Abstract
In addition to the well-characterized proteins that comprise the pre-replicative complex, recent studies suggest that chromatin structure plays an important role in DNA replication initiation. One of these chromatin factors is the histone acetyltransferase (HAT) Hbo1 which is unique among HAT enzymes in that it serves as a positive regulator of DNA replication. However, several of the basic properties of Hbo1 have not been previously examined, including its intrinsic catalytic activity, its molecular abundance in cells, and its pattern of expression in primary cancer cells. Here we show that recombinant Hbo1 can acetylate nucleosomal histone H4 in vitro, with a preference for lysines 5 and 12. Using semi-quantitative western blot analysis, we find that Hbo1 is approximately equimolar with the number of active replication origins in normal human fibroblasts but is an order of magnitude more abundant in both MCF7 and Saos-2 established cancer cell lines. Immunohistochemistry for Hbo1 in 11 primary human tumor types revealed strong Hbo1 protein expression in carcinomas of the testis, ovary, breast, stomach/esophagus, and bladder.
Keywords: DNA replication, chromatin, acetylation, proliferation, licensing
1 Introduction
DNA replication is a coordinated cell-cycle-dependent process that is highly regulated to ensure that the nuclear genome is duplicated once, and only once, per division cycle. The initiation of DNA replication is characterized by the sequential assembly of ORC, Cdc6, Cdt1, and Mcm2–7 on chromatin, beginning at telophase and continuing through G1 phase of the cell cycle, to form the pre-replicative complex (pre-RC). At the start of S phase, this assembled pre-RC is then further transformed into an active initiation complex through the actions of Cdks and other molecules (Arias and Walter, 2007; Diffley, 2004; Machida et al., 2005). Although many of the key proteins that participate in the structure and regulation of the pre-RC assembly are conserved throughout eukaryotes, any DNA sequence elements that might act in cis to specify DNA replication origins are apparently not conserved. For example, in budding yeast origins of DNA replication are specified by a well-defined autonomously replicating sequence (ARS) consensus. In contrast, higher eukaryotes appear to lack cis-regulatory elements within the most active region of the origin itself (Kalejta et al., 1998). Moreover, purified human ORC does not have a sequence-specific DNA binding activity and is capable of directing the initiation of replication from any DNA fragment in vitro, further suggesting that origin position is not specified solely by DNA sequence in vertebrate chromosomes (Vashee et al., 2003). Indeed, the ENCODE project, a pilot study for functionally analyzing 1% of the human genome, has revealed that replication timing is more closely correlated with histone modifications than it is with any DNA sequence motif (ENCODE Project Consortium, 2007).
Accumulating evidence suggest that chromatin structure may participate in providing the complex regulation necessary to specify replication origin activity (Tabancay and Forsburg, 2006). For example, the chromatin remodeling complex ISWI-ACF, is required for DNA replication of heterochromatin (Collins et al., 2002). In the case of histone modifications, histone acetylation positively regulates replication in Xenopus and in Drosophila follicle cells (Aggarwal and Calvi, 2004; Danis et al., 2004). Furthermore, replication timing, both at budding yeast origins and at the human β-globin origin, is also regulated by histone acetylation (Aparicio et al., 2004; Goren et al., 2008; Vogelauer et al., 2002). Finally, the histone deacetylase Sir2p negatively regulates pre-RC formation in budding yeast (Pappas et al., 2004).
We have been studying the histone acetyltransferase Hbo1, a member of the MYST family of HAT enzymes (Avvakumov and Cote, 2007). Hbo1 is the catalytic subunit of a multiprotein complex that includes the tumor suppressor proteins Ing4/5 and the regulatory short isoform of the Jade-1 protein (Avvakumov and Cote, 2007; Foy et al., 2008). Hbo1 interacts with Orc1 and Mcm2 (Burke et al., 2001; Iizuka and Stillman, 1999) and is required for DNA replication licensing in mammalian cells and in Xenopus oocyte extracts (Iizuka et al., 2006). Consistent with this role, the enzymatic activity of Hbo1 is activated in telophase by polo-like kinase 1 (Plk1) (Wu and Liu, 2008) and is regulated by cyclin dependent kinases (Zong et al., 2005). Importantly, Hbo1 is specifically detected at DNA replication origins by chromatin immunoprecipitation and may function as a coactivator of the licensing factor Cdt1 (Johmura et al., 2008a; Miotto and Struhl, 2008).
Hbo1 also appears to have important intra-S phase roles in replication as well (Doyon et al., 2006). We recently discovered that tumor suppressor p53 physically interacts with Hbo1 and inhibits its HAT activity in vitro and in cells (Iizuka et al., 2008). Indeed, we found that exogenous cell signals, such as hyperosmotic shock, inhibit the HAT activity of Hbo1, and the loading of Mcm2–7 into the pre-RC, through a p53-dependent pathway, suggesting that p53 and Hbo1 act in opposition to regulate replication licensing and cell fate in the face of cytotoxic stress. Because Hbo1 is a newly recognized player in these regulatory pathways several of its basic features remain unknown, including its intrinsic HAT activity, its abundance in cells, and its expression in primary human cancers. Here we report the results of studies designed to address these questions. These results will facilitate further studies of how Hbo1 affects chromatin structure, regulates DNA replication, and functions in growth control.
