Abstract
Dysregulation of the Hedgehog signaling pathway is central to the development of certain tumor types, including medulloblastoma and basal cell carcinoma (BCC). Patched1 (Ptch1) and Suppressor of fused (Sufu) are two essential negative regulators of the pathway with tumor suppressor activity. Ptch1+/− mice are predisposed to developing medulloblastoma and rhabdomyosarcoma, while Sufu+/− mice develop a skin phenotype characterized by basaloid epidermal proliferations. Here, we have studied tumor development in Sufu+/−Ptch1+/− mice to determine the effect of compound heterozygosity on the onset, incidence, and spectrum of tumors. We found significantly more (2.3-fold) basaloid proliferations in Sufu+/−Ptch1+/− compared to Sufu+/− female, but not male, mice. For medulloblastoma, the cumulative one-year incidence was 1.5-fold higher in Sufu+/−Ptch1+/− compared to Ptch1+/− female mice but this strong trend was not statistically significant. Together this suggests a weak genetic interaction of the two tumor suppressor genes. We noted a few rhabdomyosarcomas and pancreatic cysts in the Sufu+/−Ptch1+/− mice, but the numbers were not significantly different from the single heterozygous mice. Hydrocephalus developed in ∼20% of the Ptch1+/− and Sufu+/−Ptch1+/− but not in Sufu+/− mice. Interestingly, most of the medulloblastomas from the Sufu+/−Ptch1+/− mice had lost expression of the remaining Ptch1 wild-type allele but not the Sufu wild-type allele. On the contrary, Sufu as well as Gli1 and Gli2 expression was upregulated in the medulloblastomas compared to adult cerebellum in Ptch1+/− and Sufu+/−Ptch1+/− mice. This suggests that Sufu expression may be regulated by Hedgehog pathway activity and could constitute another negative feedback loop in the pathway.
Keywords: Hedgehog proteins, basal cell carcinoma, rhabdomyosarcoma, hydrocephalus, pancreas cyst
INTRODUCTION
The Hedgehog (Hh) signaling pathway regulates a variety of developmental processes, and perturbations in the pathway are involved in many developmental disorders as well as specific malignant tumors [1-3]. The Hh family of proteins are ligands to the 12-span transmembrane receptor Patched (Ptch1), which when unbound inhibits the 7-span transmembrane protein Smoothened (Smo) [4]. When Hh binds to Ptch1 this inhibition is relieved and Smo can transduce the signal, ultimately leading to activation of the Gli transcription factors. Several pathway components downstream of Smo regulate the activity of Gli. Among them is Suppressor of Fused (Sufu), which has an essential negative regulator function in the pathway, possibly through nuclear-cytoplasmic shuttling of the Gli proteins [5]. In the mouse, targeted null mutations of either Sufu [6,7] or Ptch1 [8,9] lead to embryonic lethality in midgestation with a similar phenotype.
The role of the Hh pathway in tumorigenesis was first established in the nevoid basal cell carcinoma syndrome (NBCCS), also called Gorlin syndrome, where germline mutations in PTCH1 underlie the pathological changes. Besides developing basal cell carcinoma (BCC), individuals with this syndrome are susceptible to developing other tumors, including medulloblastomas (MB) [10-12]. MB is a primitive neuroectodermal tumor that arises in the cerebellum, presumably from granule-cell precursors (GCPs) in the external granular layer (EGL) [13]. It is one of the most common brain tumors in children. Mutations in PTCH1, SUFU and SMO have been found in approximately 25% of sporadic human MBs [13]. Up to 30% of mice, heterozygous for a null mutation in Ptch1, develop MB within the first 10 months [8,9,14] and the incidence is increased to >95% on a tumor suppressor p53 null background [15]. Ptch1+/− mice also develop rhabdomyosarcoma (RMS), a soft tissue tumor, with a frequency of 2-9 %, depending on the genetic background [8,9]. Sufu+/− mice do not spontaneously develop MB [6,7], unless on a p53 null background [16], and rarely RMS. However, the Sufu+/− mice do develop a skin phenotype with basaloid epidermal lesions, alopecia, aberrant sebaceous gland morphology, and increased pigmentation [6]. The basaloid lesions histologically resemble follicular hamartomas. Similar basaloid proliferations are also found in Ptch1+/− mice but to a much lesser extent [17].
Thus, Ptch1 and Sufu heterozygous mutant mice each show increased susceptibility for tumor development, with an overlapping but different tumor spectra. We set out to investigate whether compound heterozygosity for Ptch1 and Sufu would lead to an increased frequency, a different spectrum, or the earlier onset of tumors compared to single heterozygous mice. Here, we found that the frequency of basaloid proliferations was significantly increased in the compound Sufu+/−Ptch1+/− female mice and a strong trend towards an increase of the MB incidence. This suggests a weak genetic interaction of the two tumor suppressor genes.
