Abstract
Serengeti lions frequently experience viral outbreaks. In 1994, one-third of Serengeti lions died from canine distemper virus (CDV). Based on the limited epidemiological data available from this period, it has been unclear whether the 1994 outbreak was propagated by lion-to-lion transmission alone or involved multiple introductions from other sympatric carnivore species. More broadly, we do not know whether contacts between lions allow any pathogen with a relatively short infectious period to percolate through the population (i.e. reach epidemic proportions). We built one of the most realistic contact network models for a wildlife population to date, based on detailed behavioural and movement data from a long-term lion study population. The model allowed us to identify previously unrecognized biases in the sparse data from the 1994 outbreak and develop methods for judiciously inferring disease dynamics from typical wildlife samples. Our analysis of the model in light of the 1994 outbreak data strongly suggest that, although lions are sufficiently well connected to sustain epidemics of CDV-like diseases, the 1994 epidemic was fuelled by multiple spillovers from other carnivore species, such as jackals and hyenas.
Keywords: African lion, canine distemper virus, network model, percolation, Serengeti, wildlife disease model
1. Introduction
Effective management of wildlife diseases depends on reliable information about transmission patterns, and, at the very least, knowing which species participate in transmission as maintenance and non-maintenance hosts (Cleaveland et al. 2007). Maintenance populations steadily maintain disease for long periods of time and can serve as disease reservoirs (Haydon et al. 2002a). They typically exceed a critical community size in which a pathogen can persist indefinitely (Bartlett 1960). Non-maintenance populations can experience transient outbreaks, which are either large epidemics that reach a significant fraction of hosts or small outbreaks that die out after only a few infections. There are two distinct classes of non-maintenance host populations: percolating populations can (but do not always) sustain large epidemics while non-percolating populations cannot (Newman 2002; Meyers et al. 2005; Bansal et al. 2007; Davis et al. 2008). Whether or not a non-maintenance population can sustain an epidemic on its own depends, in part, on contact patterns among hosts. Populations with ample opportunities for pathogen transmission will lie above the epidemic threshold where large epidemics are possible, while more sparsely connected populations will lie below the epidemic threshold where outbreaks rapidly fizzle out.
Disease control strategies should prioritize maintenance hosts (Haydon et al. 2002a). However, for direct intervention in non-maintenance populations, it is critical to determine whether or not the population is percolating or non-percolating. If a non-percolating population experiences repeated introductions of diseases from sympatric populations, it may experience a series of small outbreaks that together take a large toll on the population. Multiple spillover outbreaks such as these may superficially resemble a single epidemic wave; however, the optimal control strategies for these two scenarios are quite different. In the spillover case, control measures should focus almost exclusively on preventing new introductions of disease, whereas in the epidemic case, strategies should also target transmission within the host population. Incorrectly targeting interventions can waste precious resources and cause harm to wildlife (e.g. culling of Asian civets for SARS (Li et al. 2005) and UK badgers for bTB (Donnelly et al. 2006)).
Mathematical models have historically provided important insights into disease dynamics and management (Anderson & May 1991; Ferguson et al. 2001; Haydon et al. 2002b; Keeling & Rohani 2008). Traditional disease models can, however, be misleading: mass-action models assume that populations are fully mixed, and lattice-based spatial models assume that all contacts are spatially proximate. Endangered species often live in groups and defend territories against conspecifics (e.g. lions in prides, wolves in packs), thus exhibiting population structure that is neither fully mixed nor geographically localized. Their populations show ‘community structure’ (Cleaveland et al. 2008) in which the groups are highly intraconnected and more loosely interconnected based on complex movement and behavioural patterns. Epidemiological data corroborate that social groups are often the critical units for disease transmission in wildlife (Altizer et al. 2003).
Contact network models allow us to explicitly consider the epidemiological consequences of complex patterns of host connectivity and have demonstrated that contact heterogeneity can fundamentally influence disease dynamics (Keeling 2005; Meyers et al. 2005; Bansal et al. 2006; Ferrari et al. 2006). However, network modelling often suffers from a paucity of good data on contact patterns, particularly for non-human hosts. Very few studies of free-ranging wildlife provide adequate empirical information to parametrize a network model (Cross et al. 2005); but the long-term dataset of the Serengeti Lion Project (SLP), which includes decades of daily observations of behaviour and movement, is a unique exception (Packer et al. 2005).
We used the SLP data to infer the contact network structure of an African lion (Panthera leo) population and built one of the most detailed, biologically realistic epidemiological network models of a wildlife population to date (but see Cross et al. (2005)). The model incorporates pride composition, movement of nomads (roaming lions) and contact rates between prides and nomads into a stochastic susceptible–exposed–infectious–recovered (SEIR) network framework. Disease-causing contacts between lions from different groups are assumed to include chases, fights, mating, close proximity and sequential and simultaneous feeding events. We then used this model to ask whether lions alone can sustain epidemics of contact-borne infectious diseases without repeated introductions from other species and, specifically, whether an observed 1994 canine distemper virus (CDV) epidemic could have been propagated exclusively by lion-to-lion transmission. The 1994 epidemic spread discontinuously throughout the study area, infected 17 of 18 study prides and took 35 weeks to spread across the entire ecosystem (Roelke-Parker et al. 1996; Cleaveland et al. 2007; Craft et al. 2008). Lions, hyenas (Crocuta crocuta), bat-eared foxes (Otocyon megalotis) and domestic dogs (Canis lupus familiaris) were all infected with the same strain of CDV (Haas et al. 1996; Roelke-Parker et al. 1996; Carpenter et al. 1998), thus supporting the possibility of cross-species disease transmission. Some studies have argued that the lions experienced repeated introductions from other carnivore species and that multihost epidemics could produce a pattern of disease spread similar to the 1994 CDV outbreak (Cleaveland et al. 2008; Craft et al. 2008). In contrast, Guiserix et al. (2007) claimed that, once CDV was introduced into the lion population, the lions probably sustained the outbreak themselves without subsequent transmission events from other species.