2 Materials and Methods
2.1 Recombinant protein and HAT assay
His-tagged Hbol (Iizuka et al., 2008) was expressed in bacteria BL21(DE3)-RIL (Strata-gene) and purified on heparin sepharose (Amersham Pharmacia) and Ni-NTA-agarose (Qiagen). HAT assays were performed as reported previously (Iizuka and Stillman, 1999), except for the use of chicken core histones in this study. HAT assays performed on nucleosomal substrates included 40 mM NaCl. Human Hatl holoenzyme, purified from 293 cells, was a gift from Alain Verreault. (Verreault et al., 1998).
2.2 Acetylation site mapping
Following acetylation of nucleosomes, histones were separated by reverse-HPLC on a SMRT system. Fractions corresponding to H4 were pooled, dried, and subjected to microsequencing after deblocking as described previously (Mizzen et al., 1999).
2.3 Adenoviruses
Information on the construction of the adenoviruses used in this study is available upon request. Replication-deficient recombinant viruses were created as described (Hardy et al., 1997) and stocks were maintained as reported previously (Nevins et al., 1997). Viruses were purified by cesium chloride density gradient centrifugation and used at a multiplicity of infection (m.o.i.) = 100.
2.4 Cell Lines, Antibody, and Immunoprecipitation
A549, MCF-7, and Saos-2 cell lines were purchased from the ATCC. HFF2/T cells are human foreskin fibroblasts immortalized by human telomerase reverse-transcriptase (Tatsumi et al., 2006). The anti-Hbo1 antibody used in these studies was prepared as described previously (Iizuka et al., 2006). Anti-acetylated histone H4 antibody was also described previously (Lin et al., 1989). Anti-α-tubulin and anti-H2B antibody were purchased from Sigma and Upstate Biotechnology, respectively. For immunoprecipitation, cells were lysed with buffer (20 mM Tris-Cl pH 8.0, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40) and the lysate was incubated with antibodies and with protein A-Sepharose, followed by extensive washing with the lysis buffer (Harlow and Lane, 1988).
2.5 Immunohistochemistry
Two independent tissue microarrays were screened for Hbo1 expression. The first panel (H.F.F.) contained carcinomas of the testis (N=8), breast (N=25), prostate (N=19), lung (N=24), ovary (N=20), kidney (N=13), liver (N=9), colon, (N=26), stomach/esophagus (N=13), pancreas (N=5), and bladder (N=6). A formalin-fixed, paraffin-embedded section of the tissue microarray was placed in citrate buffer (pH 6.0) and heated in a microwave oven for 20 min before application of a polyclonal antibody to Hbo1 (1:200 dilution). After incubation with the primary antibody, and addition of the biotinylated secondary antibody, avidin-biotin immunoperoxidase was applied. Diaminobenzidine was used as the chromogen. Sections were then counterstained with hematoxylin. Nuclear staining was graded for intensity (negative, weak, and strong).
The second tissue microarray (Y.T. and T.F.) contained malignant tumors of the testis (N=9), breast (N=24), prostate (N=1), lung (N=12), ovary (N=7), kidney (N=11), liver (N=7), colon (N=27), stomach/esophagus (N=41), pancreas (N=3), and bladder (N=12). A formalin-fixed paraffin-embedded section of the tissue microarray was immunostained with anti-Hbo1 antibody (1:25 dilution) by the use of a Histofine Simple Stain MAX-PO kit (Nichirei, Tokyo, Japan) according to the manufacturer’s instructions. Antigen retrieval was performed by microwave oven (20 min in citric acid buffer (pH 6.0)). Nuclear staining was evaluated as follows: negative, <10% of cells stained; 10–50% of cells stained; and >50% of cells stained.
3 Results and discussion
3.1 Hbo1 is a nucleosomal histone H4 acetyltransferase
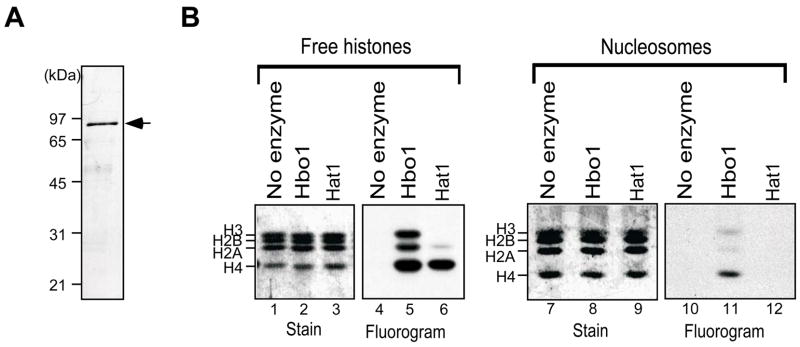
We and others have reported previously that protein complexes containing Hbo1 possess HAT activity (Doyon et al., 2006; Iizuka and Stillman, 1999). However, because initial attempts to detect HAT activity using recombinant Hbo1 were unsuccessful for both the human and Drosophila enzymes (Grienenberger et al., 2002; Iizuka and Stillman, 1999), it remained unclear whether the activities of those complexes were intrinsic to Hbo1. A formal possibility was that the activity associated with Hbo1-containing complexes is actually due to one or more other enzymes. Here we show that HAT activity is, in fact, intrinsic to Hbo1. His-tagged Hbo1 was expressed in bacteria and purified to homogeneity by affinity chromatography (Fig. 1A). When presented with a mixture of chicken free (non-nucleosomal) core histones, purified His-Hbo1 acetylated H4, H3, and to a lesser extent, H2A. This overall specificity is similar to that reported for other MYST family members such as Esa1 and Tip60, MOZ, and Sas3 (Champagne et al., 2001; Smith et al., 1998; Takechi and Nakayama, 1999; Yamamoto and Horikoshi, 1997). However, when presented with a nucleosomal substrate, His-Hbo1 acetylated H4 almost exclusively (Fig. 1B, lane 11). As a control for the quality of our nucleosomal substrate, we tested human Hat1 holoenzyme which is active on free histone H4 but inactive on nucleosomal H4 (Verreault et al., 1998). Hat1 acetylated free histone H4 but it did not display detectable HAT activity in nucleosomal assays (Fig. 1B, lane 12), indicating that the nucleosomal substrate remained intact during these reactions.