MATERIALS AND METHODS
Mouse strains and genotyping
To generate offspring with the desired genotypes (wt, Sufu+/−, Ptch1+/−, and Sufu+/−Ptch1+/−), the Sufu knockout mouse strain B6;129X1/SvJ-Sufutm1Rto [6] and the Ptch1 knockout mouse strain B6;129-Ptch1tm1Mps [8] obtained from the Jackson Laboratory (stock 003081) were intercrossed. The mice used in this study were backcrossed to C57BL/6J for four or more generations prior to intercrossing. Genotyping of the mice was done as described in [6]. The mice were housed in an SPF barrier facility according to local and national regulations, and the study was approved by the Stockholm South Animal Ethics Committee.
Histology and X-gal staining
For histological analysis, tissues were fixed overnight in 4% paraformaldehyde (PFA), or for skull, for a week in Bouin's solution (Sigma, St Louis, MI) to allow for visualizing the brain in situ within the skull cavity. To detect β-galactosidase activity in skin, tissues were fixed in 2% PFA in PBS for 30 min at room temperature. After fixation, tissues were rinsed twice in PBS for 30 min and then once in rinse buffer (2 mM MgCl2, 0.01% NP40 in PBS) for 30 minutes. Tissues were then stained for 24-36 hrs with X-gal reaction mixture [5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 1 mg/ml X-galactoside 5-bromo-4-chloro-3-indlyl-beta-D-galactopyranoside (X-gal) in rinse buffer]. Tissues were then transferred to 70% ethanol, paraffin embedded and sectioned at 6 μm. These sections were stained with eosin or used for immunohistochemistry. For the PAS staining, diastase was used to remove glycogen.
Immunohistochemistry
The following primary antibodies were used: anti-K5 (1:1000, PRB-160P, Covance Inc., Princeton, NJ); anti-K6 (1:2000, PRB-169P, Covance Inc.); anti-K17 (1:2500, kind gift from Drs P. Coulombe and K. McGowan, Johns Hopkins University School of Medicine, Baltimore, MD); anti-K10 (1:2500, PRB-159P, Covance Inc.); anti-Caspase 3 (cleaved, Asp175) (1:200, 9664S, Cell Signaling Tecnology Inc., Danvers, MA); anti-phospho-Histone H3 (1:750, 06-570, Millipore Corp., Billerica, MA); anti-Ki67 (1:3000, NCL-Ki67p, Leica Microsystems, Biosystems Div., Wetzlar, Germany); anti-Cyclin D1 (SP4) (1:200, Thermo Fisher Scientific Inc., Fremont, CA); anti-GABA-A-recα6 subunit (1:3000, Millipore Corp.); anti-PDGFRα (C-20) (1:1000, sc-338, Santa Cruz Biotechnology Inc., Santa Cruz, CA); anti-Desmin (1:100, D1033, Sigma); anti-Sufu (1:200, C81H7, Cell Signaling Tecnology Inc.); anti-β-catenin (1:2000, Epitomics Inc., Burlingame, CA); anti-E-cadherin (1:10000, BD, Franklin Lakes, NJ). The biotinylated secondary antibodies were either α-rabbit IgG (1:200, BA-1000, Vector Laboratories, Burlinggame, CA) or α-goat IgG (1:200, BA-5000, Vector Laboratories). Streptavidin-conjugated peroxidase (Invitrogen Corp., Carlsbad, CA) together with DAB as substrate (Invitrogen Corp.) were used to visualize the bound antibodies, followed by counterstaining in hematoxylin. Mouse primary antibodies were used in combination with Histomouse™-SP (Invitrogen Corp.). For all antibodies, antigen retrieval was performed for 20 min at 97.5°C in 10 mM sodium citrate pH 6.0.
Quantitative real-time reverse transcriptase PCR
Total RNA from frozen or RNAlater-preserved (Applied Biosystems, Foster City, CA) tissue from ventral skin and cerebella was prepared using RNeasy spin columns (Qiagen Inc., Valencia, CA) with on-column DNaseI treatment. One μg of RNA was reverse transcribed using Superscript (Invitrogen Corp.) with oligo(dT)15 primers (Promega Corp., Madison, WI) in a volume of 30 μl. Real-time PCR reactions were performed with a 10 μl mixture containing Power SYBR Green PCR master mix (Applied Biosystems), 0.3 μl cDNA and 0.3 μM forward and reverse primers. Samples were analyzed on a 7500 Fast Real-Time PCR System (Applied Biosystems) in duplicate, and ΔCt and ΔΔCt values were calculated using the average of Hprt and Gapdh as endogenous controls. An alternative approach involved the TaqMan Gene Expression Assay for analyzing mouse Sufu expression (assay ID #Mm00489385_m1), using TaqMan Universal PCR master mix (Applied Biosystems) and the endogenous rodent control GAPDH (#4308313). Data is represented as the mean of each sample assayed in duplicate ± s.e.m. All tissues were from one-year-old mice except MBs, which were from mice of different ages. Real-time PCR primer sequences are listed in Supplementary Table SI.
Statistical analysis
Statistical analyses were performed using Prism 5.0 (GraphPad Software, Inc.). Differences in tumor incidence, weight, and number of skin proliferations were analyzed using Fisher's exact test, unpaired t-test, and one-way ANOVA corrected for multiple comparisons using Tukey's post-test, respectively.