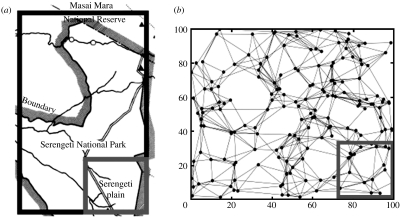
In addressing the plausibility of lion-to-lion transmission, we tackled larger issues about extrapolating disease dynamics from a geographically restricted study area (figure 1a) to a greater ecosystem. By taking samples from comparable areas or ‘subsets’ of our model ecosystems (figure 1b), we identified several unexpected discrepancies between sample data and ecosystem-wide disease dynamics, which are likely to arise in many wildlife disease field studies. In contrast to prior studies of the 1994 CDV outbreak (Guiserix et al. 2007), we analysed the field data in light of these discrepancies.
Figure 1.
The ecosystem and study area (subset) in both the Serengeti and the model. (a) The Serengeti ecosystem (outer rectangle, suitable lion habitat; inner square, SLP study area). Lions are essentially absent from outside the park, especially on the eastern and southern boundary of the SLP study area (Packer 1990; Ikanda & Packer 2008). (b) A simulated lion population based on the estimates of territory locations and adjacencies from SLP data (outer rectangle, model ecosystem; inner square, sampled subset). Nodes represent prides and edges indicate prides with adjacent territories.
2. Material and methods
(a) Modelling lion population structure
Lions live in gregarious groups (prides) composed of related females and their dependent offspring. Prides are territorial and infrequently contact their neighbours (Packer et al. 1992); inter-pride encounters can be deadly (Schaller 1972; McComb et al. 1993; Grinnell et al. 1995). When prides grow too large, young females split off and form a neighbouring pride (Pusey & Packer 1987) and are more tolerant of theirnon-pride relatives than lions from unrelated prides (VanderWaal et al. in press). Coalitions of males can reside in more than one pride (Bygott et al. 1979) and distribute their time between neighbouring prides (Schaller 1972). By contrast, nomads do not maintain a territory and move throughout the ecosystem (Schaller 1972). Lions from different social groups interact during territorial defence and at kills. Nomads can be seen as long distance disease dispersers, while shared males increase disease transmission between neighbouring prides. A quantitative summary of lion population structure is given in table 1 (Craft 2008).
Table 1.
Demographic, contact and epidemiological parameters for Serengeti lion prides. (For details about these parameters see Craft (2008).)
| estimated quantities | distributionsa | reference | |
|---|---|---|---|
| demographic parameters | |||
| A: area of ecosystem | 10 000 km2 | n.a. | Packer (1990) |
| NP: no. of prides in ecosystem | 180 | NP˜U(150,200)* | Packer (1990) |
| XP: pride sizes (no. of females and cubs over three months old) | XP∼gamma(k,θ) with θ=4.707, k=2.226; mean pride size=10.48 | SLP data 1991–1992 | |
| η: fraction of prides sharing males | 0.882 | η∼N(0.882,0.078) | SLP data 1992 |
| ς: rate at which coalitions switch prides | 0.25 switches/day | ς∼N(0.25,0.070) | B. Kissui (2003, unpubl. data) |
| Madj: territory adjacency model | b; mean number adjacent prides=7.36 | SLP data 1991–1992 | |
| Ψ: proportion of adjacent prides recently split from a common pride | 0.063 | Ψ∼N(0.063,0.021) | SLP data 1985–1987 |
| NN: no. of nomads | 180 | NN∼U(150–200)* | SLP data 1992 |
| XN: nomad group sizes | XN∼log-N (μ,σ) with μ=0.292, σ=0.446; mean group size=1.51 | SLP data 1992 | |
| Mnomad: nomad migration model horizontal (x) and vertical (y) displacements in km per two weeks are given by gamma distributions | dispx: gammax(kx,θx) with kx=0.382, θx=2.85c dispy: gammay(ky,θy) with ky=0.714, θy=1.743d | M. E. Craft (2006, unpubl. data) | |
| contact parameters | |||
| CP: rate of pride–pride contacts | 4.55 contacts/two weeks | CP∼N(4.55,0.573) | SLP data 1985–1987 |
| Mcontact: contact weighting model | e | SLP data 1985–1987 | |
| α: Mcontact intercept | 3.265 | α∼N(3.265,0.371) | SLP data 1985–1987 |
| βd: Mcontact network distance coefficient | 1.698 | βd∼N(1.698,0.264) | SLP data 1985–1987 |
| βs: Mcontact recent split coefficients | 0.696 | βd∼N(0.696,0.220) | SLP data 1985–1987 |
| CN: rate of pride–nomad group contacts | 7.136 contacts/two weeks | CN∼N(7.136,1.018) | SLP data 1985–1987 |
| G: pride group size during contact G′=log(G+1) | with , ; mean group size=3.65 | SLP data 1985–1987 | |
| epidemiological parameters | |||
| ϵ: incubation period (days) | ϵ∼Exp(λ) with λ=1/7 | n.a. | Appel (1987) |
| ι: infectious period (days) | ι∼Exp(λ) with λ=1/14 | n.a. | Greene & Appel (2006) |
| T: per contact transmission probability | free parameter | [0,0.3] | |
Confidence intervals marked with an asterisk (*) are best guesses made by M.E.C. and C.P.