Figure 1. Hbo1 per se is a nucleosomal H4 acetyltransferase.
(A) Purified His-Hbo1 protein (arrow) was resolved by SDS-PAGE and stained with Coomassie Blue. The migration of molecular weight markers (kDa) is shown. (B) Chicken free core histone mixture (lanes 1–6) and chicken mononucleosomes (lanes 7–12) were incubated with [3H]-acetyl-CoA and no added enzyme (lanes 1, 4, 7, and 10) or plus purified His-Hbo1 (lanes 2, 5, 8, and 11) or plus human Hat1 (lanes 3, 6, 9, and 12). Reaction products were then resolved by 15% SDS-PAGE and detected by Coomassie Blue and fluorography.
3.2 Mapping of acetylation site on nucleosomal H4
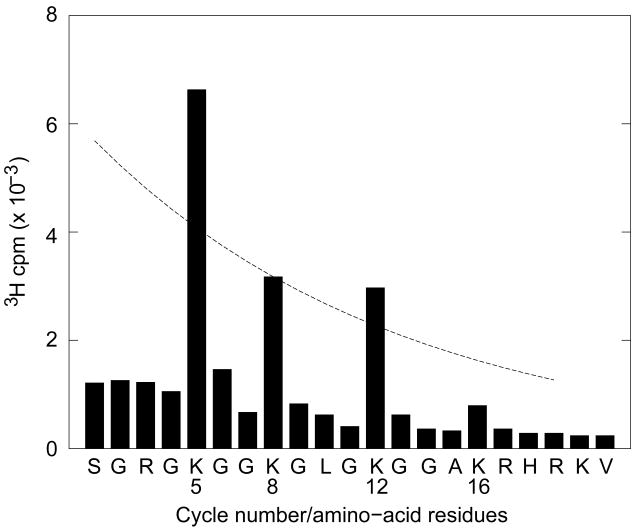
We mapped the amino acid residues in nucleosomal H4 that are acetylated by His-Hbo1 by microsequencing. As shown Fig. 2, acetylation was detected at lysines 5, 8, 12, and 16. After compensating for the effect of repetitive yield (~92–96%, data not shown) (Sobel et al., 1995), on the release of 3H cpm during sequential Edman degradation these data suggest that lysines 5 is preferentially modified, relative to the other sites, lysine 12 is preferentially modified relative to lysine 8, and lysine 16 is very poorly modified under these conditions. Preferential acetylation at lysines 5 and 12 of histone H4 correlates with the deposition of newly-synthesized histones Sobel et al. (1995), and this same specificity is displayed by the hHat1 holoenzyme (Verreault et al., 1998).
Figure 2. Acetylation site mapping of nucleosomal H4 by Hbo1.
(Chicken mononucleosomes were incubated with purified His-Hbo1 and [3H]-acetyl-CoA. Histones were then recovered from the reaction by acid extraction and purified by RP-HPLC. Purified H4 was then treated to deblock the α-amino group and microsequenced. The dashed line illustrates the experimental repetitive yield of 92% normalized to the recovery of [3H]-acetyl-Lys8.
3.3 Hbo1 is a sole HAT catalytic subunit in Hbo1 protein complexes
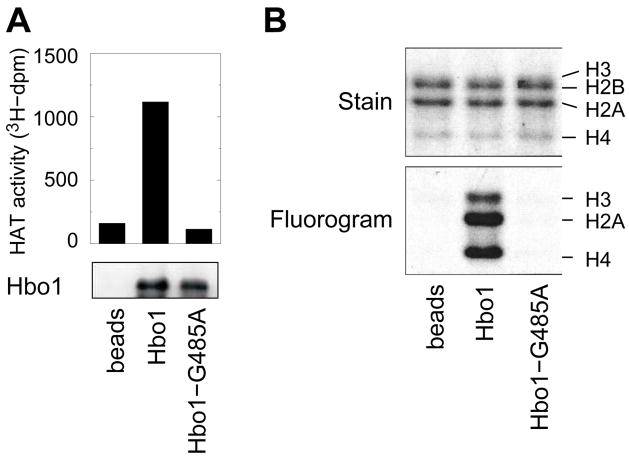
To ascertain whether Hbo1 was responsible for all of the HAT activity associated with anti-Hbo1 immunoprecipitates, we compared the HAT activity of complexes immunoprecipitated from A549 cells following infection with adenoviruses directing the expression of either wild-type Hbo1 or Hbo1-G485A, a catalytically inactive mutant (Iizuka et al., 2008). The absence of HAT activity associated with complexes containing Hbo1-G485A, compared to that of an equivalent amount of complexes containing wild type Hbo1 (Fig. 3A), suggests that Hbo1 is the sole HAT catalytic component of Hbo1 complexes. The immunoprecipitated wild type Hbo1 complexes preferentially acetylated free histone H4 and H2A, with less efficient activity against histone H3, while there was no detectable histone signal in the Hbo1-G485A reactions (Fig. 3B). Identical results were obtained when we tested HeLa, MCF7, and Saos-2 cells (data not shown). Two classes of Hbo1-containing complexes have been identified in HeLa cells by affinity purification: one containing ING4 and the other containing ING5 (Doyon et al., 2006). Interestingly, the ING5-TAP preparations contained both Hbo1 and MOZ/MORF as catalytic subunits. Thus, our results indicate that Hbo1 and MOZ/MORF are likely mutually exclusive in their association with ING5 or, alternatively, MOZ/MORF is inactive when present with Hbo1.