RESULTS
Overall survival and growth of Sufu+/−Ptch1+/− mice is not significantly different from Ptch1+/− mice
To study possible genetic co-operativity of the tumor suppressors Sufu and Ptch1 in the mouse, we intercrossed mice heterozygous for the Sufu [6] and Ptch1 [8] loss-of-function alleles, to generate Sufu+/−Ptch1+/− mice. We followed a cohort of Sufu+/−Ptch1+/− mice (n = 56) up until one year of age or until severe symptoms developed, after which they were sacrificed and necropsies performed. For comparison, Sufu+/− (n = 25), Ptch1+/− (n = 49) and wild-type (wt; n = 12) littermate control mice were similarly followed and analyzed. We found that the overall survival of the Sufu+/−Ptch1+/− mice (29%) was not significantly different from the Ptch1+/− mice (39%) at one year of age (Figure 1A). At that age, 18% (10/56) of the Sufu+/−Ptch1+/− mice remained seemingly healthy and free of any gross tumors compared to 33% (16/49) of the Ptch1+/− mice. The major contributors to morbidity, affecting the survival of the Sufu+/−Ptch1+/− mice, were the development of MB and hydrocephalus (HC), which was similar to that found in the Ptch1+/− mice (Figure 1B). By contrast, none of the 25 Sufu+/− or 12 wt mice developed any tumors or HC during the same one-year period. One Sufu+/− mouse developed renal amyloidosis.
Figure 1.
Survival, growth, and incidence of tumors and other lesions in Sufu+/−Ptch1+/− mice. (A) Kaplan-Meier survival plot of wt, Sufu+/−, Ptch1+/−, and Sufu+/− Ptch1+/− mice followed until one year of age. (B) Cumulative one-year incidence of medulloblastoma (MB), rhabdomyosarcoma (RMS), pancreatic lesions (PL), and hydrocephalus (HC) in female (F) and male (M) Ptch1+/− [F (n=26); M (n=23)] and Sufu+/−Ptch1+/− [F (n=29); M (n=27)] mice. Numbers above the bars represents number of mice with the lesions. No wt or Sufu+/− mouse suffered any of these lesions. (C) Mean weight of female and male wt, Sufu+/−, Ptch1+/−, and Sufu+/−Ptch1+/− mice between 4 to 20 weeks of age. (D) Age distribution of the MB occurrence in Ptch1+/−and Sufu+/−Ptch1+/− mice. (E) Number of epidermal proliferations scored per hematoxylin-eosin stained sagittal section from the plantar surface of the hind paw from one-year old mice of the indicated genotypes. The data represents the average ± standard deviation of the number of proliferations. One-way ANOVA with Tukey's post-test was used for statistical analysis. P-values * = < 0.05, ** = < 0.01, *** = 0.001, and ns = not significant. Note that zero proliferations were encountered in the wt control mice.
Ptch1 has been implicated in body size regulation [18] and Ptch1+/− mice are on average 10% larger than wt controls [8]; some individuals with Gorlin syndrome also show increased body size [11]. To investigate whether loss of one Sufu allele would act synergistically with Ptch1+/−, we weighed healthy mice between 4 and 20 weeks of age. We found that both female and male Sufu+/−Ptch1+/− mice were significantly larger than either Sufu+/− or wt mice of the corresponding sex, but not compared to Ptch1+/− mice, which are equally large [Figure 1C; P = 0.002 (females) and 0.015 (males) when comparing Sufu+/−Ptch1+/− to wt mice at 10 weeks of age]. Since Sufu+/−mice are equal in size to the wt controls of the corresponding sex and age, this suggests that Sufu may not be involved in body size regulation. Alternatively, Sufu, in contrast to Ptch1, may be haplosufficient for that function.
The incidence of medulloblastoma is not significantly altered in Sufu+/−Ptch1+/−mice compared to Ptch1+/− mice
Ptch1+/− mice have been reported to develop MBs at a frequency of up to 30% [8,9,19]. To study whether this frequency would be increased in Sufu+/−Ptch1+/− mice, we examined the brains of all mice in the study, whether they exhibited symptoms or not. We found that 48% (27/56) of Sufu+/−Ptch1+/− mice developed MB within 1 year of age compared to 37% (18/49) of Ptch1+/− mice. This increase was, however, not statistically significant (P = 0.3231). However, the MB incidence was significantly higher in Sufu+/−Ptch1+/− females versus males (P = 0.0016). Sufu+/−Ptch1+/− mice developed symptoms due to MB at the earliest around 1 month of age, similar to the age of onset seen in the Ptch1+/− mice (Figure 1D). The mean age for MB development was also similar for Sufu+/−Ptch1+/− (24 weeks) and Ptch1+/− mice (22 weeks) (Figure 1D).