SAB=the no. of prides located in the union of two regions: (1) the semicircle with straight-edge centered at A that runs through B, and (2) the semicircle with straight-edge centred at B that runs through A.
Horizontal displacement is the difference between randomly chosen right and left displacement, both distributed gammax(kx, θx).
Similarly, vertical displacement is the difference between randomly chosen variables, both distributed gammay(ky, θy).
wc(A,B) is the estimated probability that pride A will contact pride B per daylight hour of observation of A; dt(A,B) is the territory distance between the prides.
Our network model places NP=180 prides and NN=180 nomads at uniform random locations in a square region representing A=10 000 km2 of the high lion density area of the Serengeti (figure 1). The location of each pride is represented by a single point or centroid (geographical centre of its territory). Prides are assigned to be adjacent to one another according to the estimated adjacency model (Madj), and these adjacencies form the edges of the territory network (example in figure 1b). A fraction (Ψ) of adjacent pairs are randomly assigned to have recently ‘split off’ from one another. Each pride is given a size (XP) drawn from a best-fit gamma distribution. Contacts between prides occur at an average of Cp=4.55 contacts per two-week period per pride, as estimated from a study in which 16 lionesses were observed continuously for a total of 2213 hours (Packer et al. 1990). Contacts between pairs of prides occur stochastically at rates that are weighted by a logistic function of their territory distance and whether they recently split (Mcontact).
Coalitions of resident males and nomads are treated separately from prides of females and cubs. Male coalitions are represented as single units that increase connectivity between prides. Each territorial coalition belongs to either one or two prides; an estimated fraction η of all prides share their territorial coalition with one of their adjacent prides, and each remaining pride has a territorial coalition to itself. If a territorial coalition is associated with two prides, it will switch between prides with probability , where h is a small time step and ς is the rate at which territorial males switch prides.
Nomadic lions are given group sizes (XN) randomly generated from an estimated distribution and are assumed to migrate via a variance gamma process (Mnomad) (Madan et al. 1998; Glasserman 2004). Each group is initially assigned to the territory of a randomly selected pride, and at any point thereafter, resides in or around the territory of exactly one pride. In any small time step h, a group of nomads will migrate from the territory of its current pride (i) to that of another pride (j) with probability given by
where F( ) is the cumulative distribution function for displacement over a two-week period; dij is the distance between the centroids of territories i and j; α is the average pride territory width ; and ci is a normalizer. Nomads are assumed to contact their local pride at a uniform rate derived from the average rate of pride–nomad contacts per pride (CN).
When a pride contacts another pride or nomadic group, only a subset of the pride is actually involved in the interaction (G), and the number of lions involved is drawn randomly from an estimated distribution that depends on the size of that pride. Specifically, the log of group size increases approximately linearly with pride size (table 1). When nomads contact prides, all members of the nomadic group are assumed to be present.
(b) Modelling epidemiological dynamics
We model disease dynamics using a stochastic SEIR approach. Lions frequently contact all other lions in their nomadic group or pride (Packer et al. 1990), so we assume that any given pride or group of nomads moves through the four disease classes as a unit, as in a Levins-type patch model (Levins 1969; Hanski & Gilpin 1997). A group is considered exposed when its first member becomes infected; the group transitions stochastically from exposed to infectious at a rate of 1 of 7 per day and from infectious to recovered at a rate of 1 of 14 per day based on published estimates for domestic dogs. (Sequential infection among pride members and longer latent and infectious periods would probably slow the spread of disease, but not change the total number of infections.)
When an infected group (A) contacts a susceptible group (B), the probability of disease transmission is a function of the number of individuals involved in the interaction and a per-contact transmissibility parameter (T), given by
where pj and qk are the probabilities that the group sizes from A and B are j and k, respectively. This assumes that every lion in one group encounters every lion in the other group (recall that the expected size of a contact group is typically smaller than the size of the pride). When a susceptible coalition of territorial males resides with an infected pride, the coalition is immediately infected; and when an infected coalition of territorial males switches to a susceptible pride, it immediately infects the second pride.
Unless stated otherwise, the analysis is based on 200 simulated epidemics at 60 transmissibility values (T) between 0.0 and 0.3. For each run, a new lion population network was generated randomly, parameters were set to the values given in the estimated quantities column of table 1, and the first pride infected was chosen at random from either the subset or the population as a whole. We conducted a sensitivity analysis by running 200 replicate simulations at each of 50 transmissibility values using parameter values chosen randomly from the distributions given in the distributions column of table 1 (figure S2 in the electronic supplementary material). Statistical methods used for analysing centrality and network correlograms are described in the electronic supplementary material (text S1).
3. Results
We built an epidemiological network model, based on contact patterns within a lion population estimated from detailed SLP data (Craft 2008). The core of the model was a territory network in which prides were aggregated into single units (nodes), and edges were drawn between prides with adjacent territories, based on observed data. The territory distance between any two prides was then defined as the shortest path connecting their respective nodes. The prides contacted each other as a function of territory distance, and nomads migrated as a type of variance gamma process, contacting prides in their vicinity according to empirical estimates. In our stochastic SEIR simulations of CDV transmissions through the lion network, we monitored disease spread for the entire population and within a geographically restricted subset of 18 prides (figure 1b) resembling the study population (figure 1a).
