Summary
Retrotransposons and repetitive DNA elements in eukaryotes are silenced by small RNA-directed heterochromatin formation. In Arabidopsis, this process involves 24 nt siRNAs that bind to ARGONAUTE4 (AGO4) and facilitate the targeting of complementary loci1,2 via unknown mechanisms. Nuclear RNA Polymerase V is an RNA silencing enzyme recently shown to generate noncoding transcripts at loci silenced by 24nt siRNAs3. We show that AGO4 physically interacts with these Pol V transcripts and is thereby recruited to the corresponding chromatin. We further show that DEFECTIVE IN MERISTEM SILENCING3 (DMS3), a Structural Maintenance of Chromosomes (SMC) hinge-domain protein4, functions in the assembly of Pol V transcription initiation or elongation complexes. Collectively, our data suggest that AGO4 is guided to target loci through base-pairing of associated siRNAs with nascent Pol V transcripts.
Arabidopsis Pol V, AGO45, DMS34 and the putative chromatin remodeller, DRD16 function in the silencing of siRNA-homologous loci at one or more steps downstream of siRNA biogenesis3,7–10. Recently, we showed that DRD1 facilitates Pol V transcription of noncoding RNAs at target loci, revealing a functional relationship between these two activities3. However, the functional relationships, if any between AGO4, DMS3 and Pol V transcription are unclear.
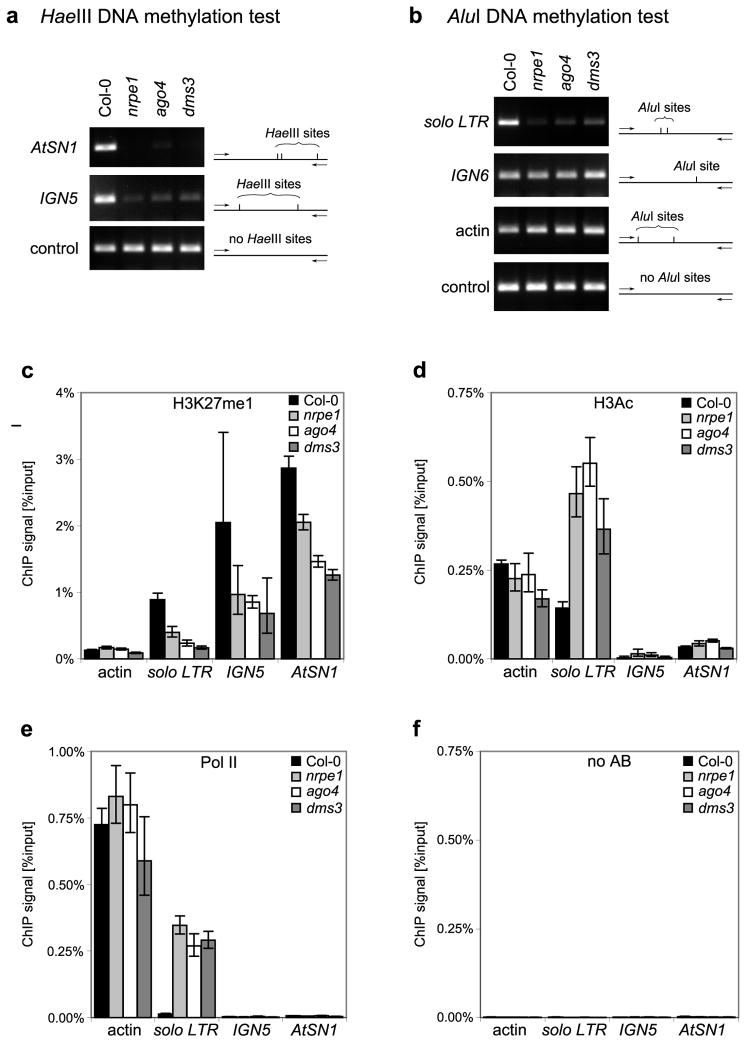
Mutations disrupting NRPE1 (the Pol V largest subunit), AGO4 or DMS3 cause similar losses of RNA-directed DNA methylation at AtSN1 retrotransposons, INTERGENIC REGION 5 (IGN5) and a retroelement solo LTR locus (Figs. 1A and 1B). Likewise, histone H3 lysine 27 monomethylation (H3K27me1), a characteristic of silenced heterochromatin, is reduced at these loci in nrpe1, ago4 and dms3 mutants compared to wild-type plants (ecotype Col-0) (Fig 1C). These results indicate that Pol V, AGO4 and DMS3 collaborate in the establishment of repressive chromatin modifications. At the solo LTR locus transcribed by RNA polymerase II (Pol II), chromatin immunoprecipitation (ChIP) shows that levels of diacetylated histone H3 (H3Ac2; acetylated on lysines 9 and 14), a mark of active chromatin, increases in the mutants (Fig 1D), coincident with increased Pol II occupancy of the locus (Fig. 1E). At IGN5 and AtSN1, which lack associated Pol II (Fig. 1E), no increase in histone H3 acetylation is observed in the mutants (Fig. 1D). AtSN1 elements are thought to be transcribed by Pol III, therefore differences in H3 acetylation at the solo LTR and AtSN1 loci may reflect the different polymerases involved.