Figure 3. Hbo1 is the sole acetyltransferase in the Hbo1 protein complex.
A549 cells were infected with adenoviral vectors expressing either wild type (“Hbo1”) or catalytically dead (“Hbo1-G485A”) Hbo1 and immunoprecipitates were prepared using anti-Hbo1 antibody. The immunoprecipitates were assayed for HAT activity in reactions containing [3H]-acetylCoA and chicken core histones. (A) HAT activity. The incorporation of [3H]-acetate is plotted in the top panel. HAT reactions were spotted on P81 filters, washed, and assayed by scintillation counting. The recovery of Hbo1 in the immunoprecipitates, as assayed by western blot, is shown in the bottom panel. The left lane (“beads”) represents a mock immunoprecipitate prepared without primary antisera from cells infected with the wild type Hbo1 adenovirus. (B) Substrate specificity. HAT reactions were separated by electrophoresis on a 15% SDS-PAGE gel. The top panel shows the histone content of the reactions by Coomassie staining. The bottom panel shows the [3]-labeled histones visualized by fluorography.
3.4 Global H4 acetylation is regulated by Hbo1
In budding yeast, the HAT enzyme Esa1 regulates global acetylation of histone H4 (Clarke et al., 1999) and is involved in DNA repair (Bird et al., 2002). Tip60 is the closest mammalian counterpart of Esa1, with which it shares the presence of a chromodomain motif, evolutionarily conserved protein complex subunits, and a role in DNA repair (Doyon et al., 2004; Ikura et al., 2000; Murr et al., 2006; Yang, 2004). However, unlike Esa1, inhibiting Tip60 expression by siRNA did not alter global acetylation of histone H4. Instead, the global balance of histone H4 acetylation appears to be regulated by Hbo1, at least in 293T and MCF7 cells (Doyon et al., 2006). This unexpected result prompted us to examine whether Hbo1 might control global H4 acetylation using different methods to deplete Hbo1 expression. HeLa and MCF7 cells were infected with adenoviruses expressing either anti-sense Hbo1 RNA or catalytically-inactive Hbo1 mRNA, and extracts then were assayed by immunoblotting with an antibody (“penta”) directed against H4 N-terminal tail acetylated lysines (Fig. 4). The level of acetylation of H4 is reduced in both HeLa and MCF7 cells infected with the antisense-Hbo1 virus, as compared to those infected with the control virus (Fig. 4A). Similarly, the dominant-negative, catalytically-inactive mutant Hbo1, when overexpressed, causes reduced H4 acetylation in both HeLa and MCF7 cells (Fig. 4B). Because H2B expression is equivalent in these experiments (Fig. 4C), decreased H4 acetylation in these experiments is not due to a quantitative loss of core histones. Thus, while it is possible that Hbo1 may have currently unrecognized roles in DNA damage repair, our results suggest that the dual roles of Esa1 in global histone H4 acetylation and DNA double-strand break repair in budding yeast have become divided between two different MYST family enzyme complexes in mammalian cells during evolution.
Figure 4. Hbo1 regulates global H4 acetylation.
