Abstract
Background
The European seabass (Dicentrarchus labrax), one of the most extensively cultured species in European aquaculture productions, is, along with the gilthead sea bream (Sparus aurata), a prospective model species for the Perciformes which includes several other commercially important species. Massive mortalities may be caused by bacterial or viral infections in intensive aquaculture production. Revealing transcripts involved in immune response and studying their relative expression enhances the understanding of the immune response mechanism and consequently also the creation of vaccines. The analysis of expressed sequence tags (EST) is an efficient and easy approach for gene discovery, comparative genomics and for examining gene expression in specific tissues in a qualitative and quantitative way.
Results
Here we describe the construction, analysis and comparison of a total of ten cDNA libraries, six from different tissues infected with V. anguillarum (liver, spleen, head kidney, gill, peritoneal exudates and intestine) and four cDNA libraries from different tissues infected with Nodavirus (liver, spleen, head kidney and brain). In total 9605 sequences representing 3075 (32%) unique sequences (set of sequences obtained after clustering) were obtained and analysed. Among the sequences several immune-related proteins were identified for the first time in the order of Perciformes as well as in Teleostei.
Conclusion
The present study provides new information to the Gene Index of seabass. It gives a unigene set that will make a significant contribution to functional genomic studies and to studies of differential gene expression in relation to the immune system. In addition some of the potentially interesting genes identified by in silico analysis and confirmed by real-time PCR are putative biomarkers for bacterial and viral infections in fish.
Background
The European seabass Dicentrarchus labrax is one of the most extensively aquacultured fish species in the Mediterranean, resulting in steadily increasing pressure on producers. Consequently, it is important to acquire new techniques and knowledge in order to improve aquaculture practices. Detailed information concerning growth, health, disease resistance and flesh quality benefit from the molecular as well as from the physiological point of view can provide illuminating new findings leading to improved aquaculture techniques. Several efforts have been made up till now to enrich the genomic resources in aquaculture production in the Mediterranean (chiefly for the gilthead sea bream Sparus aurata and for the European seabass Dicentrarchus labrax), e.g. Marine Genomics Europe (Network of Excellence) (CT-2003-505403), [1-5] as well as in the Atlantic (e.g. Atlantic halibut Hippoglossus hippoglossus, Salmon Salmo salar) e.g. [6-13]. These studies focused mainly on non-challenged tissues in order to obtain a first unigene catalogue. Aquaculture production however is affected by viral and pathogenic bacteria, particularly in respect of D. labrax which has been shown to be the species most sensitive to pathogenic bacteria such as Vibrio anguillarum [14] and to viral infections such as Nodavirus [15,16]. There are several commercial vaccines which provide protection against infection from V. anguillarum although the mechanism of immune response still remains unknown. Nodavirus can cause massive mortalities [17] and cannot be controlled so far because the production of commercial vaccines here is still in its infancy. In the present study we have generated a collection of EST sequences from tissues of European seabass infected with V. anguillarum and Nodavirus. Within this collection we were able to isolate immune relevant genes, and have gone on to compare gene expression in different tissues after viral and pathogenic bacteria infection. Additionally we determined in silico differential expression between the two infections. In this context the construction and analysis of a total of ten cDNA libraries are described; six cDNA libraries were from tissues of the European seabass infected with V. anguillarum (liver, spleen, head kidney, peritoneal exudate, gill and intestine) with peritoneal exudate, gill and intestine as target organs for V. anguillarum infections, and four cDNA libraries were from tissues of the European seabass infected with Nodavirus (liver, spleen, head kidney and brain) with the brain as target organ of the virus. Comparisons between the predicted European seabass peptide data set and the zebrafish, medaka, stickleback, tetraodon and human proteomes were performed. Genes showing in silico differential expression between Nodavirus infection and V. anguillarum infection were further analysed by real-time PCR.
Results
Summary of ESTs from the cDNA libraries infected with Nodavirus and V. anguillarum
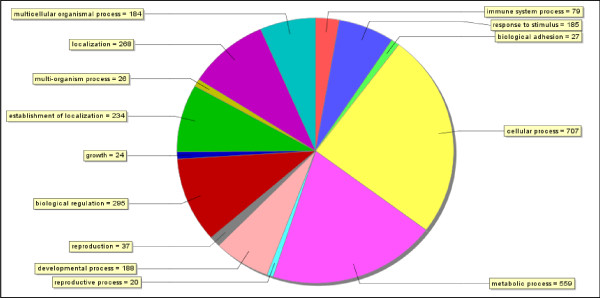
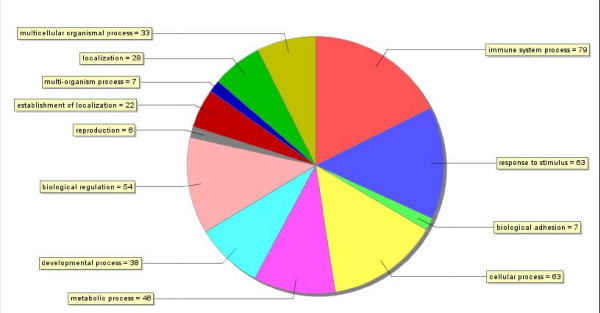
The amplified libraries contained insert size from approximately 0.5 to 2.0 kb. Single pass sequencing was performed resulting in 9605 high quality sequences. All sequences were submitted to the EST database (dbEST http://www.ncbi.nlm.nih.gov/projects/dbEST/ with the accession numbers FK939975 – FK944381, FL484477 – FL488763 and FL501471 – FL502381. A set of 3075 unique sequences was generated. Among the unique sequences (3075) [see Additional file 1] 371 [12%, see Additional file 2] sequences contained Simple Sequence Repeats (SSR). Cluster analyses performed for each library separately (Table 1a and Table 1b) revealed redundancy rates which varied from 72% (28% unique sequences) in intestine cDNA library infected with V. anguillarum to 37% (63% unique sequences) in spleen cDNA library infected with V. anguillarum (Table 1a). The set of unique EST sequences was annotated with Blast2GO which carries out BLASTX searches and attempts to assign function and GO classification. Out of the 3075 unique EST sequences submitted to GO2Blast for annotation and GO classification, 1521 sequences fell into 14 categories of biological process function at GO annotation level 2 (Fig. 1), where two categories, cellular process and metabolic process, were predominant. The category "immune system process" was represented by 79 transcripts.
Table 1.
Summary of sequences derived of cDNA libraries of D. labrax tissue infected with V. anguillarum (a) and Nodavirus (b)
| A | |||||
| Tissue | singletons | contigs | unique | total sequences | % of unique sequences |
| Liver | 503 | 190 | 693 | 2140 | 32.38 |
| Spleen | 326 | 38 | 364 | 651 | 63.14 |
| Kidney | 266 | 66 | 332 | 911 | 36.24 |
| Peritoneal exudate | 386 | 103 | 489 | 827 | 59.13 |
| Gill | 92 | 15 | 107 | 343 | 31.19 |
| Intestine | 88 | 4 | 92 | 326 | 28.22 |
| B | |||||
| Tissue | singletons | contigs | unique | total sequences | % of unique sequences |
| Liver | 253 | 127 | 380 | 1126 | 33.75 |
| Spleen | 698 | 118 | 816 | 1284 | 63.55 |
| Brain | 617 | 41 | 658 | 1099 | 59.87 |
| Kidney | 321 | 55 | 376 | 934 | 40.26 |
Figure 1.
Summary of GO category Biological process of unique ESTs obtained from seabass cDNA libraries infected with Nodavirus and Vibrio anguillarum.