Figure 1. Pol V, AGO4 and DMS3 work non-redundantly in heterochromatin formation.
(A, B) DNA methylation analysis at the AtSN1, IGN5 and solo LTR loci in nrpe1, ago4 and dms3 mutants. Genomic DNA was digested with HaeIII (A) or AluI (B) methylation-sensitive restriction endonucleases followed by PCR. Sequences lacking HaeIII sites (actin 2; panel A) or AluI sites (IGN5, panel B) served as controls to show that equivalent amounts of DNA were tested in all reactions.
(C–D) ChIP analysis of H3K27me1 (C) and H3Ac2 (D) levels in nrpe1, ago4 and dms3 mutants. Histograms show means +/− standard deviations obtained from three independent amplifications.
(E) ChIP analysis of Pol II binding to chromatin in nrpe1, ago4 and dms3 mutants. Histograms show means +/− standard deviations obtained from three independent amplifications.
(F) Control ChIP reactions performed in the absence of antibody reveal background signal levels.
AGO4 and Pol V colocalize in a nucleolus-associated Cajal body7,8 that is distant from the target loci subjected to siRNA-mediated silencing. These observations have suggested that AGO4-siRNA complexes might guide Pol V to the target loci7,8. To test this hypothesis, we asked whether production of Pol V transcripts is AGO4-dependent. At intergenic regions IGN5 and IGN63, Pol V transcripts are lost or substantially reduced in the Pol V mutant (nrpe1) but not in the ago4 mutant (Fig. 2A); in fact, IGN5 transcript levels increase by ~50% in ago4 (Fig. 2B). This increase in transcript levels is Pol V-dependent, as shown by analysis of the nrpe1 ago4 double mutant (Fig. 2A). In the rdr2 (rna-dependent rna polymerase2) mutant, which abolishes 24 nt siRNA biogenesis11,12, or in an rdr2 ago 4 double mutant, Pol V transcript levels are unaffected compared to wild-type (Col-0) plants. We conclude that AGO4-siRNA complexes are dispensable for Pol V transcription at target loci, arguing against the hypothesis that AGO4-siRNA complexes guide Pol V to target loci. The functional significance of AGO4 and Pol V colocalization in Cajal bodies is unclear but could reflect independent protein processing/assembly or storage functions that are unrelated to RISC (RNA-induced silencing complex) assembly.
Figure 2. AGO4 is not required for Pol V transcription.
(A) Strand specific RT-PCR of Pol V transcription at IGN5 and IGN6 in ago4 and rdr2 mutants as well as nrpe1 ago4, and rdr2 ago4 double mutants. Wt sibling is a wild type sibling of the ago4 mutant identified in a segregating family. Actin RT-PCR products and ethidium bromide-stained rRNAs resolved by agarose gel electrophoresis serve as loading controls. To control for background DNA contamination, a reaction using IGN5 top strand primers but no reverse transcriptase (no RT) was performed. No RNA (0 μg) controls are provided for all primer pairs.
(B) Densitometric analysis of RT-PCR data for the ago4 mutant presented in panel A. The histogram provides mean band intensities relative to wild type Col-0, +/− standard deviations obtained from three independent experiments.
