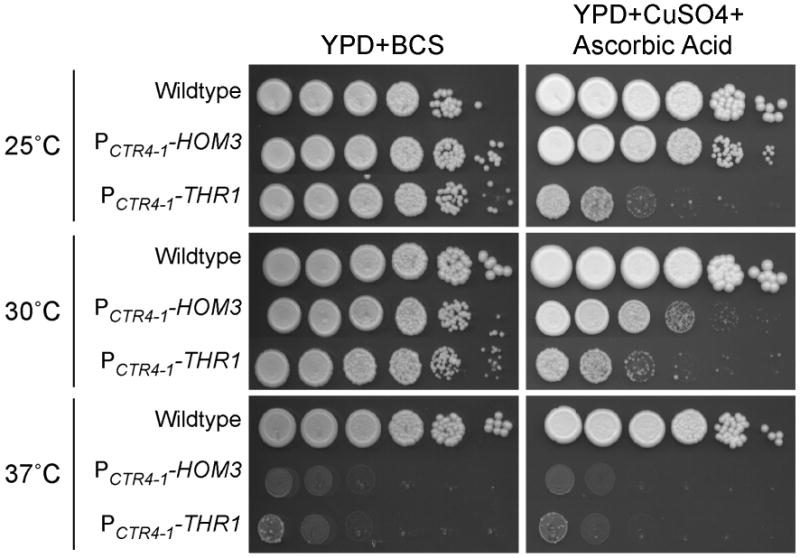
Temperature-dependent phenotypes of PCTR4-1-HOM3 (H99-76), PCTR4-1-THR1 (H99-73), and the wildtype (H99) strains in gene-repressing (CuSO4 + ascorbic acid) and gene-inducing (BCS) conditions. Ten-fold dilutions of strains were plated and incubated for three days.
Essential phenotype is dependent on nitrogen source and amino acid form
The lack of growth of the PCTR4-1-HOM3 strain on rich media (YPD) in promoter-repressing conditions is likely not due to an inability of C. neoformans to supplement the methionine auxotrophy of this strain, since C. neoformans met2, met3, and met6 mutants all grow on YPD, and their methionine auxotrophy can be supplemented by methionine or methionine dipeptides, both when ammonium or proline are the nitrogen source (Nazi et al., 2007; Pascon et al., 2004; Yang et al., 2002). Since both PCTR4-1-HOM3 and PCTR4-1-THR1 strains were unable to grow under promoter-repressing conditions, the growth deficiency on YPD may instead be because C. neoformans is unable to satisfy the threonine auxotrophy in this medium.
Growth of the PCTR4-1-HOM3 and PCTR4-1-THR1 strains was compared by spot dilutions on minimal media containing ammonium (SD) or proline (SD(Pro)) as the nitrogen source, supplemented with various combinations of homoserine, methionine and threonine amino acids or dipeptides, in PCTR4-1 repression conditions (Figure 5). While no colony formation of strains was observed in PCTR4-1 repression conditions in the absence of amino acid supplements, residual growth similar to that present on YPD in repressing conditions was observed, which we attribute to a basal level of gene expression still occurring, and/or utilization of cell reserves accumulated during the pre-growth in gene-expressing conditions. Strains grew no better when the amino acids methionine and threonine were added to either SD or SD(Pro) media, than on media lacking amino acids. Strains were also unable to grow when SD media was supplemented with threonine and methionine dipeptides, but growth was enhanced above background levels when the threonine and methionine dipeptides were added to SD(Pro) media. Furthermore, growth of the PCTR4-1-THR1 strain required only threonine dipeptides, while the PCTR4-1-HOM3 strain growth required both methionine and threonine dipeptides, or homoserine, providing further evidence that these genes indeed confer homoserine kinase and aspartate kinase activities, respectively.
Figure 5.