EST matches with known function
Out of the 3075 EST sequences, 1246 (~ 41%) had a positive hit after submission to BLASTX database search. Among those EST sequences with a known function, 128 homologues were found to be involved in the immune response and 79 of these were grouped into the GO category "immune system process". The remaining 49 transcripts were manually determined to be involved in the immune response (see Additional file 3). Immune related transcripts isolated for the first time for seabass amounted to 115 (Table 2). Among transcripts of interest, the transcript encoding for an important antimicrobial protein, hepicidin, was isolated. Aligning EST sequences grouped into one contig can provide additional data. In the case of hepcidin it is probable that different isoforms are grouped together. Alignments of other cDNA sequences either showed alternative polyadenylation or they showed in silico polymorphism of microsatellite DNA as for instance the transcript coding for cysteine-rich protein 1-I (see Additional file 4).
Table 2.
Transcripts isolated for the first time in D. labrax and grouped to the GO category" immune system process"
| Contig ID | BLASTX Hit | Accession number |
| Contig_265, Contig_3 | alpha globin | Q9PVM4 BAA86218 |
| Contig_681 | alpha-1-microglobulin bikunin precursor | CAA45294 |
| Contig_3061 | b-cell leukemia lymphoma 6 | XP_001340785 |
| Contig_1838 | bcl2 adenovirus e1b 19 kda interacting protein 3 | NP_001012245, AAR83676 |
| Contig_276 | beta-2 microglobulin | ABB60035, ABB60037 |
| Contig_773 | beta-2 microglobulin precursor | AAC64994 |
| Contig_2731 | blood thirsty | AAX12162 |
| Contig_2210 | blood thirsty | XP_699830 |
| Contig_1617 | c1 inhibitor | NP_001117851, CAD58653 |
| Contig_936 | cathepsin s | AAQ01147 |
| Contig_2779 | cc chemokine | AAY79324 |
| Contig_616 | ccaat enhancer binding protein (c ebp)beta | BAB40971 |
| Contig_1810 | cd59-like protein 2 | NP_001117969, AAT94063 |
| Contig_1441 | cell division cycle 42 | NP_956159, AAH48035, AAH75761, AAX20139, CAM56524, AAI64988 |
| Contig_2676 | chemokine (c-c motif) ligand 13 | BAC20610 |
| Contig_2058 | chemokine (c-c motif) ligand 21b | AAT52146, ABA54959 |
| Contig_241 | chemokine (c-c motif) ligand 25 | ABC69050 |
| Contig_2858 | chemokine (c-x-c motif) ligand 12b (stromal cell-derived factor 1) | NP_840092, AAN64414, AAS92649, AAI09418 |
| Contig_524 | chemokine (c-x-c motif) ligand 9 | ABC69049 |
| Contig_627 | chemokine (c-x-c motif) receptor 4 | ABP48751 |
| Contig_525, Contig_1672 | chemokine cxc-like protein | ABC69049 |
| Contig_983 | complement c4-2 | CAD45003 |
| Contig_2306 | complement component 1 q subcomponent | ABV57766 |
| Contig_513 | complement component 1 qb chain | XP_001110783 |
| Contig_986 | complement component 5 | BAC23058 |
| Contig_1044 | complement component 7 | BAA88899 |
| Contig_2558 | complement component alpha polypeptide | NP_001118096, CAH6548 |
| Contig_1413 | complement component c3 | BAA88901 |
| Contig_1114 | complement component c4 | CAD45003 |
| Contig_2843 | complement component c5-1 | BAC23057 |
| Contig_2534, Contig_2600 | complement component c9 | P79755, AAC60288 |
| Contig_1499 | complement component factor h | NP_001117882, CAF25505 |
| Contig_1496 | complement component gamma polypeptide | NP_001117880, CAF22027 |
| Contig_927 | complement component1, q gamma polypeptide | XP_544508 |
| Contig_2432 | complement component beta subunit | Q9PVW7, BAA86877 |
| Contig_2605 | complement component q subcomponent binding protein | EDM05067 |
| Contig_1536 | complement component r subcomponent | AAR20889 |
| Contig_868 | complement factor b | CAD21938 |
| Contig_1607 | complement factor d preproprotein | XP_001117186 |
| Contig_2013 | complement factor h | NP_001117876, CAF05664, CAF05665 |
| Contig_1481 | complement factor h-related 1 | AAA92556 |
| Contig_1375 | cornichon homolog | O35372, AAC15828 |
| Contig_1198 | c-reactive protein | NP_999009, O19062 BAA21473 |
| Contig_1657 | c-type lectin | BAE45333 |
| Contig_1596 | cu zn superoxide dismutase | AAW29025 |
| Contig_395 | deah (asp-glu-ala-his) box polypeptide 16 | NP_956318, AAH45393, AAI65206 |
| Contig_2814 | ets-1 transcript variant ets-1 delta(iii-vi) | AAY19514 |
| Contig_626 | ferritin heavy chain | NP_001117129, P49946, AAB34575 |
| Contig_280 | fth1 protein | CAL92185 |
| Contig_2392 | g-protein couplededg6 | NP_001112363 |
| Contig_2662 | heat shock 10 kda protein 1 (chaperonin 10) | AAV37068 |
| Contig_2975 | heat shock 70 kda protein 4 | AAH65970 |
| Contig_2939 | heme oxygenase1 | ABL74501 |
| Contig_275 | hemoglobin alpha chain | CAP69820 |
| Contig_731 | Hephaestin | NP_579838 |
| Contig_1713 | hypoxanthine phosphoribosyltransferase 1 | NP_001002056, AAH71336 |
| Contig_1745, Contig_1876 | integrin beta 2 | BAB39130, NP_990582, CAA50671 |
| Contig_2452 | interleukin 18 receptor accessory protein | XP_001371334 |
| Contig_2261 | interleukin 1 type i | XP_416914 |
| Contig_1718 | interleukin 1 type ii | NP_001015713, AAH89644 |
| Contig_1506 | interleukin 2 receptor gamma chain | CAJ38407 |
| Contig_1835 | interleukin enhancer binding factor 3 | AAH47175 |
| Contig_966 | interleukin-1 receptor type ii | ABP99035 |
| Contig_852 | interleukin-1 receptor type ii | CAL30143 |
| Contig_2196 | loc559360 protein | AAI51869 |
| Contig_2044 | macrophage migration inhibitory factor | ABG54276 |
| Contig_1348 | major histocompatibility complex class i a chain | BAD13369 |
| Contig_17 | mflj00348 protein | BAD90390 |
| Contig_889 | mhc class i alpha antigen | ABB04088 |
| Contig_1349 | mhc class i antigen | BAD13366 |
| Contig_2797 | mitochondrial ribosomal protein s18b | NP_001106610, AAI52129, AAI55448 |
| Contig_3008 | natural resistance-associated macrophage protein | AAG31225 |
| Contig_576 | neurofibromatosis 1 | AAD15839 |
| Contig_1716 | novel protein vertebrate complement component 3 | NP_001116778, CAQ13357 |
| Contig_1327 | otuubiquitin aldehyde binding 1 | NP_001002500, AAH76301 |
| Contig_2661 | Proteasome activator subunit 1 (pa28 alpha) | ABE60902, ABK41199 |
| Contig_1948 | protein kinase alpha | AAI51472 |
| Contig_2319 | protein tyrosinereceptorc | XP_547374 |
| Contig_2009 | purinergic receptorg-protein13 | XP_001516794 |
| Contig_1715 | rhamnose binding lectin | NP_001117668, BAA92256 |
| Contig_934 | ribosomal protein s19 | P61155, AAP20214 |
| Contig_2680 | sam domain- and hd domain-containing protein 1 | XP_001097562 |
| Contig_684 | serum amyloid p-component | P12246 AAA40093, CAA34774, AAH61125, AAY88178, BAE25796, BAE38344, EDL39002 |
| Contig_2165 | sffv proviral integration 1 | NP_035485 |
| Contig_1473 | sh2 containing inositol-5-phosphatase | XP_687502 |
| Contig_2675 | skin mucus lectin | BAD90686 |
| Contig_840 | strawberry notch homolog 2 | EDL31603 |
| Contig_1540 | tnf superfamily member 14 | NP_001118039, ABC84585 |
| Contig_469 | transcription factor 3 isoform cra_b | NP_571169, CAA54305 |
| Contig_2299 | transforming growth beta receptor ii (70 80 kda) | XP_534237 |
| Contig_2692 | trypsin 10 | BAF76146 |
| Contig_1814 | vascular endothelial growth factor | NP_001038320, AAY89335 |
| Contig_1660 | x-box binding protein 1 | AAQ08005 |
Similarity relationships
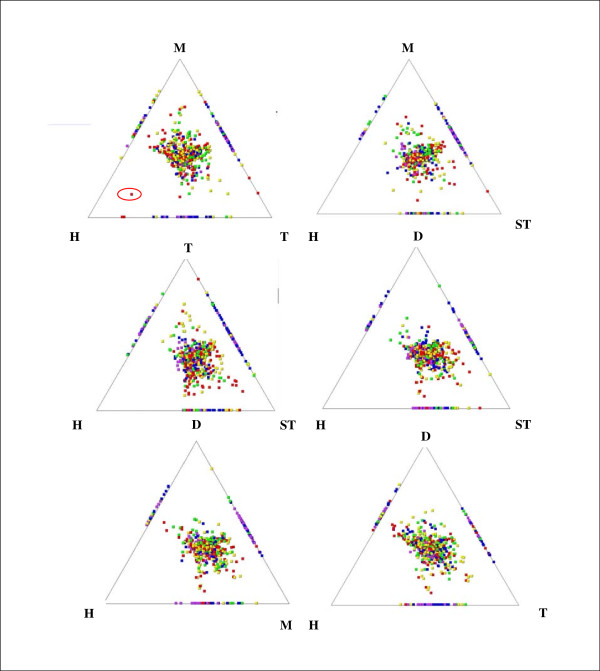
Figs. 2 and 3 show SimiTri representation of predicted seabass transcripts compared to Danio rerio, Homo sapiens, Oncorhynchus mykiss, Gasterosteus aculateus and Tetraodon nigroviridis proteomes. Of 3075 isolated unique transcripts 1040, 1051, 1122 1159, 1103 had Blast hits with a score > 50 against the H. sapiens, D. rerio, O. mykiss, G. aculateus and T. nigroviridis protein databases respectively.