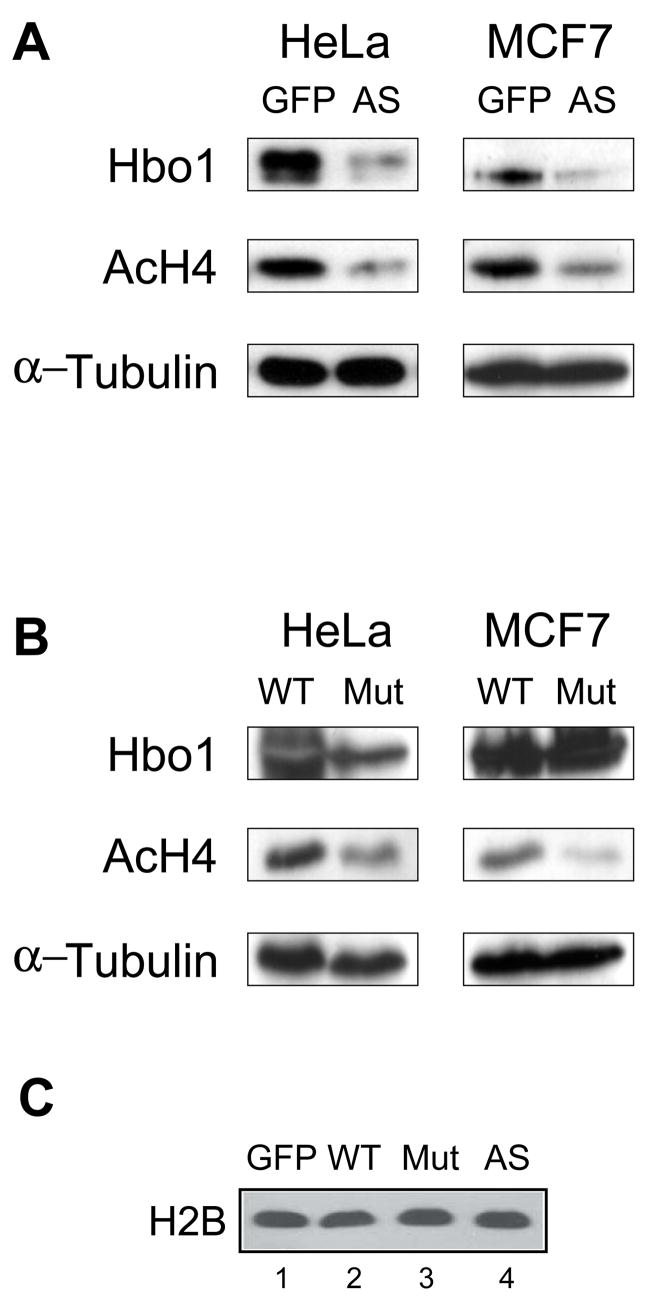
(A) Whole cell extracts infected with Ad-GFP (“GFP”) and Ad-antisense Hbo1 (“AS”) for 48 hr were analyzed by western blotting with antibodies to Hbo1, acetylated histone H4, and α-tubulin. (B) Whole cell extracts infected with Ad-Hbo1 (“WT”) and Ad-catalytically inactivated Hbo1 (“Mut”) for 48 hr were analyzed by western blotting with antibodies to Hbo1, acetylated histone H4, and α-tubulin. (C) Chromatin fractions of virus-infected MCF7 cells were separated on a protein gel, transferred to a membrane, were assayed for Histone H2B by immunoblotting to examine integrity of core histones.
3.5 Quantification of Hbo1 in cell lines
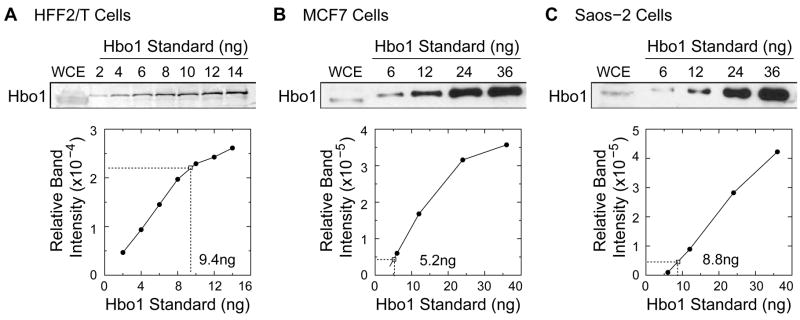
Endogenous Hbo1 protein is almost entirely chromatin-bound and interacts with ORC, Mcm, Cdt1, and geminin for DNA replication licensing (Burke et al., 2001; Doyon et al., 2006; Iizuka et al., 2006; Iizuka and Stillman, 1999; Johmura et al., 2008a; Miotto and Struhl, 2008). Thus, we were interested in determining whether there was sufficient Hbo1 in the cell to interact with all origins or whether Hbo1 might be limiting. To get an insight into this stoichiometry, we examined the amounts of Hbo1 by semi-quantitative immunoblotting experiments in asynchronous populations of normal human fibroblasts immortalized with telomerase (Tatsumi et al., 2006), and in two established human cancer cell lines, MCF7 and Saos-2. As shown in Fig. 5, cell lysates were compared against a standard of known amounts of purified His-tagged Hbo1 protein. From this comparison, we estimate that 1.0 × 106 normal HFF2/T cells contain approximately 3.2 ~ 6.4 ng of Hbo1 protein, in independent experiments, representing roughly 2.8 ~ 5.3 × 104 molecules per cell. The average length of a mammalian replicon is estimated to be 50–-250 kb (Huberman and Riggs, 1968). and so assuming 100 kb per replicon, there are approximately 0.9 ~ 1.8 molecules of Hbo1 per origin. These results suggest that in normal cells Hbo1 is in a critical concentration range and its activity may need to be modulated if it is to participate in both DNA replication licensing and in transcription regulation (Contzler et al., 2006; Foy et al., 2008; Georgiakaki et al., 2006; Grienenberger et al., 2002; Miotto et al., 2006; Sharma et al., 2000). In contrast, Hbo1 is an order of magnitude more abundant in the established human cancer cells examined. As shown in Fig. 5b and 5c, we estimate that 1.0 × 106 MCF7 and Saos-2 cells express approximately 26 ng and 44 ng of Hbo1 protein respectively, representing 19 ~ 32 × 104 molecules per cell. Therefore, in these cancer cell lines chromatin-bound Hbo1 is present at about 6 ~ 11 molecules per replication origin. This increase in Hbo1 concentration per cell has potential significance for both cellular regulation of replication licensing and response to transcription signaling pathways.