To test an alternative hypothesis, that AGO4-siRNA complexes are recruited to chromatin in a Pol V-dependent manner, we assayed AGO4 associations with target loci using ChIP (Fig. 3). In wild-type (Col-0) plants, solo LTR, IGN5, AtSN1 and IGN6 loci are all enriched upon AGO4-ChIP, whereas only background levels are observed in ago4 or nrpe1 mutants or in control ChIP reactions lacking anti-AGO4 antibody (Fig. 3A). These findings indicate that AGO4 interacts with target locus chromatin, and does so in a Pol V-dependent manner. AGO4-chromatin interactions are not diminished by mutation of DRM2 (Fig. 3A), the de novo DNA methyltransferase that carries out siRNA and AGO4-dependent cytosine methylation13,14. Collectively, these data indicate that Pol V, but not pre-existing DNA methylation, is required to recruit AGO4 to chromatin.
Figure 3. Pol V transcription is necessary for AGO4-chromatin interactions.

(A) ChIP data showing AGO4 binding to chromatin at solo LTR, IGN5, AtSN1 and IGN6 loci in ago4, nrpe1 and drm2 mutants. DNA purified from input chromatin samples, chromatin subjected to the immunoprecipitation procedure in the absence of antibody (no AB) and chromatin immunoprecipitated using anti-AGO4 antibody (αAGO4) was amplified by PCR using locus-specific primers. Primers amplifying the Actin 2 locus served as an internal control.
(B) ChIP data showing AGO4 binding to chromatin at solo LTR, IGN5, AtSN1 and IGN6 loci in nrpe1 mutant, nrpe1 mutant transformed with a wild type NRPE1 transgene (NRPE1 wt) and nrpe1 mutant transformed with an NRPE1 active site mutant transgene (NRPE1 ASM).
(C) Immunoblot detection of AGO4 in protein extracts of wild-type (Col-0), ago4, nrpe1, or nrpe1 transformed with either a wild type NRPE1 transgene (NRPE1 wt) or an NRPE1 active site mutant transgene (NRPE1 ASM). Ponceau S staining revealed equal loading of lanes. 100% and 50% sample loadings demonstrate that the assay is semi-quantitative.
To test whether Pol V enzymatic activity is required for AGO4 binding to chromatin we examined AGO4-chromatin associations in nrpe1 mutants that had been transformed with either a full-length, wild-type NRPE1 transgene or an equivalent transgene bearing point mutations within the Metal A motif of the active site (NPRE1 ASM transgene). The active site point mutations do not affect NRPE1 stability or its association with the second-largest subunit but eliminate Pol V transcripts and Pol V biological activity3,15. Whereas the wild-type NRPE1 genomic transgene (NRPE1 wt) restored AGO4 interaction with the solo LTR, IGN5, AtSN1 and IGN6 loci in the nrpe1 mutant background (Fig. 3B), the active-site mutant (NRPE1 ASM) failed to do so. Immunoblotting ruled out the trivial explanation that AGO4 protein levels might be differentially affected by the nrpe1 mutation or the NRPE1 transgenes (Fig. 3C) and also demonstrates that the antibody specifically recognizes AGO4, which is absent in the ago4 mutant. Collectively, the data indicate that Pol V transcriptional activity is required to recruit AGO4 to chromatin.