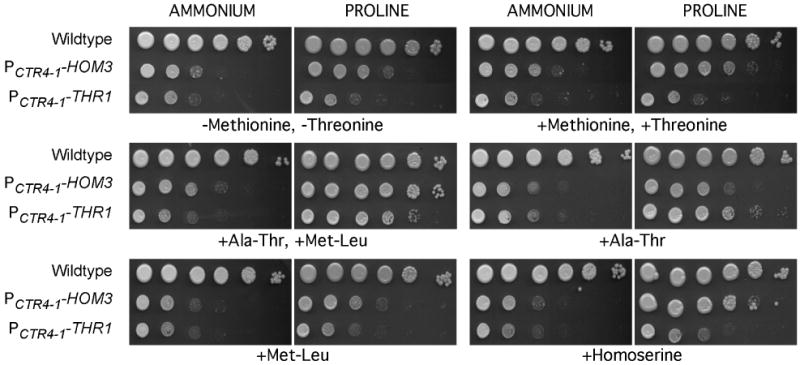
Auxotrophic supplementation of PCTR4-1-HOM3 (H99-76) and PCTR4-1-THR1 (H99-73) strains. Strains were serially diluted ten-fold, and plated on SD+CuSO4+Ascorbic acid or SD(Pro)+CuSO4+Ascorbic acid that was supplemented with various combinations of methionine, threonine, Ala-Thr, Met-Leu or homoserine, as indicated. Plates were incubated for three days at 30 °C.
The demonstration that threonine dipeptides, but not amino acids, supplement Cryptococcal threonine auxotrophy, and only in the presence of proline, but not ammonium, as a sole nitrogen source, is similar to results observed previously for supplementation of other C. neoformans auxotrophies (Kingsbury et al., 2004a; Kingsbury et al., 2004b). Combined, these results indicate that C. neoformans possesses less amino acid permeases, permeases that have a lower transport velocity, or more are subject to ammonium repression. While YPD medium contains significant levels of peptides, our results indicate that the lack of growth in this media is likely due to nitrogen repression, and/or transport of other peptides out competing transport of threonine-containing peptides.
Temperature determines severity of phenotype
While the inability of a Cryptococcal amino acid auxotroph to grow on YPD at 30 °C has not been previously demonstrated, it has been shown that SPE3-lys9 (lysine auxotrophic) and ilv2 (isoleucine and valine auxotrophic) mutants die in YPD at 37 °C (Kingsbury et al., 2004a; Kingsbury et al., 2004b). We therefore determined whether the incubation temperature affects growth of PCTR4-1-HOM3 and PCTR4-1-THR1 strains on YPD. Interestingly, reduction of the incubation temperature to 25 °C allowed the growth of PCTR4-1-HOM3 strain in repressing conditions, while the PCTR4-1-THR1 strain was no better able to grow at this temperature than at 30 °C (Figure 4).
Differences in the growth phenotype between the two strains could be attributable to either homoserine kinase having a role in addition to threonine biosynthesis in C. neoformans, or the block in the biosynthetic pathway caused by inhibition of homoserine kinase leading to the accumulation of a toxic intermediate. Consistent with this, elevated levels of the intermediate predicted to accumulate, homoserine, is toxic to mammalian (Rees et al., 1994) and bacterial cells (Kotre et al., 1973; O'Barr T & Everett, 1971). Moreover, the accumulation of a toxic intermediate has been hypothesized to be responsible for toxic effects associated with other amino acid biosynthetic mutants (Arevalo-Rodriguez et al., 2004; Kingsbury et al., 2004a; Pascon et al., 2004; Suliman et al., 2007), and may explain deleterious phenotypes associated with THR1 and THR4 mutation in S. cerevisiae. In S. cerevisiae, the threonine pathway is regulated positively in response to threonine starvation, by upregulation of gene transcription and eliminating feedback inhibition (Hinnebusch, 1992; Martin-Rendon et al., 1993; Mountain et al., 1991; Ramos & Calderon, 1992). If the pathway is similarly regulated in C. neoformans, threonine starvation conditions, such as what Thr1p-inhibited C. neoformans would likely encounter in vivo, should result in increased flux through the threonine biosynthetic pathway, thus increased toxic intermediate accumulation, and hence intensified growth defects.