(a) Edge effects
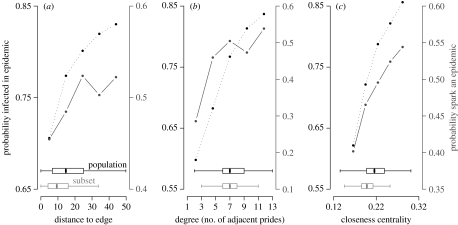
We use two network quantities to characterize the location of a pride in the overall network. The degree of a pride is the number of directly adjacent neighbouring prides; and the closeness centrality of a pride is the reciprocal of the pride's average minimum path length to all other prides in the network, which intuitively correlates with the likelihood that disease will reach the pride from elsewhere in the ecosystem. In our model, the subset prides were biased towards the physical and network boundaries of the ecosystem, having lower average distance to the ecosystem boundary, degree and closeness centrality than the population as a whole (figure 2, horizontal box plots).
Figure 2.
Epidemiological risk versus the geographical and network location of a pride. At T=0.10, (a) distance to edge, (b) degree and (c) closeness centrality all positively correlate with each other, with the probability that a pride will become infected during an epidemic (dotted lines, y-axis (left)) and that the pride will spark an epidemic if it is the first to be infected (solid lines, y-axis (right)). An epidemic is defined as any outbreak that reaches at least 50% of prides. Each graph is based on 1400 simulations. Box plots show the distributions of these values for the entire population (black) and the subset (grey), excluding outliers beyond the median ±1.5×IQR (means of entire population and subset, respectively: (a) 16.69 and 10.71; (b) 7.34 and 6.99; (c) 0.219 and 0.200). At higher transmissibilities most prides get infected, hence the edge effect is less pronounced.
We investigated the relationship between these metrics and the probability that a pride (i) will become infected during an epidemic and (ii) can spark a large-scale epidemic in an immunologically naive population (figure 2, dotted and solid lines, respectively). Both of these epidemiological risks increase with distance to edge, degree and centrality of a pride. To compare the relative importance of these factors on epidemiological risk, we performed a multivariate logistic regression, which indicated that that degree and closeness centrality account for the variation in the probability that a pride becomes infected (p<0.001; table S1 in the electronic supplementary material). In other words, the network structure may be the reason that distance to edge correlates with the probability of infection. These patterns explain the lower disease burden in the subset when compared with the overall population (figure 3b).
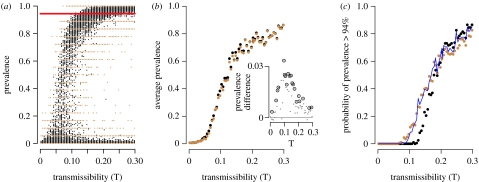
Figure 3.
The prevalence of CDV in the population and subset as a function of transmissibility. (a) Prevalence over a range of transmissibility values in the entire population of 180 prides (black) and the subset of 18 prides (brown). Each point represents the results of a single simulation. The red line is the prevalence observed in the 1994 CDV outbreak, as estimated from 18 prides in the SLP study area. (b) Average prevalence in the population (black) and subset (brown) over a range of transmissibility values. (Inset) Difference between overall prevalence and subset prevalence. Dotted circles are statistically significant (paired t-test, p<0.05). (c) Probability of a large outbreak (more than 94% prides infected) over a range of transmissibility values for the population (black) and subset (brown), compared to null expectations for the subset based on a hypergeometric model (blue line). The null values were generated by drawing a single hypergeometrically distributed random number for each simulation, with parameters N=180, n=18, m= total number of prides infected in the simulation. Probabilities were averaged across all simulations at each transmissibility value.
(b) Small sample size
During the 1994 CDV epidemic, 17 of the 18 prides (94%) in the 2000 km2 SLP study area became infected. Based on the edge effect, we initially assumed that the overall prevalence in the ecosystem should have been greater than or equal to this value. Instead, the model subset was more likely to experience an outbreak with greater than or equal to 94 per cent of prides infected than the overall population (figure 3c). This discrepancy has a simple combinatoric explanation; with only 18 prides monitored, observed prevalence levels could only take on a few discrete values (i.e. 17 prides, 94%; 18 prides, 100%). Consider a simple model in which (i) prides infected during an epidemic are randomly distributed throughout the ecosystem and (ii) subsets are random samples of 18 prides from the set of 180 prides. Then, subset prevalences should follow a hypergeometric distribution with parameters N=180, m=number infected prides overall and n=18. This null model closely predicts the observed differences (figure 3c, blue line), even though it ignores spatial clustering of disease and the contiguity of prides in the subset.
(c) Spatial scale
In the model, CDV epidemics typically spread wavelike across the ecosystem. Specifically, the shorter the distance between prides in the territory network, the higher the correlation between their times of infection (figure S1a in the electronic supplementary material). The wavelike pattern is more pronounced when measured by network distance rather than geographical distance (not shown). When viewed through the narrow lens of the subset, however, there is a lower correlation for directly adjacent prides and almost no correlation among more distant prides (figure S1a in the electronic supplementary material).
To compare correlograms across transmissibility values, we calculated correlations for directly adjacent prides (a network distance of one) and the slope of the correlogram for network distances between one and three (figure S1b,c in the electronic supplementary material). For outbreaks that originated in the subset (as observed in the 1994 epidemic), correlations between adjacent prides increased with transmissibility, but correlations were lower in the subset than across the entire population. The rate at which the correlations declined with network distance was similar in the subset and population and relatively uniform across all transmissibility values. Thus, figure S1a in the electronic supplementary material (which is based on T=0.1725) is representative of the spatio-temporal patterns observed across the entire range of transmissibilities, with little apparent correlation in the subset despite a wavelike spread overall.