Figure 2.
Similarity of D. labrax ESTs to the proteomes of Homo sapiens (H), Danio rerio (D), Oryzias latipes (M), Gasterosteus aculateus (ST) and Tetraodon nigroviridis (T). SimiTri plots show the graphical similarity i) between putative D. labrax peptides and H. sapiens, O. latipes and T. nigroviridis, cytochrome b is circled in red ii) between putative D. labrax peptides and H. sapiens, O. latipes and G. aculateus, iii) between putative D. labrax peptides and H. sapiens, D. rerio and T. nigroviridis, iv) between putative D. labrax peptides and H. sapiens, D. rerio, G. aculateus, v) between putative D. labrax peptides and H. sapiens, D. rerio, O. latipes, as well as vi) between putative D. labrax peptides and H. sapiens, D. rerio, T. nigroviridis.
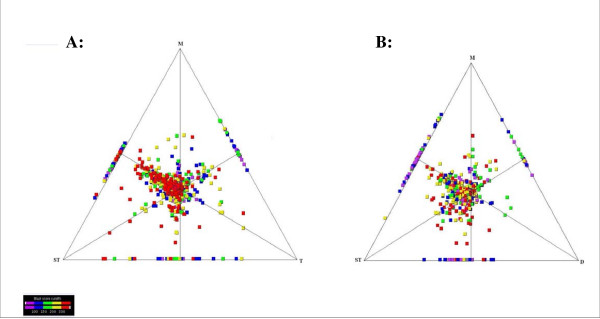
Figure 3.
Similarity of D. labrax ESTs to the proteomes of Danio rerio (D), Oryzias latipes (M), Gasterosteus aculateus (ST) and Tetraodon nigroviridis (T). SimiTri plots show the graphical similarity A: between putative D. labrax peptides and G. aculateus, O. latipes and T. nigroviridis, B: between putative D. labrax peptides and G. aculateus, O. latipes and D. rerio.
Expression analysis
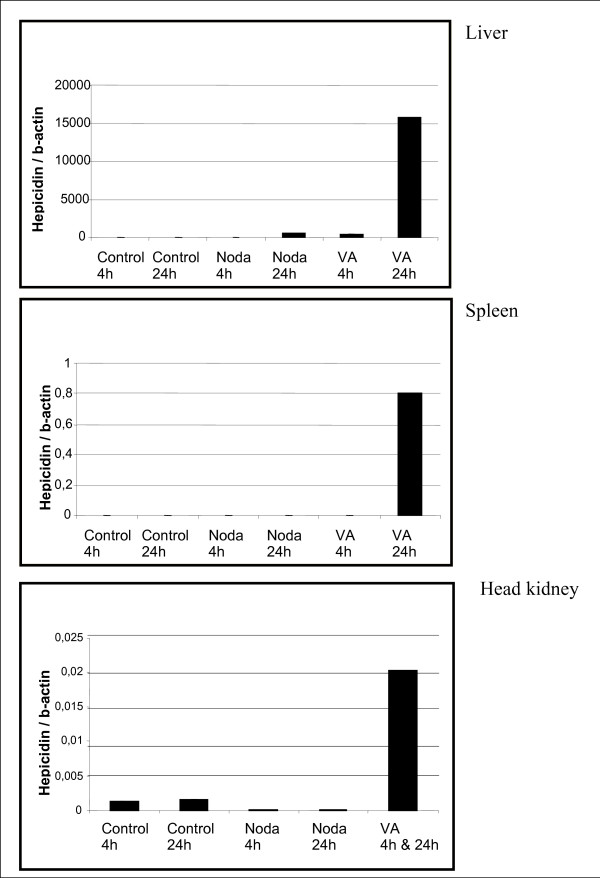
The test to compare multiple cDNA libraries with each other [22] revealed that the genes with the value > 6 of the test statistic R can be confidently considered as genes with true variation with the slope of 1.081 and are therefore not significantly different from -1 at the 5% level (see Additional file 6). The hits with R > 6 are in total 109 out of 2234 contigs resulting from EST sequences of liver, spleen and head kidney infected with Nodavirus and V. anguillarum. The list of the 109 transcripts with R > 6 and their putative homologues are shown in Additional file 5. It is interesting to note that although most transcripts were abundantly expressed in both bacterial and viral infected tissues, not all of them could be considered as specific markers of a specific infection. For example, fructose-1,6-biphosphate aldolase A, hepcidin, apolipoprotein A1 precursor, ferritin heavy chain and chemokine receptor 4 transcripts were found in V. anguillarum-infected tissues, though rarely in Nodavirus-infected tissues (see Additional file 5). Conversely, fructose-1,6-biphosphate aldolase B and 14 kDa apolipoprotein transcripts were frequently observed in Nodavirus infected tissues compared with V. anguillarum-infected tissues. The above results were further validated by determining the expression of putative markers for each infection in key tissues using real-time PCR. Here also control tissues were included in order to determine the expression of untreated fish. The real-time PCR confirmed the results obtained with the in silico analysis for selected genes. Taking into account the fold inductions of the real-time PCR experiments the correlations between in silico and qPCR are uniform. For instance the transcript for hepicidin precursor revealed in silico (R = 298.16) high expression only in liver tissues infected with V. anguillarum. The real-time PCR results show higher expression in all three tissues infected with V. anguillarum. However fold induction in liver is 20,000 times more than in spleen tissue, therefore theoretically 20,000 more cDNA clones had to be sequenced to obtain the sequence for hepicidin precursor in spleen infected with V. anguillarum. This correlation of high fold induction with in silico results can be observed for each transcript examined in this study. Thus, while the mRNA levels of hepcidin were found to increase considerably 24 h post-infection in the liver, spleen and head kidney of V. anguillarum-infected fish, they increased only slightly in Nodavirus-infected fish (Fig. 4). Notably, although the mRNA levels of transferrin and ferritin, both involved in iron metabolism with spleen and liver as the two main organs, increased in the liver after infection with both pathogens, they increased only in the spleen of V. anguillarum-infected animals (Figs. 5a and 5b).