To compare the expression of Hh pathway components in adult cerebellum and in MBs, we analyzed Gli1, Gli2, Gli3, Sufu, and Ptch1 mRNA levels by quantitative real-time reverse transcriptase PCR (qRT-PCR). We found that the Gli1 mRNA levels in adult cerebellum from Sufu+/−, Ptch1+/−, and Sufu+/−Ptch1+/− mice, in most cases were comparable to wt levels (Figure 2A). However, cerebella from one Ptch1+/− and two Sufu+/−Ptch1+/− mice showed higher Gli1 levels (Figure 2A). It is possible that these samples contained tumor cells although this was not evident from the tissue histology. Gli1 levels in MBs from Ptch1+/− and Sufu+/−Ptch1+/− mice were 3-to-11-fold higher than in normal cerebellum and the levels were comparable to each other, except in one incidence of MB in a Ptch1+/− mouse that had 107-fold higher Gli1 levels (Figure 2A). The Gli2 mRNA levels were also consistently elevated 9-to-49-fold in the MBs (Figure 2B). Interestingly, the elevated Gli1 expression seen in the three seemingly normal cerebellum samples from Ptch1+/− and Sufu+/−Ptch1+/− mice was not mirrored by increased Gli2 expression (Figure 2B). With regards to Gli3 mRNA levels in the MB samples, there was no consistent alteration: some were higher and some were lower than the average non-tumor cerebellum levels (Figure 2C).
Figure 2.
Increased expression of Glil, Gli2, and Sufu, but loss of Ptchl expression in most MBs from Ptch1+/− and Sufu+/−Ptch1+/− mice. (A-F) qRT-PCR on total RNA from cerebella of one-year-old mice and MB from mice of different age for (A) Gli1; (B) Gli2; (C) Gli3; (D) Sufu; (E) Ptch1wt+mut; and (F) Ptch1wt. Wt, wild-type; S, Sufu+/−; P, Ptch1+/−; S/P, Sufu+/−Ptch1+/−. Individual samples are numbered 1 through 34. Results shown are means ± standard deviations of mRNA expressions levels relative to a wt sample arbitrarily set to one. Samples are in duplicate from four to seven individual mice in each category.
Whether expression of the remaining Ptch1 wt allele occurs in MBs from Ptch1+/−mice has been debated. Some studies suggest that expression is lost [20,21] while other studies claim that the wt allele continues to be expressed in the tumors [19,22]. To examine the expression status of the Ptch1 wt allele in the MBs from the Sufu+/−Ptch1+/− mice, we used two sets of primers to discriminate between the wt and mutant Ptch1 transcripts. The first set is located in exons 19 and 20 and detects both wt and mutant transcripts, while the second set is located in exons 2 and 3 and only detects the wt transcript since exon 2 is deleted in the Ptch1 mutant allele [8]. We found that in MBs from five out of six Sufu+/−Ptch1+/− mice lost expression of the remaining Ptch1 wt allele (Figure 2E and F). By comparison, all (6/6) MBs from Ptch1+/− mice had lost their expression. This supports the notion that loss of Ptch1 expression is an important step in MB formation in both Ptch1+/− and Sufu+/−Ptch1+/− mice.
We have previously shown that Sufu appears to be as potent as Ptch1 for Hh pathway repression judging from the similarly high Gli activity seen in both Sufu and Ptch1 null cells and embryos [6]. Therefore, loss of expression of the remaining Sufu wt allele in MBs from Sufu+/−Ptch1+/− mice could be functionally equivalent to losing the expression of the Ptch1 wt allele. This may even explain the one Sufu+/−Ptch1+/− MB sample that retained expression of the Ptch1 wt allele but still had elevated Gli1 levels (Figure 2F). To explore this possibility, we performed qRT-PCR for Sufu mRNA expression in the MB samples. In contrast to the Ptch1 mutant allele, the Sufu mutant allele does not produce any transcript since the transcriptional start site is deleted [6]. So only Sufu wt transcripts are detected by the primer set used in the qRT-PCR. We found that the Sufu mRNA levels in all the 13 MB samples from both Ptch1+/− and Sufu+/−Ptch1+/− mice were strongly elevated (5-to-41-fold) compared to normal cerebellum of the same genotype (Figure 2D). This was also confirmed with another set of primers (data not shown). The comparatively high Sufu mRNA expression was also reflected at the protein level on MB tissue sections using immunohistochemistry (Figure 3U-Y). The expression coincides with the increased Gli1 and Gli2 levels and suggests that Sufu mRNA expression may also be regulated by Hh pathway activation. Thus, an alternative explanation for the MB sample retaining Ptch1 wt mRNA expression might be a missense or nonsense point mutation in either the Ptch1 or Sufu wt allele affecting protein function.
Figure 3.
Expression of molecular markers in medulloblastomas from Ptch1+/− and Sufu+/−Ptch1+/− mice. (A-Y) Immunohistochemistry on representative paraffin-embedded tissue sections from normal adult cerebellum of wt and Sufu+/− mice and MBs from Ptch1+/− and Sufu+/−Ptch1+/− mice. (A-D) Phospho-histone H3. (E-H) Cleaved Caspase 3. (I-L) Cyclin D1. (M-P) Platelet-derived growth factor receptor α. (Q-T) γ-aminobutyric acid (GABA)A receptor, α6 subunit. (U-Y) Sufu. Brown color indicates positive immunostaining and light blue color is the hematoxylin counterstain. Scale bar = 50 μm (A-H, M-Y) and 100 μm (I-L).