Figure 5. Estimation of the number of Hbo1 molecules in cells.
Whole cell extracts derived from 2.9 × 106 HFF2/T cells (A), 2 × 105 MCF7 cells (B), and 2 × 105 Saos-2 cells (C), along with defined amounts of recombinant Hbo1 proteins, were separated in protein gels, and transferred onto nitrocellulose membrane. The filter was probed with anti-Hbo1 antibody. Signal intensities of the bands were counted and the amounts of Hbo1 protein were estimated by reference to the standard plot.
3.6 Hbo1 is overexpressed in cancer tissues
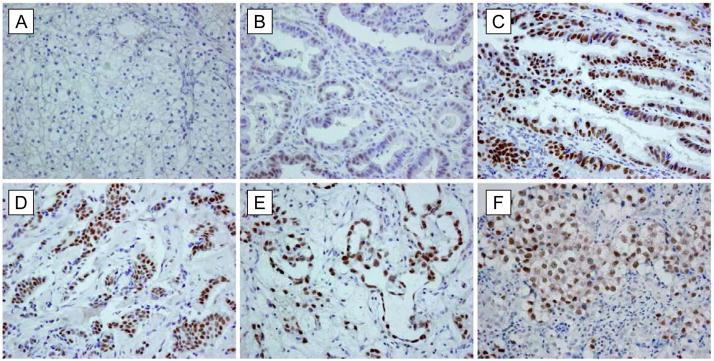
The increased Hbo1 concentrations in MCF7 and Saos-2 cell lines led us to ask whether Hbo1 might be highly expressed in primary human cancer tissues as well. The histone H4 lysine substrate specificity of Hbo1 (Fig. 2 and (Doyon et al., 2006)), parallels the H4 modification pattern observed in cancers, particularly the loss of H4 acetylation at K16 (Fraga et al., 2005). Other components of the pre-RC, such as Mcm, geminin, Cdc6, and Cdt1, are overexpressed in primary cancer tissues and they are potentially important markers of cellular proliferation (Freeman et al., 1999; Gonzalez et al., 2004; Karakaidos et al., 2004). Indeed, the Hbo1 gene maps to 17q21.3, a region where frequent allelic gains are found in breast cancers and this amplification is associated with a poor prognosis of clinical outcome (Clark et al., 2002; Hyman et al., 2002; Pollack et al., 2002). Therefore, we examined Hbo1 protein levels in primary cancers by immunohistochemistry. Initial assays of 168 tumor samples, comprising 11 cancer types, revealed a strong positive Hbo1 signal in one-third of the samples, particularly among testicular germ cell tumors, breast adenocarcinomas, and ovarian serous carcinomas (Table 1, Tissue Panel No. 1). We repeated these assays using a second independent panel of 154 tumor samples and assigned the Hbo1 expression to three classes: (1) negative Hbo1 staining; (2) Hbo1 positivity in at least 10% of tumor cells; and (3) Hbo1 positive in over 50% of tumor cells (Fig. 6). We observed positive staining in >50% of tumor cells in testicular, breast, ovarian, bladder, and stomach/esophageal carcinomas. For these five tumor types approximately 39% of the cases were positive, while 23% of these were positive in greater than 50% of the tumor cells (Table 1, Tissue Panel No. 2). Among normal tissues, testis and ovarian germ cells showed the most intense immunoreactivity (data not shown), in accord with previous Northern blot results (Iizuka and Stillman, 1999; Sharma et al., 2000). As Hbo1 modulates steroid hormone-dependent transcription (Georgiakaki et al., 2006; Sharma et al., 2000), its over-expression might functionally link DNA replication and hormone-dependent transcription affecting growth and cell proliferation. Hbo1 might be of importance in a proportion of breast carcinomas. However, over-expression of Hbo1 has been reported to have a modest inhibitory effect on H-ras induced transformation of NIH-3T3 cells (Johmura et al., 2008b). Thus, the importance of the role of Hbo1 in carcinogenesis of a variety of tumors awaits future investigation.
Table 1.
Hbo1 expression in primary cancers.
| Tissue Array No. 1 |
Tissue Array No. 2 |
||||||
|---|---|---|---|---|---|---|---|
| Tumor Type | Total Cases | No. Positive | % | Total Cases | No. >10% Positive a | No. >50% Positive b | % c |
| Testis | 8 | 6 | 75 | 9 | 6 | 4 | 44 |
| Breast | 25 | 7 | 28 | 24 | 5 | 4 | 17 |
| Ovary | 20 | 5 | 25 | 7 | 3 | 2 | 29 |
| Bladder | 6 | 1 | 17 | 12 | 5 | 2 | 17 |
| Stomach/esophagus | 13 | 1 | 8 | 41 | 17 | 10 | 24 |
| Lung | 24 | 1 | 4 | 12 | 0 | 0 | 0 |
| Prostate | 19 | 0 | 0 | 1 | 0 | 0 | 0 |
| Kidney | 13 | 0 | 0 | 11 | 0 | 0 | 0 |
| Liver | 9 | 0 | 0 | 7 | 0 | 0 | 0 |
| Colon | 26 | 0 | 0 | 27 | 2 | 1 | 0 |
| Pancreas | 5 | 0 | 0 | 3 | 0 | 0 | 0 |
Number of cases with at least 10% positive cells (includes the count of cases with >50% positive)
Number of cases with >50% positive cells
Percent of cases with >50% positive cells
Figure 6. Overexpression of Hbo1 in human primary tumors.