Figure 4.
Hepicidin precursor expression index of liver, spleen, and head kidney infected with Nodavirus or with V. anguillarum for 4 h and 24 h. Infection for 4 h and 24 h of head kidney with V. anguillarum is pooled as not enough material was available. Each bar represents the mean of two technical duplicates of cDNA originating out of three individuals pooled prior to RNA extraction.
Figure 5.
(A) Ferritin expression index of liver and spleen infected with Nodavirus or with V. anguillarum for 4 h and 24 h. (B) Transferrin expression index of liver and spleen infected with Nodavirus or with V. anguillarum for 4 h and 24 h. Each bar represents the mean of two technical duplicates of cDNA originating out of three individuals pooled prior to RNA extraction.
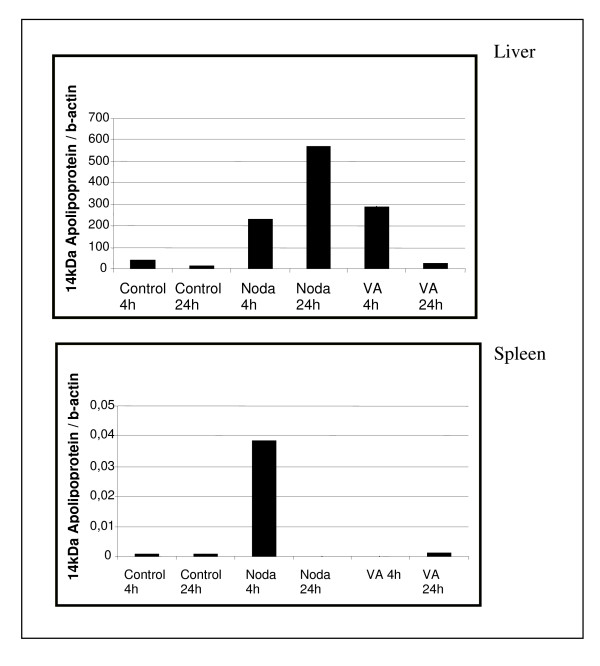
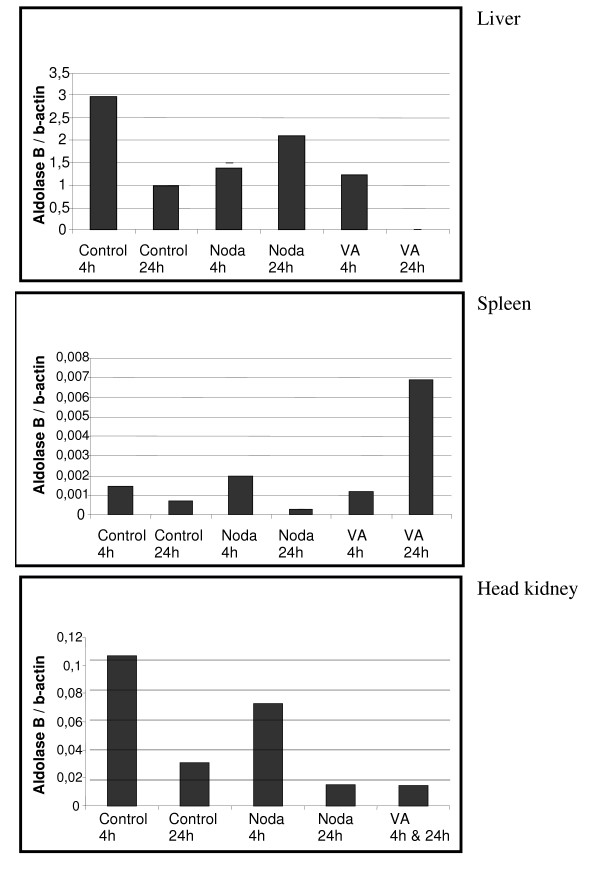
The mRNA levels of the chemokine receptor 4 were not affected or were slightly reduced in the head kidney and spleen of Nodavirus-infected fish but were considerably increased in these two tissues after V. anguillarum infection (Fig. 6). On the other hand, the mRNA levels of the 14 kDa apolipoprotein increased in the fish livers infected with both pathogens, but at 4 h and 24 h post-infection in the case of the Nodavirus and at 4 h post-infection in the case of V. anguillarum (Fig. 7). Here the expression in the liver is studied, as the liver is the major organ in the production of apoliprotein. Finally, although the mRNA levels of fructose-1,6-bisphosphate aldolase B decreased in the liver and head kidney following infection with both pathogens, they increased at 24 h post-infection in the spleen of V. anguillarum-infected fish (Fig. 8).
Figure 6.
Chemokine (c-x-c motif) receptor 4 (Cxcr 4) expression index of liver, spleen, and head kidney infected with Nodavirus or with V. anguillarum for 4 h and 24 h. Infection for 4 h and 24 h of head kidney with V. anguillarum is pooled as not enough material was available. Each bar represents the mean of two technical duplicates of cDNA originating out of three individuals pooled prior to RNA extraction. Between the two technical replicates of Noda 4 h and of VA 24 h a greater variation was detected. The values of the two replicates of Noda 4 h are 0.014 and 0.00095 and the values of the two replicates of VA 24 h are 0.03 and 0.009.
Figure 7.
14 KDa apolipoprotein expression index of liver infected with Nodavirus or with V. anguillarum for 4 h and 24 h. Each bar represents the mean of two technical duplicates of cDNA originating out of three individuals pooled prior to RNA extraction.
Figure 8.
Fructose-1,6-bisphosphatase aldolase B expression index of liver, spleen, and head kidney infected with Nodavirus or with V. anguillarum for 4 h and 24 h. Infection for 4 h and 24 h of head kidney with V. anguillarum is pooled as not enough material was available. Each bar represents the mean of two technical of cDNA originating out of three individuals pooled prior to RNA extraction.
Discussion
Although viral and bacterial infections are among the key challenges in fish aquaculture, nevertheless today the immune response of fish against V. anguillarum and Nodavirus remains largely unknown. Identification of genes involved in the immune response as well as the detection of differentially expressed genes between the two infections can make a significant contribution to future research leading to a better understanding of the biological system of immune response after fish infection. In the present study ten cDNA libraries, six from tissues infected with V. anguillarum and four from tissues infected with Nodavirus were analysed. Analysis of EST sequences coming from infected tissues will enhance the construction of an immune specific microarray chip containing already known transcripts involved in immune-related biological processes, such as the immune response as well as transcripts for which no annotation is available so far. Furthermore, transcripts indicating a higher expression level in one of the infections can be taken for future functional studies at RNA or DNA level as well as at protein level.
Over the past 30 years cDNA cloning for gene discovery and transcriptome analysis has become a very important molecular technique. Various techniques have been developed to address several scientific issues such as the cloning of rare transcripts, the construction of libraries with a wider cloning range, etc. (for review [26]). Construction of non-normalized libraries in the present study gave a first insight into the tissue-specific manner of transcript abundance according to their origin. Besides the possibility of identifying higher expressed transcripts, the percentages of unique sequences can also be assessed. In this study the redundancy of the cDNA libraries of liver, spleen and kidney infected with Nodavirus and V. anguillarum was in agreement with all three tissues (~33%, ~63% and ~38% respectively). This result is in line with other cDNA libraries of various fish species where the redundancy ranges between 40% and 60% depending on the tissues of origin [e.g. [12,13]]. Besides the identification and characterization of ESTs for components of the immune system, detection of microsatellite sequences will help in the completion of quantitative trait locus (QTL) scans currently being performed. Microsatellite sequences, also called Simple Sequence Repeats (SSR), are frequent in non-coding regions and are used as molecular markers. Detection of SSR within ESTs (exonic microsatellites or EST-SSRs) presents a shortcut to obtaining microsatellite markers. Since EST-SSRs are exonic they have two advantages over intergenic microsatellites. First, it is expected that their flanking regions are more conserved, so that the primers can be used even in related species, and second, it is assumed that they are in strong linkage disequilibrium with functionally important sites. Therefore they are frequently used in population genomics or in mapping of genes of economic significance identified as candidate markers for QTL and/or quantitative trait nucleotide (QTN). For EST similarity search in the present study a homologue of a known gene is defined as a cDNA whose similarity to a gene of any other organism in the database exceeds a certain fixed threshold. The identification of orthologues is outside the scope of this study. In total 1246 (41%) were assigned to a known transcript, with 79 (6%) categorized to the GO category "immune system process". Separate examination of these 79 transcripts by GO annotation reveals their involvement in 11 other categories of biological function (Fig. 9), with three dominant categories of response to stimulus, cellular process, and biological regulation. This collection should provide the base material for further research into understanding the immune response of European seabass as well as for the isolation of putative biomarkers.