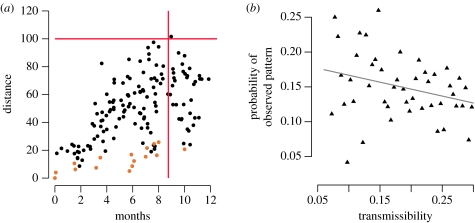
When we plotted distance from the first infected pride (pride zero) against the time of infection during a typical simulation (figure 4a), we observed relatively continuous expansion overall, but a discontinuous pattern within the subset fuelled by repeated introduction from elsewhere. For outbreaks initiated within the subset that infected at least 17 of the 18 prides, the probability of at least one reintroduction was 0.970 (s.d.=0.093); and the average number of subset prides with greater than or equal to 75 per cent chance of infection from outside the subset was 1.96 (s.d.=1.73). Thus, the spatial pattern of infections within the subset generally appeared patchy in the midst of a wavelike epidemic.
Figure 4.
Spatial spread of CDV. (a) Representative example of a simulated epidemic that began in the subset and swept through the entire population, occasionally returning to the subset. Points indicate the time and distance from first infection of each infected pride (brown, subset prides; black, other prides). Red lines represent the observation that the 1994 CDV epidemic took 35 weeks to reach 100 km from the study area. (b) The probability that the observed (1994) correlogram would arise from the model across transmissibilities. This probability is the fraction of simulations that lay both below the red line in figure S1b and above the red line in figure S1c in the electronic supplementary material. Line is the least-square linear regression line (p<0.05).
(d) Model versus data: did lions sustain the 1994 outbreak themselves?
We compared the predictions of our model with three empirical observations: the discontinuous spatial spread within the study area; 94 per cent prevalence within the study area; and the slow spread of the outbreak across the entire ecosystem. We also performed a full sensitivity analysis and found that the quantitative results were largely insensitive to uncertainty in the parameter values (table 1; figure S2 in the electronic supplementary material).
The model produced spatial patterns within the subset that were similar to the 1994 outbreak (figure 4; figure S3 and Video S1 in the electronic supplementary material). Disease appeared in clusters separated from each other in time and space. Across the entire range of transmissibility values, there is a 10–20 per cent chance that epidemics will appear at least as discontinuous as observed in 1994 (figure 4b). These probabilities are highest for low values of transmissibility, where transmission between neighbouring prides is rare, and thus the time of infection for adjacent prides is relatively uncorrelated. The model also predicts outbreaks with the observed pride prevalence, especially at higher transmissibility values (figure 3a), and predicts the observed rate of geographical spread at lower transmissibility values (figure S4 in the electronic supplementary material).
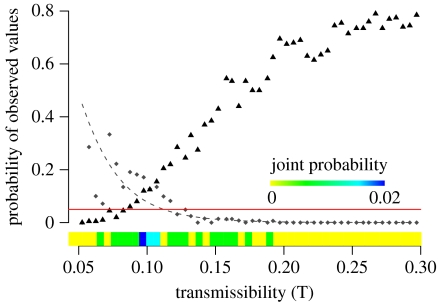
Although each of these individual patterns has a reasonable probability of occurring in a lion-to-lion epidemic, it is highly unlikely that all three could occur simultaneously (figure 5). The observed spatial spread and velocity are most likely to occur at low transmissibilities while the observed prevalence is most likely at higher transmissibilities. Only a minute fraction of simulations exhibited both the observed prevalence and velocity. The highest probability of observing both patterns is 0.02, occurring around T=0.095. We did not include the spatial analysis (figure 4; figure S1 in the electronic supplementary material) in this comparison because patchy outbreaks correlate with low velocity, and adding a spatial criterion would only reduce the joint probability further.
Figure 5.
Probability of observed epidemiological patterns in a simulated outbreak maintained solely by lion-to-lion transmission. The probability of the observed velocity (diamonds) is calculated as the fraction of simulations that took at least 35 weeks to reach 100 km. The dashed line shows the least squares regression on log–log transformed values. The probability of the observed prevalence is calculated as the fraction of simulations that infected at least 17 of the 18 prides in the subset (triangles). The red line at probability 0.05 indicates that there is a very limited range of transmissibility at which both patterns have at least a 5% of occurring. The joint probability is calculated as the fraction of simulations that exhibited both the observed velocity and prevalence.
Since the model failed to identify a range of transmission values that could have plausibly produced an epidemic that was both as large and as slow as the observed 1994 outbreak, we conclude that the assumption of strict lion-to-lion transmission must be incorrect. Thus, the actual transmission dynamics probably involved multiple introductions of disease to the lions from sympatric carnivore species.
4. Discussion
(a) Are Serengeti lions a percolating population for canine distemper virus?
Serengeti lions probably experience outbreaks of CDV and other directly transmitted viral diseases with similar infectious periods, such as feline calicivirus and parvovirus, every 4–12 years (Packer et al. 1999). Our model suggests that this population of lions is sufficiently well connected to sustain epidemics of CDV-like diseases on their own, i.e. it is a percolating population for viruses with short infectious periods. Even moderately contagious diseases (with probability of transmission per contact T ≈ 0.13) have at least a 5 per cent chance of producing an epidemic that reaches 95 per cent of all prides in the ecosystem (figure 3c); and this probability increases rapidly with transmissibility. If CDV is at least moderately infectious in lions, as suggested for domestic and wild carnivores (Appel 1987), then our model suggests that it has the potential to sweep through the entire population.