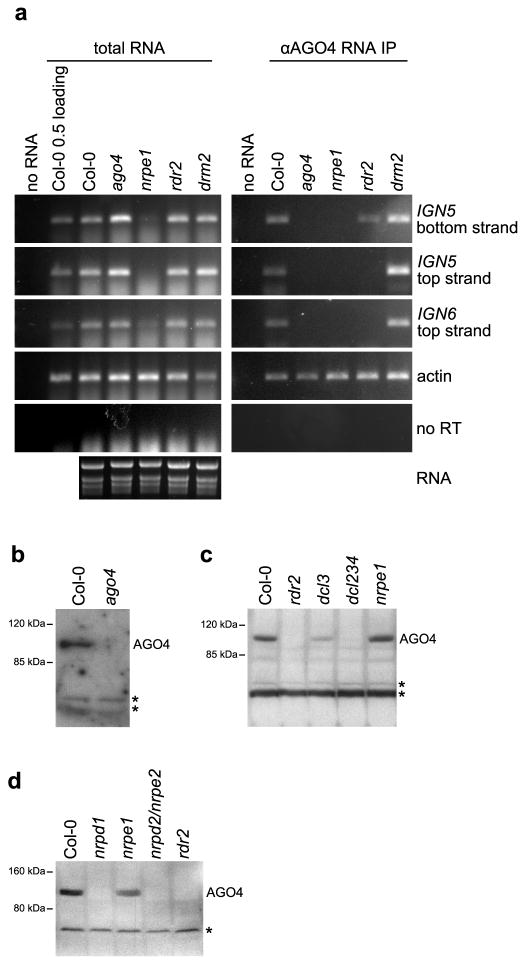
Base-pairing between AGO4-associated siRNAs and nascent Pol V transcripts could be a mechanism by which Pol V transcription recruits AGO4 to target loci. To test this hypothesis, we used RNA immunoprecipitation to ask whether AGO4 associates with Pol V transcripts in vivo. In wild-type (Col-0) plants, anti-AGO4 antibody immunoprecipitates IGN5 and IGN6 Pol V transcripts3 (Fig. 4A). Important controls show that Pol V transcripts are not immunoprecipitated in the ago4 or nrpe1 mutant backgrounds. Anti-AGO4 immunoprecipitation of IGN5 or IGN6 RNAs was also reduced or eliminated in rdr2 mutant plants, indicating that AGO4-Pol V transcript interactions are dependent on siRNAs. However, in the absence of siRNA biogenesis, as in the rdr2, nrpd1, nrpd2/nrpe2 or dcl2,3,4 mutants, AGO4 protein levels drop below the limits of immunoblot detection 7,8 (Fig. 4B–D). By contrast, AGO4 protein levels are unaffected in the nrpe1 or drm2 mutants, which act downstream of siRNA biogenesis (Fig. 4B–D). The instability of AGO4 in the absence of siRNAs complicates the interpretation of these results. Although we favor the hypothesis that siRNA-Pol V transcript base-pairing is responsible for AGO4 association with Pol V transcripts, we cannot rule out the possibility that AGO4 binds Pol V transcripts directly, with siRNAs merely being required for AGO4 stability.
Figure 4. AGO4 physically interacts with Pol V transcripts.
(A) RNA immunoprecipitation using αAGO4 antibody. Immunoprecipitated RNA isolated from the indicated mutants was digested with DNaseI and amplified by RT-PCR. Total RNA controls show that the Pol V transcripts are present in equivalent amounts in all mutants tested except nrpe1. Ethidium bromide stained rRNAs (bottom left) show that equal amounts of RNA were tested. The no reverse transcriptase (no RT) control was performed with IGN5 bottom strand primers. No RNA controls were performed for all primer pairs tested. RT-PCR amplification of actin RNA serves as a loading control.
(B) Immunoblot detection of AGO4 in protein extracts of wild-type (Col-0) plants or ago4 mutant. Stars denote nonspecific bands.
(C) Immunoblot detection of AGO4 in protein extracts of wild-type (Col-0), rdr2, dcl3, dcl234 or nrpe1 mutants. Asterisks denote nonspecific bands.
(D) Immunoblot detection of AGO4 in protein extracts of wild-type (Col-0), nrpd1 (Pol IV) nrpe1 (Pol V), nrpd2/nrpe2 (shared subunit of Pol IV and Pol V), or rdr2 mutants. Asterisks denote nonspecific bands.