The immunohistochemical staining of Hbo1 is shown for the tumor samples listed in Table 1, tissue array number 2. (A) Renal cell carcinoma, negative; (B) gastric adenocarcinoma, staining in ~10% of tumor cells; (C) gastric adenocarcinoma, staining in >50% of tumor cells; (D) breast adenocarcinoma; (E) ovarian yolk sac tumor; and (F) seminoma.
3.7 Conclusion
Here we report the enzymatic properties of recombinant Hbo1 protein. The enzyme has intrinsic HAT activity on nucleosomal H4, with a substrate preference for lysines 5 and 12. Hbo1 appears to be the sole catalytic HAT subunit in its protein complexes, and it modulates the global levels of histone H4 acetylation in cells. The number of Hbo1 molecules per cell is approximately equal to the number of DNA replication origins in normal human fibroblasts, but it is an order of magnitude more abundant in MCF7 and Saos-2 established cancer cell lines. Furthermore, Hbo1 is overexpressed in a specific subset of human primary cancers. The above data are consistent with the hypothesis that Hbo1 HAT activity is a key regulator of DNA replication and cell proliferation.
Acknowledgments
We thank Alain Verreault for Hat1 enzyme. This research was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to M.F. and the National Institute of Health to M.M.S. (GM60444).
Abbreviations
- ACF
ATP-utilizing chromatin assembly and remodeling factor
- Cdc
cell division cycle protein
- Cdk
cyclin-dependent kinase
- Cdt1
protein encoded by Cdc10 dependent transcript 1 in Schizosaccharomyces pombe
- ENCODE
the Encyclopedia Of DNA Elements
- Esa1
essential SAS2-related acetyltransferase
- HAT
histone acetyltransferase
- Hbo1
histone acetyl transferase bound to origin recognition complex-1
- hEaf
human Esa1-associated Factor
- ING
inhibitor of growth
- JADE
gene for apoptosis and differentiation in epithelia
- ISWI
Imitation Switch
- MCM
minichromosome maintenance
- MYST
gene family named according to founding members MOZ, Ybf2/Sas3, Sas2 and TIP60
- MORF
MOZ-related factor
- MOZ
monocytic leukemia zinc finger protein
- pre-RC
pre-replicative complex
- ORC
origin recognition complex
- Sir
silent information regulator
- Sas
something about silencing
- Tip60
Tat interacting protein 60 kDa
- Ybf2
synonymous with yeast Sas3
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal B, Calvi B. Chromatin regulates origin activity in Drosophila folliclecells. Nature. 2004;430:372–376. doi: 10.1038/nature02694. [DOI] [PubMed] [Google Scholar]
- Aparicio J, Viggiani C, Gibson D, Aparicio O. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4769–4780. doi: 10.1128/MCB.24.11.4769-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiplemechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and theirintimate links to cancer. Oncogene. 2007;26:5395–407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–5. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Burke TW, Cook JG, Asano M, Nevins JR. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem. 2001;276:15397–408. doi: 10.1074/jbc.M011556200. [DOI] [PubMed] [Google Scholar]
- Champagne N, Pelletier N, Yang XJ. The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene. 2001;20:404–9. doi: 10.1038/sj.onc.1204114. [DOI] [PubMed] [Google Scholar]
- Clark J, Edwards S, John M, Flohr P, Gordon T, Maillard K, et al. Identification of amplified and expressed genes in breast cancer by comparative hybridization onto microarrays of randomly selected cDNA clones. Genes Chromosomes Cancer. 2002;34:104–14. doi: 10.1002/gcc.10039. [DOI] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–26. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N, Poot RA, Kukimoto I, Garcia-Jimenez C, Dellaire G, Varga-Weisz PD. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–32. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- Contzler R, Regamey A, Favre B, Roger T, Hohl D, Huber M. Histone acetyltransferase HBO1 inhibits NF-kappaB activity by coactivator sequestration. Biochem Biophys Res Commun. 2006;350:208–13. doi: 10.1016/j.bbrc.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Danis E, Brodolin K, Menut S, Maiorano D, Girard-Reydet C, Mechali M. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol. 2004;6:721–30. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–86. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry A, Côté V, Selleck W, et al. ING Tumor Suppressor Proteins Are Critical Regulators of Chromatin Acetylation Required for Genome Expression and Perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Identification and analysis of functional elements in1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, et al. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem. 2008;283:28817–26. doi: 10.1074/jbc.M801407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Freeman A, Morris LS, Mills AD, Stoeber K, Laskey RA, Williams GH, et al. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res. 1999;5:2121–32. [PubMed] [Google Scholar]
- Georgiakaki M, Chabbert-Buffet N, Dasen B, Meduri G, Wenk S, Rajhi L, et al. Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol Endocrinol. 2006;20:2122–40. doi: 10.1210/me.2005-0149. [DOI] [PubMed] [Google Scholar]
- Gonzalez MA, Tachibana KE, Chin SF, Callagy G, Madine MA, Vowler SL, et al. Geminin predicts adverse clinical outcome in breast cancer by reflecting cell-cycle progression. J Pathol. 2004;204:121–30. doi: 10.1002/path.1625. [DOI] [PubMed] [Google Scholar]