The 1994 CDV outbreak, however, was unlikely to have been maintained by lions alone. Across the entire range of transmissibility values, a strictly lion-to-lion epidemic could not have been both as extensive and as slow moving as observed in 1994. At low rates of transmissibility, disease can spread as slowly as in 1994 but not reach the observed prevalence; the reverse is true at high rates of transmissibility (figure 5).
The most plausible explanation for this discrepancy is the absence of additional carnivore species from our model. Lions commonly contact hyenas and jackals during simultaneous or sequential feeding events (Cleaveland et al. 2008), and a single CDV variant was found to be circulating in lions, hyenas, bat-eared foxes and domestic dogs during the 1994 outbreak (Haas et al. 1996; Roelke-Parker et al. 1996; Carpenter et al. 1998). Thus, there were repeated opportunities for CDV to be introduced into the lion population. Although this conclusion contradicts a recent analysis by Guiserix et al. (2007), it is consistent with the genetic analysis and supported by observations of sick jackals at the time of the epidemic (Roelke-Parker et al. 1996). Our model suggests that lions were a ‘non-percolating’ population for this CDV epidemic and experienced transient chains of infection that ‘spilled over’ from other species.
(b) Do disease dynamics scale?
Wildlife studies can be resource and time intensive; thus, biologists regularly extrapolate from subsets of larger populations. Ecologists recognize that natural processes can vary considerably with the spatial scale of the observation (Tilman & Kareiva 1997; O'Neil & King 1998) and thus use multiscale approaches to analyse complex ecological systems. Given the difficulty of observing wildlife disease outbreaks in real time, disease ecologists are typically forced to mine sparse data without regard to sampling or scaling issues (examples include Williams et al. 1988; Woodroffe et al. 1997; Packer et al. 1999; Leendertz et al. 2004; Haydon et al. 2006).
In this study, we identified three potential sources of error that are relevant to wildlife disease ecology. The first is an edge effect, or more generally, non-random sampling with respect to the epidemiological structure of the population. Directly transmitted diseases spread primarily during contacts between neighbours or neighbouring groups; and the pattern of such interactions gives rise to a contact network. The position of a group within the network, in conjunction with the overall network structure, determines its epidemiological risk (figure 2). The contact network for Serengeti lions is highly spatial, such that contact rates are highly correlated with the number of nearby prides. Thus, prides located closest to the border of the Serengeti National Park have the fewest contacts, on average. For this reason, estimates based on samples taken from the outskirts of the park (such as the SLP study area) would tend to underestimate the overall burden of disease in the Serengeti ecosystem. Note, however, that samples from a geographical boundary will not suffer from an edge effect if the population is sufficiently well mixed that contact rates are homogeneous throughout the ecosystem.
The frequency of an epidemic in the subset can also differ significantly from the overall population, simply because of variability associated with taking a small random sample from a large population. Just by chance, the sample proportion can deviate considerably from the population proportion; the sample proportion is limited to a discrete number of values (i.e. 17/18, 18/18…). In the 1994 CDV epidemic, 94 per cent of prides in the subset were infected. At relatively low transmissibilities (T∼0.1), almost no simulated epidemics reach an overall prevalence of 94 per cent, yet a sizeable fraction infect at least 94 per cent of subset prides. Thus, at moderate transmissibilities, where few, if any, epidemics cross the 94 per cent threshold, sampling variability alone can explain the higher vulnerability of the subset to large epidemics than the overall population.
The final complication arises when sampling from a smaller geographical scale than that of disease transmission. The SLP data from the 1994 CDV outbreak suggest non-wavelike, erratic spread of disease throughout the study area, which has been seen as evidence for repeated introduction from other species (Craft et al. 2008). Although we ultimately rejected the possibility that lions sustained the 1994 outbreak by themselves, it would have been incorrect to assume that the observed spatial spread necessarily implied a similar pattern across the entire ecosystem. While contacts primarily occur between neighbouring groups, lion prides occasionally contact distant prides and migrating nomads, which reduces the correlation between the distance and the timing of infection. When the probability of transmission is low, disease may initially reach only a few prides in a given area and later return to the same vicinity via longer distance contacts. In a population with exclusively local contacts, dynamics at a small scale will become much more wavelike and more closely resemble the large-scale dynamics. On the other hand, completely mixed populations will lack scale dependencies, because they lack spatial patterns altogether. This study demonstrates that wildlife populations may not fulfil assumptions of classical epidemiological models, such as the lattice or mass-action models, and an understanding of both network structure and sampling caveats should be considered when constructing disease models for wildlife populations.
Acknowledgments
The authors thank M. Anderson, B. Kissui, A. Mosser, A. Sorensina and C. Souther for raw data or help with data extraction. We thank O. Bjornstad for assistance with the ncf-package, and A. Dobson, S. Cleaveland, K. Hampson, D. Haydon, M. Kaare, T. Lembo and E. Ernest for discussion about carnivore disease in the Serengeti. We thank the Santa Fe Institute for providing a working visit for M.E.C. and L.A.M. This research was supported by NSF grants (DEB-0225453, DEB-0343960, DEB-079097, DEB-0749097, BE-0308486, EF-0225453 and DEB-0710070) with additional funding from Lincoln Park Zoo, Sigma Xi, the U of MN's Graduate School and EEB Department, and a grant from the James F. McDonnell Foundation to L.A.M.