Figure 9.
GO categorization of only the EST sequences grouped into the category of immune system process.
Similarity relationships
Comparison of predicted seabass genes compared to the genomes of zebrafish, medaka, tetraodon, stickleback and human (Fig. 2) showed that the majority of putative proteins were located in the centre. From separate examination of the different triads a bias towards the top and right sections is revealed. This bias is not unexpected as seabass is more closely related to medaka, zebrafish, stickleback and tetraodon. However it is worth noting, that the seabass cytochrome b seems to be more similar to human cytochrome b than to the tetraodon and medaka cytochrome b as shown in Fig. 2. This was not the case with stickleback and zebrafish cytochrome b. Interestingly, results from comparisons of putative proteins of the Atlantic halibut (Hippoglossus hippoglossus) with the human, zebrafish and tetraodon protein database showed that the halibut cytochrome c oxidase subunit 3 (Cox3) is more similar to human COX3 than the zebrafish and tetraodon Cox3 [13]. Comparison of predicted proteins with only the protein database of fish genomes shows a slight bias towards medaka and stickleback looking at the triad medaka, stickleback and tetraodon (Fig. 3A) and again a slight bias towards medaka and stickleback looking at the triad stickleback, medaka and zebrafish (Fig. 3B). These results give a first insight towards the evolution of immune related genes as the relatively equal distribution indicate that sequence variation between the clade Percomorpha is comparable to that between the clade Percomorpha and Ostariophysi.
Expression analysis
For in silico expression analysis transcript appearing more than once in the cDNA libraries were selected and their relative abundance were submitted to expression analysis after Stekel et al. [22]. Validation of in silico analysis was performed by qPCR. Individual variation may be masked in this approach as pooling strategy was chosen for qPCR experiments. The differential expressed transcripts detected in the present study can be further put forward for analysis of individual expression pattern. Nonetheless in order to study individual expression pattern the sampling frame has to be extended. In the present study the pooling strategy for qPCR was chosen in order to show cross-method consistency. However since results are consistent between the two approaches, influence of between individuals variability in response to infection has been addressed to some extent. In addition total RNA for qPCR analysis was extracted out of different individuals than the once used for cDNA library construction and patterns appear to be consistent between the different samples for all the selected candidate genes, which reflect the robustness of the approach and the small, if any bias, contributed by individual outliers. In silico expression analysis revealed a number of genes for R > 6 that are considerably above the exponential curve (see Additional file 6). Genes with R > 6 can be considered as significant and thus are candidate genes for further studies. Several of those transcripts including transcripts involved in iron metabolism such as ferritin and transferrin are also reported as differential expressed genes in the catfish Ictalurus punctatus and Ictalurus furcatus infected with the gram negative bacterium Edwardsiella ictaluri [27,28]. One of the main mechanisms whereby gram-negative bacteria pathogens like V. anguillarum obtain iron is the use of free heme or heme proteins from the host tissues [29]. The heme uptake mechanisms are considered to contribute to V. anguillarum virulence in fish [29]. However, it is surprising that Nodavirus infection also resulted in the up-regulation of transferrin and ferritin expression, especially within 24 h of infection. The abundance of transferrin transcripts in Nodavirus-infected tissues may not be related to the alteration of the iron metabolism by the pathogen but rather to the ability of enzymatically cleaved forms of this protein to activate fish macrophages [30]. The specific alteration of iron metabolism by V. anguillarum infection is also supported by the higher abundance of transcripts coding for hepcidin, a major homeostatic regulator of iron metabolism [31], and for α and β chains of hemoglobin in V. anguillarum- than Nodavirus-infected livers (255 clones vs. 1 clone, respectively). The expression of Hepicidin after bacterial infection has been shown in seabass [32] as well as in several other fish species like the striped bass [33], the red sea bream [34], the catfish [35,36], the Atlantic halibut [37], the zebrafish [38], the rainbow trout [39] and the perch [40]. In this study the qPCR experiments confirmed the up-regulation of hepicidin in D. labrax after infection with V. anguillarum and showed in addition to this, that the expression of hepcidin might be considered as an excellent marker of bacterial infections, since it was up-regulated in all examined tissues of V. anguillarum-infected fish but unaffected in Nodavirus-infected tissues. Another interesting observation of the in silico gene expression analysis is the differential abundance of transcripts encoding the isoforms A and B glycolytic/gluconeogenic enzyme fructose-1,6-biphopshate aldolase in bacterial and viral infected tissues. Although the role played by this enzyme in the outcome of these infections is difficult to anticipate due to its dual role in glucose metabolism, these results suggest that the expression ratio between the two enzyme isoforms may be used as a good indicator of the type of infection in the European seabass. Thus, the up-regulation of the B isoform in the spleen exclusively by V. anguillarum might be considered another potential marker for this bacterial infection. Similarly, apolipoprotein A1 and 14 kDa apolipoprotein, two major components of high density lipoproteins (HDL) and synthesized in the fish liver [41], also show a differential expression in the liver of fish infected with V. anguillarum and Nodavirus following the time course and, therefore, they also may be good candidate indicators of the fish health status and/or the type of infection. The real-time PCR confirmed observations of in silico expression analysis and also revealed that the expression of the 14 kDa apolipoprotein and aldolase B in the spleen is an appropriate marker of Nodavirus and V. anguillarum infections, respectively. Previous studies in carp and medaka have also shown the involvement of apolipoproteins in the immune response [42,43]. Finally, the differential expression of one of the clear immune-related genes, the chemokine receptor 4, was also found to be a good putative marker for V. anguillarum infection. For assessment of variability of putative markers further studies looking at individuals, exposed to other environmental or pathogenic conditions are needed to exclude possible biological variability caused by infections.
Conclusion
In this study we generated a collection of EST sequences from tissues of the European seabass infected with V. anguillarum and Nodavirus. We compared gene expression of different tissues after viral and pathogenic bacteria infection. A collection of 3075 unigenes was generated and candidate microsatellite sequences detected. Furthermore, comparisons of D. labrax transcripts with zebrafish, human, tetraodon, medaka and stickleback were performed. The majority of putative proteins were located in the centre with a bias towards the right sections, with D. labrax as expected being more closely related to the other fish species than to human. Comparison of putative D. labrax proteins was also performed among fish species. In this case a slight bias towards stickleback and medaka was observed when comparing medaka, stickleback and tetraodon and a slight bias towards stickleback and medaka was observed when comparing medaka, stickleback and zebrafish. Furthermore, in silico analysis of differential gene expression between the two infections based on EST sequences suggests a list of genes with a presumed function in the immune response of D. labrax revealing also the importance of looking at "non-classical" immune host proteins and emphasizing the significance of EST sequences generated from cDNA libraries of infected fish tissues. In addition, we show the power of sequencing cDNA sequences for expression analysis by performing real-time PCR experiments for transcripts with high, medium and low R-value. In view of new and high throughput sequence techniques detection of differential expression by measuring in silico the abundance of each transcript will enhance significantly the era of functional genomics. Furthermore in silico analysis in this study, followed by the confirmation with real-time PCR of potentially interested genes, has revealed some of them as potential biomarkers for bacterial and viral infections in fish.