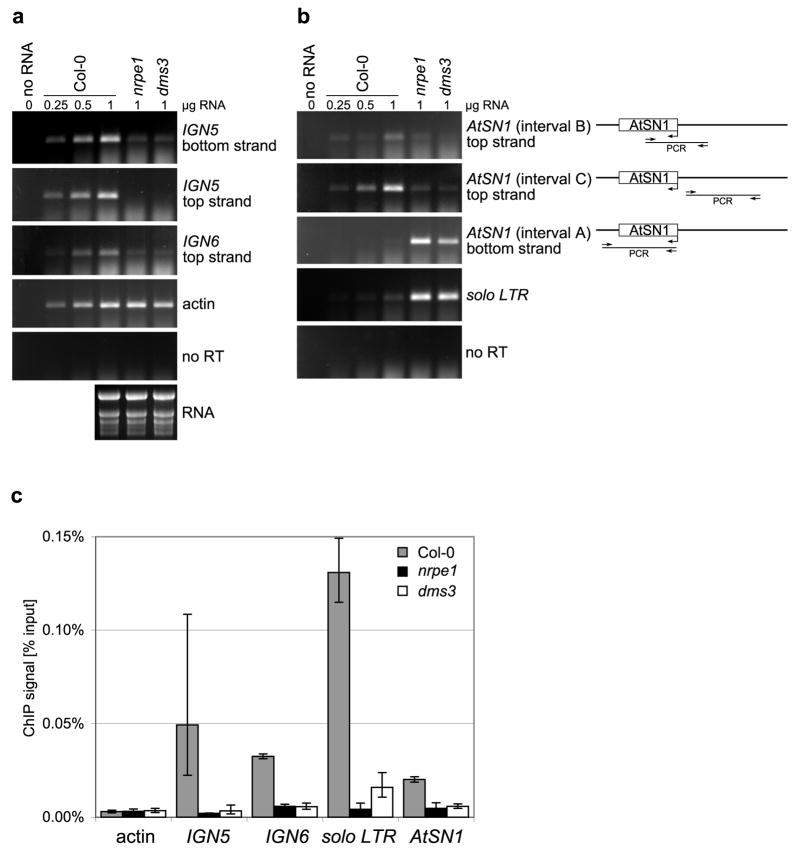
DMS3 was recently identified as a gene required for RNA-directed DNA methylation that acts at an unspecified step downstream of siRNA biogenesis4. The encoded protein shares sequence similarity with the hinge domain regions of Structural Maintenance of Chromosomes (SMC) proteins, such as the core proteins of cohesin and condensin complexes16, suggesting a chromatin-related function. We find that at IGN5, IGN6 and AtSN1 loci, Pol V transcripts are substantially reduced or absent in dms3 mutant plants, as in nrpe1 (Fig. 5A) or drd1 mutants3. Likewise, transcriptional suppression of AtSN1 and solo LTR elements is similarly disrupted in dms3 and nrpe1 mutants (Fig. 5B). Chromatin immunoprecipitation using an anti-NRPE1 antibody revealed that in the dms3 mutant, Pol V-chromatin associations are reduced to background levels resembling the actin and nrpe1 mutant controls (Fig. 5C). Collectively, the data of Figure 5 indicate that DMS3 is required for Pol V transcription, as shown previously for the chromatin remodeller, DRD13. The loss of detectable Pol V-chromatin association in dms3 or drd1 mutants suggests that these chromatin proteins participate in the assembly of Pol V transcription complexes.
Figure 5. The SMC hinge-domain protein, DMS3 is required for Pol V transcription and detectable Pol V-chromatin interactions.
(A) Strand-specific RT-PCR detection of Pol V transcripts at IGN5 and IGN6 and AtSN1 in wild-type (Col-0) and nrpe1 and dms3 mutants. Derepression of Pol II transcripts at the solo LTR and putative Pol III transcripts at AtSN1 in the nrpe1 and dms3 mutants is shown in the right panel. Actin RT-PCR products and ethidium bromide-stained rRNAs resolved by agarose gel electrophoresis serve as loading controls. To control for background DNA contamination, a reaction using IGN5 bottom strand and AtSN1 (interval B) primers, but no reverse transcriptase (no RT) was performed. No RNA (0 μg) controls are provided for all primer pairs.
(B) ChIP with anti-NRPE1 antibody in Col-0 wild-type, nrpe1 and dms3 mutants followed by real-time PCR. Histograms show means +/− standard deviations obtained from three independent amplifications.
Our results suggest that siRNAs and Pol V transcripts are produced by independent pathways that intersect to bring about heterochromatin formation and gene silencing (Fig. 6). In one pathway, Pol IV, RDR2 and DCL3 collaborate to produce 24 nt siRNAs that associate with AGO41. Independent of this pathway, DRD1 and DMS3 facilitate noncoding Pol V transcription at target loci. AGO4’s interaction with Pol V transcripts, and the fact that AGO4 association with chromatin requires the Pol V active site, suggests that siRNA-AGO4 complexes are guided to target loci by interacting with Pol V transcripts. It has also been reported that AGO4 can interact with the C-terminal domain (CTD) of NRPE1 in vitro7,17 and in vivo7, suggesting that Pol V might recruit AGO4 directly, in an RNA-independent manner. However, we have been unable to detect AGO4-Pol V associations in vivo using immunoprecipitation and subsequent immunoblotting nor by mass spectrometric analysis of affinity purified Pol V (data not shown) suggesting that any interactions between AGO4 and Pol V may be weak or transient. We suggest that AGO4 recruitment to chromatin is primarily an RNA-mediated process but may also involve protein-protein interactions.