Supplementary Material
See document
Animation of one simulated epidemic
References
- Altizer S., et al. Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu. Rev. Ecol. Evol. Syst. 2003;34:517–547. doi: 10.1146/annurev.ecolsys.34.030102.151725. [DOI] [Google Scholar]
- Anderson R.M., May R.M. Oxford University Press; Oxford, UK: 1991. Infectious diseases of humans: dynamics and control. [Google Scholar]
- Appel M. Canine distemper virus. In: Appel M.J.G., editor. Virus infections of carnivores. Elsevier Science; New York, NY: 1987. pp. 132–159. [Google Scholar]
- Bansal S., Pourbohloul B., Meyers L.A. A comparative analysis of influenza vaccination programs. PLoS Med. 2006;3:e387. doi: 10.1371/journal.pmed.0030387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S., Grenfell B.T., Meyers L.A. When individual behaviour matters: homogeneous and network models in epidemiology. J. R. Soc. Interface. 2007;4:879–891. doi: 10.1098/rsif.2007.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett M.S. The critical community size for measles in the United States. J. R. Stat. Soc. A-G. 1960;123:37–44. doi: 10.1098/rsif.2007.1100. [DOI] [Google Scholar]
- Bygott J.D., Bertram B.C.R., Hanby J.P. Male lions in large coalitions gain reproductive advantages. Nature. 1979;282:839–841. doi: 10.1038/282839a0. [DOI] [Google Scholar]
- Carpenter M.A., Appel M.J.G., Roelke-Parker M.E., Munson L., Hofer H., East M., O'Brien S.J. Genetic characterization of canine distemper virus in Serengeti carnivores. Vet. Immunol. Immunopathol. 1998;65:259–266. doi: 10.1016/S0165-2427(98)00159-7. [DOI] [PubMed] [Google Scholar]
- Cleaveland S., Mlengeya T., Kaare M., Haydon D., Lembo T., Laurenson M.K., Packer C. The conservation relevance of epidemiological research into carnivore viral diseases in the Serengeti. Conserv.Biol. 2007;21:612–622. doi: 10.1111/j.1523-1739.2007.00701.x. [DOI] [PubMed] [Google Scholar]
- Cleaveland S., Packer C., Hampson K., Kaare M., Kock R., Craft M., Lembo T., Mlengeya T., Dobson A. The multiple roles of infectious diseases in the Serengeti ecosystem. In: Sinclair A.R.E., Packer C., Mduma S., Fryxell J., editors. Serengeti III: human impacts on ecosystem dynamics. Chicago University Press; Chicago, IL: 2008. pp. 209–239. [Google Scholar]
- Craft, M. E. 2008 Predicting disease dynamics in African lion populations. PhD thesis, University of Minnesota.
- Craft M.E., Hawthorne P.L., Packer C., Dobson A.P. Dynamics of a multihost pathogen in a carnivore community. J. Anim. Ecol. 2008;77:1257–1264. doi: 10.1111/j.1365-2656.2008.01410.x. [DOI] [PubMed] [Google Scholar]
- Cross P.C., Lloyd-Smith J.O., Getz W.M. Disentangling association patterns in fission–fusion societies using African buffalo as an example. Anim. Behav. 2005;69:499–506. doi: 10.1016/j.anbehav.2004.08.006. [DOI] [Google Scholar]
- Davis S., Trapman P., Leirs H., Begon M., Heesterbeek J.A.P. The abundance threshold for plague as a critical percolation phenomenon. Nature. 2008;454:634–637. doi: 10.1038/nature07053. [DOI] [PubMed] [Google Scholar]
- Donnelly C.A., et al. Positive and negative effects of widespread badger culling on tuberculosis in cattle. Nature. 2006;439:843–846. doi: 10.1038/nature04454. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M., Donnelly C.A., Anderson R.M. Transmission intensity and impact of control policies on the foot and mouth epidemic in Great Britain. Nature. 2001;413:542–548. doi: 10.1038/35097116. [DOI] [PubMed] [Google Scholar]
- Ferrari M.J., Bansal S., Meyers L.A., Bjornstad O.N. Network frailty and the geometry of herd immunity. Proc. R. Soc. B. 2006;273:2743–2748. doi: 10.1098/rspb.2006.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasserman P. Springer; New York, NY: 2004. Monte Carlo methods in financial engineering. [Google Scholar]
- Greene C.E., Appel M.J. Canine distemper. In: Greene C.E., editor. Infectious diseases of the dog and cat. W.B. Saunders; Philadelphia, PA: 2006. pp. 25–27. [Google Scholar]
- Grinnell J., Packer C., Pusey A.E. Cooperation in male lions: kinship, reciprocity or mutualism? Anim. Behav. 1995;49:95–105. doi: 10.1016/0003-3472(95)80157-X. [DOI] [Google Scholar]
- Guiserix M., Bahi-Jaber N., Fouchet D., Sauvage F., Pontier D. The canine distemper epidemic in Serengeti: are lions victims of a new highly virulent canine distemper virus strain, or is pathogen circulation stochasticity to blame? J. R. Soc. Interface. 2007;4:1127–1134. doi: 10.1098/rsif.2007.0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas L., Hofer H., East M., Wohlsein P., Leiss B., Barrett T. Canine distemper virus infection in Serengeti spotted hyaenas. Vet. Microbiol. 1996;49:147–152. doi: 10.1016/0378-1135(95)00180-8. [DOI] [PubMed] [Google Scholar]
- Hanski I., Gilpin M.E. Academic Press; San Diego, CA: 1997. Metapopulation biology: ecology, genetics, and evolution. [Google Scholar]