- Goren A, Tabib A, Hecht M, Cedar H. DNA replication timing of the human beta-globin domain is controlled by histone modification at the origin. Genes Dev. 2008;22:1319–24. doi: 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger A, Miotto B, Sagnier T, Cavalli G, Schramke V, Geli V, et al. The MYST domain acetyltransferase Chameau functions in epigenetic mechanisms of transcriptional repression. Curr Biol. 2002;12:762–6. doi: 10.1016/s0960-9822(02)00814-x. [DOI] [PubMed] [Google Scholar]
- Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps ML. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71:1842–9. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual Manual. Cold Spring Harbor Press; Cold Spring Harbor, N.Y.: 1988. [Google Scholar]
- Huberman JA, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–41. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–5. [PubMed] [Google Scholar]
- Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol. 2006;26:1098–108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Sarmento OF, Sekiya T, Scrable H, Allis CD, Smith MM. Hbo1 Links p53-dependent stress signaling to DNA replication licensing. Mol Cell Biol. 2008;28:140–53. doi: 10.1128/MCB.00662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Johmura Y, Osada S, Nishizuka M, Imagawa M. FAD24 acts in concert with histone acetyltransferase HBO1 to promote adipogenesis by controlling DNA replication. J Biol Chem. 2008a;283:2265–74. doi: 10.1074/jbc.M707880200. [DOI] [PubMed] [Google Scholar]
- Johmura Y, Suzuki M, Osada S, Nishizuka M, Imagawa M. FAD24, a regulator of adipogenesis and DNA replication, inhibits H-RAS-mediated transformation by repressing NF-kappaB activity. Biochem Biophys Res Commun. 2008b;369:464–70. doi: 10.1016/j.bbrc.2008.02.036. [DOI] [PubMed] [Google Scholar]
- Kalejta RF, Li X, Mesner LD, Dijkwel PA, Lin HB, Hamlin JL. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol Cell. 1998;2:797–806. doi: 10.1016/s1097-2765(00)80294-4. [DOI] [PubMed] [Google Scholar]
- Karakaidos P, Taraviras S, Vassiliou LV, Zacharatos P, Kastrinakis NG, Kougiou D, et al. Overexpression of the replication licensing regulators hCdt1 and hCdc6 characterizes a subset of non-small-cell lung carcinomas: synergistic effect with mutant p53 on tumor growth and chromosomal instability–evidence of E2F-1 transcriptional control over hCdt1. Am J Pathol. 2004;165:1351–65. doi: 10.1016/S0002-9440(10)63393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Leone JW, Cook RG, Allis CD. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989;108:1577–88. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y, Hamlin J, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Miotto B, Sagnier T, Berenger H, Bohmann D, Pradel J, Graba Y. Chameau HAT and DRpd3 HDAC function as antagonistic cofactors of JNK/AP-1-dependent transcription during Drosophila metamorphosis. Genes Dev. 2006;20:101–12. doi: 10.1101/gad.359506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008;22:2633–8. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen CA, Brownell JE, Cook RG, Allis CD. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol. 1999;304:675–96. doi: 10.1016/s0076-6879(99)04041-0. [DOI] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Nevins JR, DeGregori J, Jakoi L, Leone G. Functional analysis of E2F transcription factor. Methods Enzymol. 1997;283:205–19. doi: 10.1016/s0076-6879(97)83017-0. [DOI] [PubMed] [Google Scholar]
- Pappas D, Jr, Frisch R, Weinreich M. The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev. 2004;18:769–781. doi: 10.1101/gad.1173204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–8. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Zarnegar M, Li X, Lim B, Sun Z. Androgen receptor interacts with a novel MYST protein, HBO1. J Biol Chem. 2000;275:35200–8. doi: 10.1074/jbc.M004838200. [DOI] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, et al. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci U S A. 1998;95:3561–5. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–41. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabancay AP, Jr, Forsburg SL. Eukaryotic DNA replication in a chromatin context. Curr Top Dev Biol. 2006;76:129–814. doi: 10.1016/S0070-2153(06)76005-7. [DOI] [PubMed] [Google Scholar]
- Takechi S, Nakayama T. Sas3 is a histone acetyltransferase and requires a zinc finger motif. Biochem Biophys Res Commun. 1999;266:405–10. doi: 10.1006/bbrc.1999.1836. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Sugimoto N, Yugawa T, Narisawa-Saito M, Kiyono T, Fujita M. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J Cell Sci. 2006;119:3128–40. doi: 10.1242/jcs.03031. [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer B, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Wu ZQ, Liu X. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc Natl Acad Sci U S A. 2008;105:1919–24. doi: 10.1073/pnas.0712063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–8. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–76. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Li Z, Liu L, Hong Y, Yun X, Jiang J, et al. Cyclin-dependent kinase 11(p58) interacts with HBO1 and enhances its histone acetyltransferase activity. FEBS Lett. 2005;579:3579–88. doi: 10.1016/j.febslet.2005.05.039. [DOI] [PubMed] [Google Scholar]