We also compared the growth of PCTR4-1-HOM3 and PCTR4-1-THR1 strains at 37 °C. While some background growth of both strains was still present at 30 °C in repressing conditions, growth was completely eliminated at 37 °C in repressing conditions. Surprisingly, growth was also eliminated in induction conditions, which may indicate that there is a greater difference between expression from the CTR4-1 promoter and endogenous THR1 and HOM3 expression at 37 °C, compared with 30 °C. To examine this, we compared HOM3, THR1, and CTR4 transcript levels following a 5 hr incubation of the PCTR4-1-HOM3, PCTR4-1-THR1 and wildtype strains, in promoter-induction and repression conditions, at 25 °C and 37 °C (Figure 6). Following normalization to the GPD housekeeping gene, we observed no obvious temperature-dependent changes in THR1, HOM3, or CTR4 transcript levels from the PCTR4-1-HOM3, PCTR4-1-THR1 and wildtype strains, respectively, grown in promoter-induction conditions, thus the CTR4 (and CTR4-1) promoter is not regulated by temperature. Furthermore, although HOM3 transcript levels were barely detectable in both temperatures, THR1 and HOM3 transcription levels in the wildtype did not appear to be enhanced at 37 °C compared with 25 °C. Results also show that HOM3 and THR1 transcripts expressed from PCTR4-1-HOM3 and PCTR4-1-THR1 strains in induction conditions were at higher than wildtype levels at both 30 °C and 37 °C. One possible explanation for the lack of growth in induction conditions is that higher expression of these genes might result in growth impairment by perturbing metabolic flux, interfering with general cell metabolism, which may be accentuated at higher temperatures. Given our SPE3-lys9 and ilv2 findings, the increased severity of growth defects at 37 °C in repressing conditions may be due to decreased threonine transport at this temperature.
Figure 6.
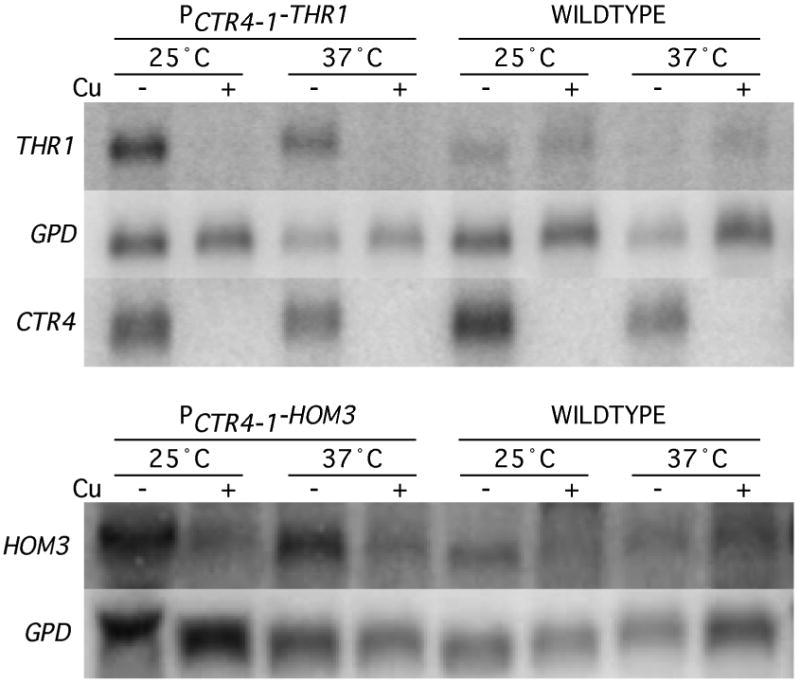
Northern hybridization analysis comparing THR1, HOM3, CTR4 and GPD transcript abundance. RNA was prepared from H99 (wildtype), H99-73 (PCTR4-1-THR1), and H99-76 (PCTR4-1-HOM3) strains, grown in PCTR4-1-repressing (YPD + CuSO4 + ascorbic acid; + Cu) or PCTR4-1-inducing (YPD + BCS; - Cu) conditions, at 25 °C or 37 °C.