Methods
Experimental condition and tissues collection
Two infections, one with Nodavirus strain 475-9/99 isolated from diseased sea bass [from the Instituto Zooprofilattico Sperimentale delle Venezie (Italy) [16]] and one with V. anguillarum strain R-82 (serogoup 01) [from the University of Santiago (Spain) [14]] were performed with seabass as previously described [14,16]. Tissues were taken 4 and 24 h post-infection. Three tissue types (spleen, liver and head kidney) of each experimental condition as well as peritoneal exudate, gill, intestine from V. anguillarum infection and brain from Nodavirus infection were selected and immediately frozen with liquid nitrogen. The experiments described comply with the guidelines of the European Union Council (86/609/EU) for the use of laboratory animals and have been approved by the Bioethical Committee of the University of Murcia (Spain) and the CSIC National Committee on Bioethics.
In brief; For Nodavirus infection fish were injected intramuscularly with 100 μl of nodavirus suspension in Minimum Essential Medium (MEM) (5.9 × 106 TCID50 ml-1) and placed at 25°C. Mock-infected control fish were injected with the medium alone, and maintained under the same experimental conditions. Three fish from each experimental and control groups were sampled 4 and 24 hours post-infection. Animals were sacrificed by anesthetic (MS-222) overdose and dissected. For the present study brain, spleen, head kidney and liver were sampled. For V. anguillarum infection fish were injected intraperitoneally (i.p.) with 1 ml of phosphate-buffered saline (PBS) alone or containing either 2 × 106 live or 108 formalin-killed V. anguillarum R82 cells (serogroup 01). Under these experimental conditions, about half of the fish were moribund at 24 h post-infection and all of them died within 48 h post-infection. Head kidney (bone marrow equivalent of fish) and peritoneal exudate cells were obtained 4 h and 24 h after bacterial challenge.
RNA extraction
Total RNA was extracted using the NucleoSplin RNA II extraction kit (Machinery Nagel, Dueren, Germany). RNA quality was checked on EtBr stained agarose gels and RNA concentrations and purity were measured using a NanoDrop spectrophotometer. For library construction equal amounts of total RNA extracted out of infected tissues (4 h and 24 h) were pooled. For qPCR experiments total RNA was freshly extracted out of infected tissues originating from three different individuals pooled prior to RNA extraction (liver, spleen and head kidney) with 4 h and 24 h post-infection.
cDNA library construction
All libraries were constructed from total RNA using the Creator SMART cDNA library construction kit (BD Bioscience-Clontech, Mountain View, Canada) using the LD PCR based method. Between 20 and 22 PCR cycles were performed before size separation of inserts. cDNA fragments > 600 bp were selected and directionally ligated at the restriction site Sfi1 of the pDNR-lib vector (BD Clontech) or the pal 32 vector. Plasmids were transformed into E. coli strain DH10B (Invitrogen) by electroporation. The libraries were tested for the presence and the size of insert by PCR using two primer pairs. For the libraries constructed with pal 32 vector, the primer pair pal 32 FOR: 5'-CTCGGGAAGCGCGCCATT-3' and pal 32, REV: 5'-TAATACGACTCACTATAGGGC-3' were used. For the libraries constructed with pDNR-lib vector pDNR FOR: 5'-TAAAACGACGGCCAGTA-3' pDNR REV: 5'-GAAACAGCTATGACCATGTTC-3' were used. The products were run on an EtBr stained agarose gel.
DNA sequencing
After plasmid preparation, dideoxy-temination DNA cycle sequencing was performed using the BigDye 3.1 sequencing method and the pDNR FOR (5'-TAAAACGACGGCCAGTA-3') primer. The sequences were run on an ABI 3730 XL sequencer at MPI Molecular Genetics, Berlin.
Sequence analysis
The raw sequence reads were quality-trimmed and vector- and poly-A-clipped using PREGAP4 [18]. Clustering (grouping of clones related to one another by sequence homology) was performed using the software SeqManII (DNAstar Inc.). After clustering the term 'contig' is used to describe the sequence obtained from one cluster (the sequences of a cluster can be collapsed into a single, non-redundant sequence) and the term 'singleton' describes sequences appearing only once in the entire dataset. The set of sequences obtained by merging contigs and singletons are named as unique sequences.
Simple Sequence Repeats (SSR) in EST sequences
In silico mining for repeat motifs within the obtained unique sequences was perfomed with the programme Msatfinder http://www.genomics.ceh.ac.uk/msatfinder/[19].
Homology search and GO annotation
Gene Ontology (GO) category (Biological process) was assigned after BLASTX search of 3075 unique EST sequences using BLAST2GO. Threshold cutoff was at E-value 1e-3 and the alignment length of 33 amino acids (aa).
Similarity relationships
The unique sequences from all seabass libraries were submitted to BLASTX similarity searches [20] against the zebrafish, tetraodon, stickleback, medaka and human predicted proteomes (downloadable from http://www.ensembl.org/index.html). For each database the highest BLAST scores (bit score values) in excess of 50 were retained. Relative similarities between triads were visualized as a triangular plot generated by the SimiTri software [21].
Expression analysis
In silico
All sequences of each cDNA library were submitted to BLASTX and BLASTN searches [20]. Transcripts appearing more than once in the cDNA libraries were selected for in silico expression analysis after Stekel et al. [22]. In brief, this method allows the comparison of gene expression in any number of libraries in order to identify differential expressed genes. The method uses a single statistical test to describe the extent to which a gene is differentially expressed between libraries by a log likelihood ratio statistic and tends asymptotically to a χ2 distribution [22]. For real-time PCR experiments transcripts with high, medium and low R-value were selected.
Real-time PCR
Gene expression was assessed by real-time PCR (qPCR) in spleen, head kidney and liver at 4 h and 24 h post-infection. RNAs out of three animals pooled prior to RNA extraction were isolated as described above and were used to obtain cDNA by the Superscript II Reverse Transcriptase and oligo (dT)12–18 primer (Invitrogen) following the manufacturer's instructions. Quantitative PCR assays were performed using the 7300 real-time PCR System (Applied Biosystems) with specific primers (Table 3). Each primer (0.5 μl; 10 μM) and the cDNA template (1 μl) were mixed with 12.5 μl of SYBR green PCR master mix (Applied Biosystems) in a final volume of 25 μl. The standard cycling conditions were 95°C for 10 min. followed by 40 cycles of 95°C 15 s and 60°C 1 min. For all reactions two technical duplicates were performed. The comparative CT method (2-ΔΔ CT method) was used to determine the expression level of analysed genes [23]. After evaluation of β-actin as a suitable reference gene for this study in seabass (data not shown) the expression of the candidate genes was normalized. The use of β-actin as a suitable reference gene was also shown in other fish studies [e.g. [24,25]].
Table 3.