To characterize the MBs from the Sufu+/−Ptch1+/− mice in more detail, we have analyzed the expression pattern of various cellular markers (Figure 3). As indicated by expression of phosphorylated histone H3 and cleaved Caspase 3, the MBs from the Sufu+/−Ptch1+/− mice have comparable proliferation and apoptosis rates, respectively, to the Ptch1+/− mice (Figure 3C, D, G and H). They also express comparable levels of Cyclin D1 (Figure 3K and L) and PDGFRα (Figure 3O and P), both known to be upregulated in MBs. GABA-A receptor alpha 6 subunit is normally expressed in cerebellar granule cells (Figure 3Q and R), but in MBs from both Ptch1+/− (Figure 3S) and Sufu+/−Ptch1+/− (Figure 3T) mice this expression was lost. The conclusion of these analyses is that we found no differences in the expression of molecular markers that could distinguish MBs arising in Ptch1+/− versus Sufu+/−Ptch1+/− mice.
Sufu+/−Ptch1+/− female mice show an increased number of basaloid epidermal proliferations compared to Sufu+/− female mice
Sufu+/− mice develop a skin phenotype with 100% penetrance, characterized by slow-growing basaloid proliferations in the epidermis [6]. The basaloid proliferations occur in all skin locations but appear first on paws and are most easy to detect there because of the lack of hair follicles. Therefore, to investigate the occurrence of the proliferations in the Sufu+/−Ptch1+/− mice, we quantified them on tissue sections from the plantar surface of the hind paws of one-year old mice. Interestingly, we found that the number of proliferations was significantly higher in Sufu+/−Ptch1+/− compared to Sufu+/− female, but not male, mice [P < 0.05; Figure 1E]. However, the time of appearance (∼6 months) of these microscopic proliferations was similar. In contrast, only a few basaloid proliferations were seen in the epidermis of Ptch1+/− mice and were significantly less frequent compared to both Sufu+/− and Sufu+/−Ptch1+/− mice (Figure 1E), while no epidermal proliferations were detected in the littermate wt control mice.
To further characterize the basaloid proliferations arising in the different mouse genotypes, we used immunostaining on skin tissue sections for epidermal and hair follicle keratin markers. Detection of Keratin 5 (K5), a marker of the basal cell layer of epidermis and the outer root sheath (ORS) of hair follicles, showed that the epidermal proliferations from all genotypes stain relatively homogenously for K5 (Figure 4B-D) indicating a close relationship with basal cells. Keratin 6 (K6) is normally only expressed in the epidermis of the plantar surface of the paws and in the companion cell layer of hair follicles. Here, K6 was heterogeneously expressed in a similar fashion in the proliferative regions, independent of mutant genotype (Figure 4J-L). This heterogeneity was characterized by a lack of K6 expression in the deeper dermal portions of the proliferations while the part closer to the epidermal surface stained strongly. Keratin 10 (K10), a marker of suprabasal cells, only occasionally stained cells, particularly in the inner portion of the proliferations, suggesting that some cell differentiation occurs within the proliferations (Figure 5F-H). Keratin 17 (K17), normally expressed in the ORS of the hair follicle and in occasional focal areas of the palmoplantar epidermis, was strongly expressed in the proliferations from all mutant genotypes in a similar pattern (Figure 4N-P). The number of cells expressing the proliferation marker Ki67 was low in the proliferations from all mutant genotypes, which was consistent with their slow growth (Figure 4R-T). In contrast, the G1-S-phase cell-cycle promoter and proposed Hh target gene Cyclin D1 prominently stained cells typically in the outer rim of the proliferations (Figure 4V-Y).
Figure 4.
Skin from Sufu+/−, Ptch1+/−, and Sufu+/−Ptch1+/− mice develop similar basaloid epidermal proliferations but differ in frequency depending on genotype. (A-Y) Immunohistochemistry on formalin-fixed paraffin-embedded representative skin tissue sections from paws. (A-D) Keratin 5 immunostaining. (E-H) Keratin 5 immunostaining and ß-galactosidase (ß-gal) activity as visualized by X-gal staining (dark blue). Wt and Sufu+/− mice have no LacZ gene and show only weak unspecific staining in the sebaceous glands. (I-L) Keratin 6 immunostaining. (M-P) Keratin 17 immunostaining. (Q-T) Ki67 immunostaining. (U-Y) Cyclin D1 immunostaining. Brown color indicates positive immunostaining and light blue color is the hematoxylin counter-stain. All tissues are from one-year-old mice. Scale bars = 50 μm.
Figure 5.
Characterization of the basaloid epidermal proliferations using immunohistochemistry on skin tissue sections from the paw. (A-D) Hemtoxylin-eosin staining and ß-galactosidase (ß-gal) staining. (E-H) Keratin 10 immunostaining. (I-L) β-catenin immunostaining. (M-P) E-cadherin immunostaining. All images are from one-year-old mice. Scale bars = 100 μm (A-D) and 50 μm (E-T).