Given the different niche occupation and evolutionary distance between S. cerevisiae, an ascomycete, and C. neoformans, a basidiomycete, it is not surprising to see differences in gene requirement between species, for example, the fatty acid synthesis genes FAS1 and FAS2, and RAM1 required for signaling, are essential in C. neoformans but not in S. cerevisiae and/or C. albicans (Chayakulkeeree et al., 2007; Vallim et al., 2004). However, given the highly conserved nature of the threonine biosynthetic pathway between fungi, it is surprising to see that threonine biosynthetic genes are essential in C. neoformans, and to our knowledge, this is the first documented case of threonine biosynthetic gene necessity in fungi. The essential nature of these genes, particularly at 37 °C, makes aspartate kinase and homoserine kinase excellent candidates for anti-cryptococcal drug targets.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Zhonghui Yang for some strain construction, Dr Joseph Heitman for comments on the manuscript, and Drs James Fraser and Tamara Doering, and for the generous gifts of plasmids pJAF1 and pCTR4-2, respectively. We also thank Dr Brian Wong for the C. neoformans cDNA library, which was a gift to the Duke University Mycology Research Unit. This study was funded by the National Institute of Health R01 grant GM070541 and R21 grant AI070247.
Footnotes
Publisher's Disclaimer: This is an author manuscript that has been accepted for publication in Microbiology, copyright Society for General Microbiology, but has not been copy-edited, formatted or proofed. Cite this article as appearing in Microbiology. This version of the manuscript may not be duplicated or reproduced, other than for personal use or within the rule of ‘Fair Use of Copyrighted Materials’ (section 17, Title 17, US Code), without permission from the copyright owner, Society for General Microbiology. The Society for General Microbiology disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by any other parties. The final copy-edited, published article, which is the version of record, can be found at http://mic.sgmjournals.org, and is freely available without a subscription.
References
- Arevalo-Rodriguez M, Pan X, Boeke JD, Heitman J. FKBP12 controls aspartate pathway flux in Saccharomyces cerevisiae to prevent toxic intermediate accumulation. Eukaryot Cell. 2004;3:1287–1296. doi: 10.1128/EC.3.5.1287-1296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell GW, Giaever G, Chu AM, Davis RW, Brown JM. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc Natl Acad Sci U S A. 2001;98:12608–12613. doi: 10.1073/pnas.231366398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell GW, Brown JA, Wu HI, Giaever G, Chu AM, Davis RW, Brown JM. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc Natl Acad Sci U S A. 2002;99:8778–8783. doi: 10.1073/pnas.132275199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A, Vousden KA, Binley KM, Radcliffe P, Trevethick J, Mannazzu I, Sudbery PE. A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics. 2004;166:707–719. doi: 10.1534/genetics.166.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayakulkeeree M, Rude TH, Toffaletti DL, Perfect JR. Fatty acid synthesis is essential for survival of Cryptococcus neoformans and a potential fungicidal target. Antimicrob Agents Chemother. 2007;51:3537–3545. doi: 10.1128/AAC.00442-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D'Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Williams RM, Chu AM, Davis RW. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CD, Lee MS, Spencer FA, Jensen RE. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol Biol Cell. 2006;17:213–226. doi: 10.1091/mbc.E05-06-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi AH, Saunders WS. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: P J, B J, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 319–414. [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:265–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Jones EW, Fink GR. Regulation of amino acid and nucleotide biosynthesis in yeast. In: Strathern JN, Jones EW, Broach JR, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Kingsbury JM, Yang Z, Ganous TM, Cox GM, McCusker JH. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37°C and in vivo. Microbiology. 2004a;150:1547–1558. doi: 10.1099/mic.0.26928-0. [DOI] [PubMed] [Google Scholar]