Real-time primer sequences
| Name | F/R | Sequence 5'-3' |
| β-actin | Forward | GTGCGTGACATCAAGGAGAA |
| β-actin | Reverse | GCTGGAAGGTGGACAGAGAG |
| Apoliprot | Forward | ATACGTCCTGGCACTGATCC |
| Apoliprot | Reverse | AGCCTGACCTTGCTCACTGT |
| Chemokin-R4 | Forward | TCAAAACGATGACGGACAAG |
| Chemokin-R4 | Reverse | ACACGCTGCTGTACAGGTTG |
| Transferrin | Forward | CTGGGAAGTGTGGTCTGGTT |
| Transferrin | Reverse | CAAGACCTCTTGCCCTTCAG |
| Ferritin_HC | Forward | ATGCACAAGCTCTGCTCTGA |
| Ferritin_HC | Reverse | TTTGCCCAGGGTGTGTTTAT |
| Hepcidin-Prec | Forward | CCAGTCACTGAGGTGCAAGA |
| Hepcidin-Prec | Reverse | TCAGAACCTGCAGCAGACAC |
| Aldolase-B | Forward | TGACATTGCTCAGAGGATCG |
| Aldolase-B | Reverse | AGTTGGACATGGAGGGACTG |
Authors' contributions
ES contributed to EST production, performed analysis and the conception, design and manuscript writing. PS performed infections with VA and tissue sampling and contributed to RNA extractions. LP performed qPCR experiments. VM conceived the study, contributed to the conception and design of the project and also to the manuscript writing. JM contributed to the conception and design of the project. AF contributed to conception and design of the project and helped to draft the manuscript. BN conceived qPCR experiments, Nodavirus infections and tissue sampling and contributed to conception and design of the project and helped to draft the manuscript. VT and RR created the cDNA libraries and performed the sequencing of the clones. AM contributed to conception and design of the project and helped to draft the manuscript. GK conceived the study, contributed to the conception and design of the project and also to the manuscript writing.
Supplementary Material
Appendix 1. Catalogue of all unique sequences obtained from all constructed cDNA libraries in this study.
Appendix 2. EST-SSRs identified among EST sequences of all cDNA libraries constructed in this study.
Appendix 3. EST sequences (after clustering of all 3075) grouped into GO group Immune system process.
Appendix 4. A: CDNA sequences of hepicidin precursor showing putative isoforms B: CDNA showing microsatellite sequence with in silico SSR polymorphism after alignment of 8 sequences and C: sequences of cysteine-rich protein 1-I showing putative alternative splicing polyadenylations.
Appendix 5. Calculated R-values for contigs resulting from the EST sequencing of the cDNA library of spleen, liver and kidney infected with Nodavirus and Vibrio anguillarum.
Appendix 6. The number of genes for a given value or the test statistic R is plotted as a function of R. The data falling within 1 < R < 6 are decrees exponential curve, and decreasing exponentially with R. The slope is -1.081 with significance at the 5% level of 0.013 and is therefore not significantly different from -1 at 5% significance. When R > 6, the number of genes is above this exponential curve.
Acknowledgments
Acknowledgements
The authors would like to thank Margaret Eleftheriou for carefully proofreading the manuscript. This work was supported by the European Commission's 5th Framework Programme WEALTH (Contract No. 501984, Welfare and health in sustainable aquaculture [WEALTH]).
Contributor Information
Elena Sarropoulou, Email: sarris@her.hcmr.gr.
Pilar Sepulcre, Email: mpsepul@um.es.
Laura Poisa-Beiro, Email: laurap@iim.csic.es.
Victoriano Mulero, Email: vmulero@um.es.
José Meseguer, Email: meseguer@fcu.um.es.
Antonio Figueras, Email: antoniofigueras@iim.csic.es.
Beatriz Novoa, Email: virus@iim.csic.es.
Vasso Terzoglou, Email: vassoter@her.hcmr.gr.
Richard Reinhardt, Email: rr@molgen.mpg.de.
Antonios Magoulas, Email: magoulas@her.hcmr.gr.
Georgios Kotoulas, Email: kotoulas@her.hcmr.gr.
References
- Chistiakov DA, Hellemans B, Haley CS, Law AS, Tsigenopoulos CS, Kotoulas G, Bertotto D, Libertini A, Volckaert FA. A microsatellite linkage map of the European sea bass Dicentrarchus labrax L. Genetics. 2005;170:1821–1826. doi: 10.1534/genetics.104.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov DA, Hellemans B, Tsigenopoulos CS, Law AS, Bartley N, Bertotto D, Libertini A, Kotoulas G, Haley CS, Volckaert FA. Development and linkage relationships for new microsatellite markers of the sea bass (Dicentrarchus labrax L.) Anim Genet. 2004;35:53–57. doi: 10.1046/j.1365-2052.2003.01076.x. [DOI] [PubMed] [Google Scholar]
- Chini V, Rimoldi S, Terova G, Saroglia M, Rossi F, Bernardini G, Gornati R. EST-based identification of genes expressed in the liver of adult seabass (Dicentrarchus labrax, L.) Gene. 2006;376:102–106. doi: 10.1016/j.gene.2006.02.010. [DOI] [PubMed] [Google Scholar]
- Sarropoulou E, Power DM, Magoulas A, Geisler R, Kotoulas G. Comparative analysis and characterization of expressed sequence tags in gilthead sea bream (Sparus aurata) liver and embryos. Aquaculture. 2005;243:69–81. doi: 10.1016/j.aquaculture.2004.10.012. [DOI] [Google Scholar]
- Sarropoulou E, Franch R, Louro B, Power DM, Bargelloni L, Magoulas A, Senger F, Tsalavouta M, Patarnello T, Galibert F, Kotoulas G, Geisler R. A gene-based radiation hybrid map of the gilthead sea bream Sparus aurata refines and exploits conserved synteny with Tetraodon nigroviridis. BMC Genomics. 2007;8:44. doi: 10.1186/1471-2164-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Caplice NC, Davey GC, Powell R. EST-based identification of genes expressed in the liver of adult Atlantic salmon (Salmo salar) Biochem Biophys Res Commun. 2002;293:578–585. doi: 10.1016/S0006-291X(02)00263-2. [DOI] [PubMed] [Google Scholar]
- Haugland O, Torgersen J, Syed M, Evensen O. Expression profiles of inflammatory and immune-related genes in Atlantic salmon (Salmo salar L.) at early time post vaccination. Vaccine. 2005;23:5488–5499. doi: 10.1016/j.vaccine.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Adzhubei AA, Vlasova AV, Hagen-Larsen H, Ruden TA, Laerdahl JK, Hoyheim B. Annotated expressed sequence tags (ESTs) from pre-smolt Atlantic salmon (Salmo salar) in a searchable data resource. BMC Genomics. 2007;8:209. doi: 10.1186/1471-2164-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildahl J, Galay-Burgos M, Sweeney G, Einarsdottir IE, Bjornsson BT. Identification of two isoforms of Atlantic halibut insulin-like growth factor-I receptor genes and quantitative gene expression during metamorphosis. Comp Biochem Physiol B Biochem Mol Biol. 2007;147:395–401. doi: 10.1016/j.cbpb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Hildahl J, Sweeney G, Galay-Burgos M, Einarsdottir IE, Bjornsson BT. Cloning of Atlantic halibut growth hormone receptor genes and quantitative gene expression during metamorphosis. Gen Comp Endocrinol. 2007;151:143–152. doi: 10.1016/j.ygcen.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Douglas SE, Knickle LC, Kimball J, Reith ME. Comprehensive EST analysis of Atlantic halibut (Hippoglossus hippoglossus), a commercially relevant aquaculture species. BMC Genomics. 2007;8:144. doi: 10.1186/1471-2164-8-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE, Gallant JW, Bullerwell CE, Wolff C, Munholland J, Reith ME. Winter Flounder Expressed Sequence Tags: Establishment of an EST Database and Identification of Novel Fish Genes. Mar Biotechnol (NY) 1999;1:458–0464. doi: 10.1007/PL00011802. [DOI] [PubMed] [Google Scholar]
- Bai J, Solberg C, Fernandes JM, Johnston IA. Profiling of maternal and developmental-stage specific mRNA transcripts in Atlantic halibut Hippoglossus hippoglossus. Gene. 2007;386:202–210. doi: 10.1016/j.gene.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Sepulcre MP, Sarropoulou E, Kotoulas G, Meseguer J, Mulero V. Vibrio anguillarum evades the immune response of the bony fish sea bass (Dicentrarchus labrax L.) through the inhibition of leukocyte respiratory burst and down-regulation of apoptotic caspases. Mol Immunol. 2007;44:3751–3757. doi: 10.1016/j.molimm.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Ucko M, Colorni A, Diamant A. Nodavirus infections in Israeli mariculture. J Fish Dis. 2004;27:459–469. doi: 10.1111/j.1365-2761.2004.00565.x. [DOI] [PubMed] [Google Scholar]