- Haydon D.T., Cleaveland S., Taylor L.H., Laurenson M.K. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg. Infect. Dis. 2002a;8:1468–1473. doi: 10.3201/eid0812.010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon D.T., Laurenson M.K., Sillero-Zubiri C. Integrating epidemiology into population viability analysis: managing the risk posed by rabies and canine distemper to the Ethiopian wolf. Conserv. Biol. 2002b;16:1372–1385. doi: 10.1046/j.1523-1739.2002.00559.x. [DOI] [Google Scholar]
- Haydon D.T., et al. Low-coverage vaccination strategies for the conservation of endangered species. Nature. 2006;443:692–695. doi: 10.1038/nature05177. [DOI] [PubMed] [Google Scholar]
- Ikanda D., Packer C. Ritual vs. retaliatory killing of African lions in the Ngorongoro Conservation Area, Tanzania. Endanger. Species Res. 2008;6:67–74. doi: 10.3354/esr00120. [DOI] [Google Scholar]
- Keeling M. The implications of network structure for epidemic dynamics. Theor. Popul. Biol. 2005;67:1–8. doi: 10.1016/j.tpb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Keeling M.J., Rohani P. Princeton University Press; Princeton, NJ: 2008. Modeling infectious diseases in humans and animals. [Google Scholar]
- Leendertz F.H., Junglen S., Boesch C., Formenty P., Couacy-Hymann E., Courgnaud V., Pauli G., Ellerbrok H. High variety of different simian T-cell leukemia virus type 1 strains in chimpanzees (Pan troglodytes verus) of the Tai National Park, Cote d'Ivoire. J. Virol. 2004;78:4352–4356. doi: 10.1128/JVI.78.8.4352-4356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levins R. Some demographic and genetic consequences of environmental heterogeneity for biological control. Bull. Entomol. Soc. Am. 1969;15:237–240. [Google Scholar]
- Li W., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Madan D.B., Carr P.P., Chang E.C. The variance gamma process and option pricing. Eur. Finance Rev. 1998;2:79–105. doi: 10.1023/A:1009703431535. [DOI] [Google Scholar]
- McComb K., Pusey A., Packer C., Grinnell J. Female lions can identify potentially infanticidal males from their roars. Proc. R. Soc. B. 1993;252:59–64. doi: 10.1098/rspb.1993.0046. [DOI] [PubMed] [Google Scholar]
- Meyers L.A., Pourbohloul B., Newman M.E.J., Skowronski D.M., Brunham R.C. Network theory and SARS: predicting outbreak diversity. J. Theor. Biol. 2005;232:71–81. doi: 10.1016/j.jtbi.2004.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.E.J. Spread of epidemic disease on networks. Phys. Rev. E. 2002;66:016128. doi: 10.1103/PhysRevE.66.016128. [DOI] [PubMed] [Google Scholar]
- O'Neil R.V., King A.W. Homage to St Michael: or, why are there so many books on scale? In: Peterson D.L., Parker V.T., editors. Ecological scale. Columbia University Press; New York, NY: 1998. pp. 3–15. [Google Scholar]
- Packer, C. 1990 Serengeti lion survey: report to TANAPA, SWRI, MWEKA and the Wildlife Division.
- Packer C., Scheel D., Pusey A.E. Why lions form groups: food is not enough. Am. Nat. 1990;136:1–19. doi: 10.1086/285079. [DOI] [Google Scholar]
- Packer C., Lewis S., Pusey A.E. A comparative analysis of non-offspring nursing. Anim. Behav. 1992;43:265–281. doi: 10.1016/S0003-3472(05)80222-2. [DOI] [Google Scholar]
- Packer C., Altizer S., Appel M., Brown E., Martenson J., O'Brien S.J., Roelke-Parker M., Hofmann-Lehmann R., Lutz H. Viruses of the Serengeti: patterns of infection and mortality in African lions. J. Anim. Ecol. 1999;68:1161–1178. doi: 10.1046/j.1365-2656.1999.00360.x. [DOI] [Google Scholar]
- Packer C., Hilborn R., Mosser A., Kissui B., Borner M., Hopcraft G., Wilmshurst J., Mduma S., Sinclair A.R. Ecological change, group territoriality, and population dynamics in Serengeti lions. Science. 2005;307:390–393. doi: 10.1126/science.1105122. [DOI] [PubMed] [Google Scholar]
- Pusey A.E., Packer C. The evolution of sex-based dispersal in lions. Behaviour. 1987;101:275–310. doi: 10.1163/156853987X00026. [DOI] [Google Scholar]
- Roelke-Parker M.E., et al. A canine distemper virus epidemic in Serengeti lions (Panthera leo) Nature. 1996;379:441–445. doi: 10.1038/379441a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller G.B. University of Chicago Press; Chicago, IL: 1972. The Serengeti lion; a study of predator–prey relations. [Google Scholar]
- Tilman D., Kareiva P. 30 edn. Princeton University Press; Princeton, NJ: 1997. Spatial ecology: the role of space in population dynamics and interspecific interactions. [Google Scholar]
- VanderWaal, K., Mosser, A. & Packer, C. In press. Optimal group size, dispersal decisions and post-dispersal relationships in female African lions. Anim. Behav.
- Williams E., Thorne E., Appel M., Belitsky D. Canine distemper in black-footed ferrets (Mustela nigripes) from Wyoming. J. Wildl. Dis. 1988;24:385–398. doi: 10.7589/0090-3558-24.3.385. [DOI] [PubMed] [Google Scholar]
- Woodroffe, R., Ginsberg, J. R., Macdonald, D. W. and the IUCN/SSC canid specialist group 1997 The African wild dog–status survey and conservation action plan. Gland, Switzerland: IUCN.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See document
Animation of one simulated epidemic