The mutant Ptch1 allele contains a LacZ knock-in gene [8] and since Ptch1 is a Hh target gene, X-gal staining for ß-galactosidase (β-gal) activity could be used to monitor Hh pathway activity in the skin of Ptch1+/− and Sufu+/−Ptch1+/− mice. In the Sufu+/−Ptch1+/− skin there was strong X-gal staining in part of the epidermal proliferations and in dermal regions, often in conjunction with the proliferations, that resembled dermal papilla (Figures 4H and 5D). In Ptch1+/− mice, cells that are X-gal positive are much less frequent (Figures 4C and 5C). This implies that the extent of Hh pathway activity correlates well with the manifestation of the skin phenotype. Further analyses showed that there was no difference in ß-catenin (Figure 5I-L) or Ecadherin (Figures 5M-P) immunostaining of the proliferations from the different genotypes. The ß-catenin staining was mainly membraneous indicative of no or little canonical Wnt signaling pathway activity (Figures 5J-L).
In conclusion, from the set of protein markers analyzed here, the proliferations share identity with cells of basal and hair follicle origin. There was no significant difference in expression patterns that could discriminate between the proliferations from the different genotypes. There does, however, appear to be some level of differentiation within the proliferations since not all keratin markers stained uniformly.
To compare the mRNA expression levels of Hh pathway components in skin from the different genotypes, we analyzed the levels of Gli1, Gli2, Gli3, Ptch1 and Sufu expression by qRT-PCR in ventral skin from one-year old mice. Gli1 expression was, on average, increased by 2.7 times in Sufu+/−, 4.6 times in Ptch1+/− and 4.0 times in Sufu+/−Ptch1+/− mice compared to wt mice (Figure 6A). No consistent pattern was seen for the other genes analyzed (Figure 6B-F).
Figure 6.
Increased expression of Gli1 in skin from Ptch1+/− and Sufu+/−Ptch1+/− mice. (A-F) qRT-PCR on total RNA from ventral skin of one-year-old mice for (A) Gli1; (B) Gli2; (C) Gli3; (D) Sufu; (E) Ptch1wt+mut; and (F) Ptch1wt. Wt, wild-type; S, Sufu+/−; P, Ptch1+/−; S/P, Sufu+/−Ptch1+/−. Individual samples are numbered 1 through 17. Results shown are means ± standard deviations of mRNA expressions levels relative to a wt sample arbitrarily set to one. Samples are in duplicate from three to five individual mice in each category.
Ptch1+/− and Sufu+/−Ptch1+/− mice develop cysts of the exocrine pancreas and diverticular harmartomatous lesions in the intestine and stomach
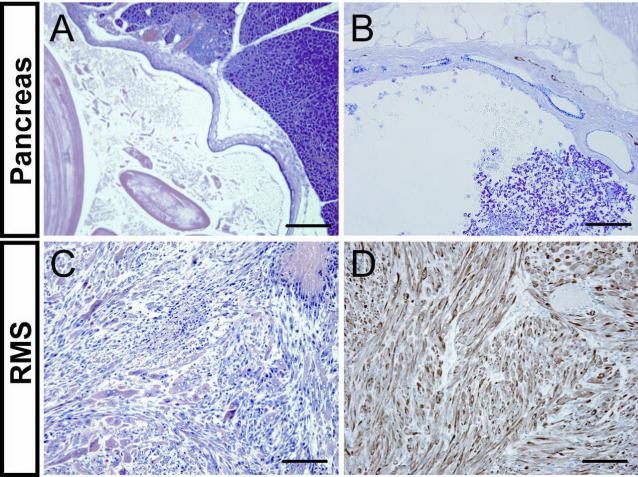
A few Ptch1+/− (1/49) and Sufu+/−Ptch1+/− (4/56) mice, but no Sufu+/− or wt mice developed macroscopically visible cystic lesions of the pancreas (Figures 1B and 7A). We used Alcian blue, which stains intestinal-type mucins, and PAS, which stains both gastric- and intestinal-type mucins, to study the differentiation of these cysts. Both PAS and Alcian blue stained the cystic epithelium (Figure 7B). Normal pancreatic parenchyma does not stain for PAS- or Alcian blue, whereas positive staining is characteristic of mucinous cystic tumors as well as adenocarcinoma of the pancreas. We also noted a few diverticular harmartomatous lesions in the intestine and stomach of Ptch1+/− mice and Sufu+/−Ptch1+/− mice (data not shown).
Figure 7.
Development of pancreatic lesions and RMS in Ptch1+/− and Sufu+/−Ptch1+/−mice. (A) H&E-stained pancreatic cyst. (B) PAS and Alcian blue-stained pancreatic cyst. (C) Hematoxylin-eosin (H&E)-stained RMS. (D) Desmin immunostaining of RMS. Scale bars =200 μm (A) and 100 μm (B-D).