- Kingsbury JM, Yang Z, Ganous TM, Cox GM, McCusker JH. A novel chimeric spermidine synthase-saccharopine dehydrogenase (SPE3-LYS9) gene in the human pathogen Cryptococcus neoformans. Eukaryot Cell. 2004b;3:752–763. doi: 10.1128/EC.3.3.752-763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury JM, Goldstein AL, McCusker JH. Role of nitrogen and carbon transport, regulation, and metabolism genes for Saccharomyces cerevisiae survival in vivo. Eukaryot Cell. 2006;5:816–824. doi: 10.1128/EC.5.5.816-824.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotre AM, Sullivan SJ, Savageau MA. Metabolic regulation by homoserine in Escherichia coli B-r. J Bacteriol. 1973;116:663–672. doi: 10.1128/jb.116.2.663-672.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Rendon E, Farfan MJ, Ramos C, Calderon IL. Isolation of a mutant allele that deregulates the threonine biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1993;24:465–471. doi: 10.1007/BF00351707. [DOI] [PubMed] [Google Scholar]
- McDade HC, Cox GM. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol. 2001;39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- Mountain HA, Bystrom AS, Larsen JT, Korch C. Four major transcriptional responses in the methionine/threonine biosynthetic pathway of Saccharomyces cerevisiae. Yeast. 1991;7:781–803. doi: 10.1002/yea.320070804. [DOI] [PubMed] [Google Scholar]
- Nazi I, Scott A, Sham A, Rossi L, Williamson PR, Kronstad JW, Wright GD. Role of homoserine transacetylase as a new target for antifungal agents. Antimicrob Agents Chemother. 2007;51:1731–1736. doi: 10.1128/AAC.01400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Barr TP, Everett KA. Effect of L-homoserine on the growth of Mycobacterium tuberculosis. Infect Immun. 1971;3:328–332. doi: 10.1128/iai.3.2.328-332.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory JJ, Griffith CL, Doering TL. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. Yeast. 2004;21:919–926. doi: 10.1002/yea.1139. [DOI] [PubMed] [Google Scholar]
- Pascon RC, Ganous TM, Kingsbury JM, Cox GM, McCusker JH. Cryptococcus neoformans methionine synthase: expression analysis and requirement for virulence. Microbiology. 2004;150:3013–3023. doi: 10.1099/mic.0.27235-0. [DOI] [PubMed] [Google Scholar]
- Payne SH, Loomis WF. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot Cell. 2006;5:272–276. doi: 10.1128/EC.5.2.272-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Toffaletti DL, Rude TH. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61:4446–4451. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C, Calderon IL. Overproduction of threonine by Saccharomyces cerevisiae mutants resistant to hydroxynorvaline. Appl Environ Microbiol. 1992;58:1677–1682. doi: 10.1128/aem.58.5.1677-1682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees WD, Grant SD, Hay SM, Saqib KM. Threonine synthesis from homoserine as a selectable marker in mammalian cells. Biochem J. 1994;299(Pt 3):637–644. doi: 10.1042/bj2990637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg KJ, Bickel S, Rowley N, Kaiser CA. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by Sec13, Lst4, Lst7 and Lst8. Genetics. 1997;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sherman F, Fink GR, Lawrence CW. Methods in yeast genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- Suliman HS, Appling DR, Robertus JD. The gene for cobalamin-independent methionine synthase is essential in Candida albicans: a potential antifungal target. Arch Biochem Biophys. 2007;467:218–226. doi: 10.1016/j.abb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvarna K, Bartiss A, Wong B. Mannitol-1-phosphate dehydrogenase from Cryptococcus neoformans is a zinc-containing long-chain alcohol/polyol dehydrogenase. Microbiology. 2000;146:2705–2713. doi: 10.1099/00221287-146-10-2705. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallim MA, Fernandes L, Alspaugh JA. The RAM1 gene encoding a protein-farnesyltransferase beta-subunit homologue is essential in Cryptococcus neoformans. Microbiology. 2004;150:1925–1935. doi: 10.1099/mic.0.27030-0. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Yang Z, Pascon RC, Alspaugh A, Cox GM, McCusker JH. Molecular and genetic analysis of the Cryptococcus neoformans MET3 gene and a met3 mutant. Microbiology. 2002;148:2617–2625. doi: 10.1099/00221287-148-8-2617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