Figure 6.
A model for Pol V and siRNA-dependent heterochromatin formation. DMS3 and DRD1 mediate the assembly of Pol V initiation and/or elongation complexes and the production of Pol V transcripts. AGO4-siRNA complexes recognize target loci via base-pairing of siRNAs with nascent Pol V transcripts. AGO4 subsequently recruits chromatin modifying activities including the de novo DNA methyltransferase DRM2 and histone modifying enzymes via unknown mechanisms.
In fission yeast, artificial tethering of the RNA Induced Transcriptional Silencing (RITS) complex to ura4 pre-mRNAs is sufficient to induce heterochromatin formation at the normally euchromatic ura4+ locus18. These, and other, results are consistent with the hypothesis that fission yeast silencing complexes are guided to chromatin via associations with nascent Pol II transcripts19. Our findings suggest that plants and yeast are fundamentally similar in their use of RNA guidance mechanisms for recruiting Argonaute-containing transcriptional silencing complexes to target loci. It is intriguing that plants should have evolved a unique RNA polymerase, Pol V whose specialized role appears to be the generation of noncoding RNAs that can serve as scaffolds for Argonaute recruitment.
Methods
Plant strains
Arabidopsis thaliana nrpe1 (nrpd1b-11) was described previously8. The dms3–4 mutant (SALK_125019C) of locus At3g49250 was obtained from the Arabidopsis Biological Resource Center. The dcl2, dcl3, dcl4 triple mutant (dcl234) was provided by Todd Blevins. The ago4-1 mutant (Ler ecotype background) was kindly provided by Steve Jacobsen (UCLA) and was introgressed into the Col-0 background by 3 rounds of backcrossing.
Antibodies
Anti-Pol II (anti-NRPB2) was described previously20. Anti H3K27me1 antibody #883521 was provided by Thomas Jenuwein. Antibody against diacetyl-H3 (K9 and K14) was obtained from Millipore (cat. #06599, lot #JBC1349702). Rabbit anti-NRPE1 antibody has been described9. Rabbit anti-AGO4 was raised against a C-terminal portion of the protein (amino acids 573–924) expressed in bacteria.
RNA and DNA analysis
RNA isolation, RT-PCR and real-time quantitative PCR were performed as described3 except that real time quantitative PCR analysis of the IGN5 locus was performed using the following oligonucleotides: A195: ACATGAAGAAAGCCCAAACCA and A196: GGCCGAATAACAGCAAGTCCT. Densitometric analysis of DNA resolved by agarose gel electrophoresis was performed using ImageJ (http://rsbweb.nih.gov/ij/).
ChIP and RNA IP
ChIP and RNA IP were performed as described3 except that for ChIP with anti-AGO4, RNase A was added during IP, washes with TE buffer were omitted, immune complexes were eluted with 100 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS for 10 min at room temperature and a second elution at 65°C was performed. Crosslinking was reversed at 65°C for 1h in the presence of 40 μg Proteinase K (Invitrogen). DNA was purified by extraction with phenol:chloroform and ethanol precipitation. DNA recovery was assayed by PCR using 1.5u Platinum Taq (Invitrogen).
Supplementary Material
Acknowledgments
Our work is supported by NIH grant GM077590. The content of the paper is the sole responsibility of the authors and does not necessarily reflect the views of the NIH.
References
- 1.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–96. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 2.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–48. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA Polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanno T, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–5. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 5.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–8. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Kanno T, et al. A SNF2-like protein facilitates dynamic control of DNA methylation. EMBO Rep. 2005;6:649–55. doi: 10.1038/sj.embor.7400446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li CF, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 8.Pontes O, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 9.Pontier D, et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–40. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanno T, et al. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–5. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 11.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasschau KD, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–7. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 14.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haag JR, Pontes O, Pikaard CS. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS ONE. 2009;4:e4110. doi: 10.1371/journal.pone.0004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peric-Hupkes D, van Steensel B. Linking cohesin to gene regulation. Cell. 2008;132:925–8. doi: 10.1016/j.cell.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 17.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–44. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–86. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 19.Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 20.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–22. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Peters AHFM, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.