Ptch1+/− and Sufu+/−Ptch1+/− mice develop rhabdomyosarcoma with similar frequency
Ptch1+/− mice develop RMS with a frequency of 2-9 % depending on genetic background [8,9]. In our study, we found that the frequency of RMS is similar in Ptch1+/− and Sufu+/−Ptch1+/− mice (6 and 7 %, respectively; Figures 1B and 7C). There was no difference in histology or immunohistochemical staining with Desmin, a skeletal muscle marker, between the RMS from the two different mutant strains (Figure 7D). By comparison, no RMS was detected in Sufu+/− or wt mice.
Hydrocephalus occurs with similar frequency in Ptch1+/− and Sufu+/−Ptch1+/− mice
From two weeks to around three months of age approximately 20% of the Sufu+/−Ptch1+/− mice developed hydrocephalus (HC). A similar frequency (22%) was seen in the Ptch1+/− mice (Figure 1B). No Sufu+/− or wt mice developed HC, indicating that this phenotype is linked to the Ptch1 mutant allele. A number of mice with HC also had MB. In this case the cause of HC is probably secondary to tumor growth due to blockage of the flow of cerebrospinal fluid. However, in the majority of hydrocephalus cases, no detectable tumor growth was seen in the cerebellum.
DISCUSSION
In this study we have investigated the consequences of carrying heterozygous germline loss-of-function mutations at both the Sufu and Ptch1 loci by studying Sufu+/−Ptch1+/− mice. Since Ptch1+/− mice are predisposed to developing MB and RMS [8,9], and Sufu+/− mice are prone to developing basaloid skin proliferations [6], our hypothesis was that Sufu+/−Ptch1+/− compound mice would have an increased frequency, severity, and/or earlier onset of these and possibly other tumors. For MB development, the cumulative one-year incidence was increased in Sufu+/−Ptch1+/− mice compared to Ptch1+/− mice. However, this difference was not statistically significant. The average time of onset for MB was similar in the two mouse strains. There was no difference in the expression of molecular markers for cellular growth or apoptosis within the MB tumors, and the Gli1 and Gli2 mRNA levels were similarly elevated. Interestingly, the incidence of MB in Sufu+/−Ptch1+/−female mice was almost twice that of Sufu+/−Ptch1+/− males. The same tendency was seen in Ptch1+/− mice of different sex but this difference was not statistically significant. In humans, MB is more common in males [13]. The reason for this sex bias is unknown.
We found that the expression of the remaining Ptch1 wt allele was lost in all MBs from Ptch1+/− mice, and in all except one Sufu+/−Ptch1+/− mouse. This result is in concordance with the studies suggesting that loss of the Ptch1 wt allele is an early and necessary event in MB development in Ptch1+/− mice [20,21] and also in Ptch1+/−Ptch2−/− mice [23]. The rate of MB development in Ptch1+/− mice can be significantly accelerated by treatment with ionizing radiation at postnatal day 1 and 4 but not at postnatal day 10 or at 3 months [24,25] or by breeding onto a p53 null background [15]. Increased frequency of inactivating the remaining Ptch1 wt allele is most likely the driving factor behind the earlier onset and higher incidence of MB in these cases. An alternative way of gaining constitutive Hh pathway activity in MBs from the Sufu+/−Ptch1+/− mice, other than by loss of Ptch1 expression, would possibly be through loss of Sufu expression. However, we found no evidence for that in our study. On the contrary, in MBs from Sufu+/−Ptch1+/− mice, mRNA expression from the remaining Sufu wt allele was considerably higher than in normal Sufu+/−Ptch1+/− adult cerebellum and mirrors the high level of Sufu expression seen in the EGL of the early postnatal cerebellum [26]. Perhaps Sufu, like Ptch1, is also part of a negative feedback loop in response to Hh pathway activity. However, there are no obvious Gli binding sites in the immediate upstream Sufu promoter region so in that case it might be an indirect effect. Regardless, the increased Sufu expression was apparently not sufficient to inhibit the Hh pathway activity in the MBs from the Ptch1+/− and Sufu+/−Ptch1+/− mice as inferred from the high Gli1 expression in those tumors.
Exposing Sufu+/− mice to ionizing radiation at 2 months of age did not result in any MB up to 12 months after exposure [7], which fits with the data from irradiating Ptch1+/− mice at a similar age [24]. However, breeding Sufu+/− onto a p53 null background gives a dramatic increase in MB incidence [16] and K. Heby-Henricson, B. R., R. T. and S. T. (unpublished results)], with a frequency similar to Ptch1+/−Trp53−/− mice [15]. Somatic loss or silencing of the remaining Sufu wt allele, like Ptch1, probably needs to occur in a narrow window before or during the expansion of GCPs in the EGL to result in MB. Taken together, this indicates that the Ptch1 locus is more sensitive to a genetic or epigenetic change than the Sufu locus in GCPs, at least on a Trp53 wt background. Tissue-specific homozygous ablation of Sufu in the developing cerebellum will reveal whether loss of Sufu is sufficient to promote a high incidence of MB development in mice.