- Poisa-Beiro L, Dios S, Montes A, Aranguren R, Figueras A, Novoa B. Nodavirus increases the expression of Mx and inflammatory cytokines in fish brain. Mol Immunol. 2008;45:218–225. doi: 10.1016/j.molimm.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Frerichs GN, Rodger HD, Peric Z. Cell culture isolation of piscine neuropathy nodavirus from juvenile sea bass, Dicentrarchus labrax. J Gen Virol. 1996;77:2067–2071. doi: 10.1099/0022-1317-77-9-2067. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- Thurston MIFD. Msatfinder: detection and characterization of microsatellites. 2005. http://www.genomics.ceh.ac.uk/msatfinder/
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J, Blaxter M. SimiTri – visualizing similarity relationships for groups of sequences. Bioinformatics. 2003;19:390–395. doi: 10.1093/bioinformatics/btf870. [DOI] [PubMed] [Google Scholar]
- Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000;10:2055–2061. doi: 10.1101/gr.GR-1325RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Tang R, Dodd A, Lai D, McNabb WC, Love DR. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim Biophys Sin (Shanghai) 2007;39:384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerslev HC, Pettersen EF, Jakobsen RA, Petersen CB, Wergeland HI. Expression profiling and validation of reference gene candidates in immune relevant tissues and cells from Atlantic salmon (Salmo salar L.) Mol Immunol. 2006;43:1194–1201. doi: 10.1016/j.molimm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Harbers M. The current status of cDNA cloning. Genomics. 2008;91:232–242. doi: 10.1016/j.ygeno.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Peatman E, Baoprasertkul P, Terhune J, Xu P, Nandi S, Kucuktas H, Li P, Wang S, Somridhivej B, Dunham R, Liu Z. Expression analysis of the acute phase response in channel catfish (Ictalurus punctatus) after infection with a Gram-negative bacterium. Dev Comp Immunol. 2007;31:1183–1196. doi: 10.1016/j.dci.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Peatman E, Terhune J, Baoprasertkul P, Xu P, Nandi S, Wang S, Somridhivej B, Kucuktas H, Li P, Dunham R, Liu Z. Microarray analysis of gene expression in the blue catfish liver reveals early activation of the MHC class I pathway after infection with Edwardsiella ictaluri. Mol Immunol. 2008;45:553–566. doi: 10.1016/j.molimm.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Stork M, Di Lorenzo M, Welch TJ, Crosa LM, Crosa JH. Plasmid-mediated iron uptake and virulence in Vibrio anguillarum. Plasmid. 2002;48:222–228. doi: 10.1016/S0147-619X(02)00111-7. [DOI] [PubMed] [Google Scholar]
- Stafford JL, Belosevic M. Transferrin and the innate immune response of fish: identification of a novel mechanism of macrophage activation. Dev Comp Immunol. 2003;27:539–554. doi: 10.1016/S0145-305X(02)00138-6. [DOI] [PubMed] [Google Scholar]
- Ganz T, Nemeth E. Regulation of iron acquisition and iron distribution in mammals. Biochim Biophys Acta. 2006;1763:690–699. doi: 10.1016/j.bbamcr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Rodrigues PN, Vazquez-Dorado S, Neves JV, Wilson JM. Dual function of fish hepcidin: response to experimental iron overload and bacterial infection in sea bass (Dicentrarchus labrax) Dev Comp Immunol. 2006;30:1156–1167. doi: 10.1016/j.dci.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Lauth X, Babon JJ, Stannard JA, Singh S, Nizet V, Carlberg JM, Ostland VE, Pennington MW, Norton RS, Westerman ME. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J Biol Chem. 2005;280:9272–9282. doi: 10.1074/jbc.M411154200. [DOI] [PubMed] [Google Scholar]
- Chen SL, Xu MY, Ji XS, Yu GC, Liu Y. Cloning, characterization, and expression analysis of hepcidin gene from red sea bream (Chrysophrys major) Antimicrob Agents Chemother. 2005;49:1608–1612. doi: 10.1128/AAC.49.4.1608-1612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B, Peatman E, Li P, He C, Liu Z. Catfish hepcidin gene is expressed in a wide range of tissues and exhibits tissue-specific upregulation after bacterial infection. Dev Comp Immunol. 2005;29:939–950. doi: 10.1016/j.dci.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Hu X, Camus AC, Aono S, Morrison EE, Dennis J, Nusbaum KE, Judd RL, Shi J. Channel catfish hepcidin expression in infection and anemia. Comp Immunol Microbiol Infect Dis. 2007;30:55–69. doi: 10.1016/j.cimid.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Park KC, Osborne JA, Tsoi SC, Brown LL, Johnson SC. Expressed sequence tags analysis of Atlantic halibut (Hippoglossus hippoglossus) liver, kidney and spleen tissues following vaccination against Vibrio anguillarum and Aeromonas salmonicida. Fish Shellfish Immunol. 2005;18:393–415. doi: 10.1016/j.fsi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Lin B, Chen S, Cao Z, Lin Y, Mo D, Zhang H, Gu J, Dong M, Liu Z, Xu A. Acute phase response in zebrafish upon Aeromonas salmonicida and Staphylococcus aureus infection: striking similarities and obvious differences with mammals. Mol Immunol. 2007;44:295–301. doi: 10.1016/j.molimm.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Gerwick L, Corley-Smith G, Bayne CJ. Gene transcript changes in individual rainbow trout livers following an inflammatory stimulus. Fish Shellfish Immunol. 2007;22:157–171. doi: 10.1016/j.fsi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Rossi F, Chini V, Cattaneo AG, Bernardini G, Terova G, Saroglia M, Gornati R. EST-based identification of genes expressed in perch (Perca fluviatilis, L.) Gene Expr. 2007;14:117–127. doi: 10.3727/105221607783417600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Kawazoe I, Nakaya M, Kikuchi K, Aida K, Watabe S. The novel sequences of major plasma apolipoproteins in the eel Anguilla japonica. Biochim Biophys Acta. 2001;1531:132–142. doi: 10.1016/s1388-1981(01)00099-3. [DOI] [PubMed] [Google Scholar]
- Concha MI, Smith VJ, Castro K, Bastias A, Romero A, Amthauer RJ. Apolipoproteins A-I and A-II are potentially important effectors of innate immunity in the teleost fish Cyprinus carpio. Eur J Biochem. 2004;271:2984–2990. doi: 10.1111/j.1432-1033.2004.04228.x. [DOI] [PubMed] [Google Scholar]
- Villarroel F, Bastias A, Casado A, Amthauer R, Concha MI. Apolipoprotein A-I, an antimicrobial protein in Oncorhynchus mykiss: evaluation of its expression in primary defence barriers and plasma levels in sick and healthy fish. Fish Shellfish Immunol. 2007;23:197–209. doi: 10.1016/j.fsi.2006.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 1. Catalogue of all unique sequences obtained from all constructed cDNA libraries in this study.
Appendix 2. EST-SSRs identified among EST sequences of all cDNA libraries constructed in this study.
Appendix 3. EST sequences (after clustering of all 3075) grouped into GO group Immune system process.
Appendix 4. A: CDNA sequences of hepicidin precursor showing putative isoforms B: CDNA showing microsatellite sequence with in silico SSR polymorphism after alignment of 8 sequences and C: sequences of cysteine-rich protein 1-I showing putative alternative splicing polyadenylations.
Appendix 5. Calculated R-values for contigs resulting from the EST sequencing of the cDNA library of spleen, liver and kidney infected with Nodavirus and Vibrio anguillarum.
Appendix 6. The number of genes for a given value or the test statistic R is plotted as a function of R. The data falling within 1 < R < 6 are decrees exponential curve, and decreasing exponentially with R. The slope is -1.081 with significance at the 5% level of 0.013 and is therefore not significantly different from -1 at 5% significance. When R > 6, the number of genes is above this exponential curve.