In humans, data indicate that although inactivating mutations in SUFU have been reported in MB [27], studies screening for SUFU mutations in MB indicate that it is a relatively rare event [28] compared to mutations in PTCH1 [13]. Thus, the data from mouse and human appear to agree that there is differential sensitivity to genetic or epigenetic changes between the two loci that leads to MB development.
We found that the basaloid skin phenotype developing in the Sufu+/− mice was significantly escalated in the Sufu+/−Ptch1+/− female but not male mice. Again, the reason for this sex difference is unknown. We found no evidence for loss of expression of the remaining Sufu or Ptch1 wt alleles by qRT-PCR in the skin lesions. However, since the skin samples (in contrast to the more homogenous MB samples) are likely to contain a significant amount of normal heterozygous skin cells surrounding the lesions, this data is inconclusive. Nevertheless, we believe that it is unlikely that the development of these microscopic skin lesions is dependent upon homozygous inactivation of the Sufu or Ptch1 wt alleles for the following reasons: first, the number of microscopic skin lesions per mouse are too numerous to make it probable that a cell in each lesion would have suffered an inactivation event (assuming that each lesion has a clonal origin); second, the appearance of the lesions first becomes microscopically detectable after several months and progress so slowly that the mice normally suffer relatively mild clinical skin symptoms, even after one year of age. In contrast, homozygous conditional Ptch1 inactivation in the skin using inducible K6-Cre mice results in rapid BCC-development in 100% of the mice before 16 weeks [29]. Our prediction is that homozygous conditional inactivation of Sufu in the skin will result in a similar outcome. Therefore, we favor the hypothesis that the skin lesions spontaneously developing in the Sufu+/−, Sufu+/−Ptch1+/−, and to a lesser extent in the Ptch1+/− mice, are due to haploinsufficiency rather than homozygous loss of function. Why, then, does presumptive haploinsufficiency give a stronger skin phenotype in Sufu+/− than in Ptch1+/− mice? One possibility is that Ptch2 is playing a complementary role to Ptch1 in the skin to suppress Hh activity. Support for this notion is that Ptch1+/− mice on a Ptch2−/− background have an increased frequency of BCCs compared to Ptch1+/− mice on a wt background [23]. It is intriguing that the Ptch1 locus appears to be more sensitive to inactivating the remaining Ptch1 wt allele in cerebellar tissue than in the skin of Ptch1+/− mice. This may be connected to the very high proliferation rate of the GCPs in the early postnatal cerebellum.
With regards to RMS development, we found no difference in the frequency between Ptch1+/− and Sufu+/−Ptch1+/− mice. In humans, combined haploinsufficiency for the two tumor suppressor genes PTCH1 and SUFU was suggested to be important for rhabdomyoma and RMS development [30]. In mice, at least with a C57BL/6J genetic background used in this study, this does not appear to be the case.
We found a few pancreatic lesions in Sufu+/−Ptch1+/− mice and one in a Ptch1+/−mouse. Similar types of lesions were also found in mice expressing an oncogenic Smoothened allele inserted into the Rosa26 locus [31]. This is in general concordance to observations made in humans, where aberrant Hh pathway activity has been linked to pancreatic neoplasms [32].
The relatively high incidence (∼20%) of HC seen here in Ptch1+/− and Sufu+/−Ptch1+/− mice is intriguing. In a previous study, the reported incidence of HC in Ptch1+/− was 5% [19]. The mice in this study were on a C57BL/6J background where the wt incidence of HC is normally in the order of 1-3% [33]. All mice with HC had the mutant Ptch1 allele, while no wt or Sufu+/− mice with HC were seen. So clearly there is a link to the Ptch1 mutant allele that makes these mice more prone to HC development. One possibility is that all cases of HC are secondary to small MBs, escaping detection during microscopic examination of tissue sections from these mice. An interesting alternative explanation is that there is a developmental defect in those Ptch1+/− mice with HC. For example, there could be a structural defect in the ventricles leading to obstruction of the normal flow of cerebrospinal fluid or a functional defect in keeping fluid homeostasis.
In summary, we have shown that compound Sufu+/−Ptch1+/− female mice have a statistically significant increase in the number of basaloid skin proliferations and a trend for an increase of MB compared to that observed in Sufu+/− and Ptch1+/− mice, respectively. This suggests a weak genetic interaction between the two tumor suppressor genes. We find it interesting that the phenotypes of the Sufu+/− and Ptch1+/− mice are not more similar to each other, especially when the germline homozygous loss-of-function mutants are virtually identical. The basis for this difference requires further studies and the use of conditional tissue-specific Ptch1 and Sufu KO mice will hopefully shed more light on this issue.
Supplementary information
ACKNOWLEDGEMENTS
We thank B.-M. Skog, Å. Bergström, G. Brolin, and C. Lundmark for technical assistance, and M. Kasper for technical advice on qRT-PCR.
Grant support: Swedish Research Council (S. Teglund), the Swedish Cancer Society and NIH/NCI MMHCC (R. Toftgård), and the Wallenberg Consortium North (B. Rozell).
Abbrevations
- BCC
basal cell carcinoma
- MB
medulloblastoma
- NBCCS
nevoid basal cell carcinoma syndrome
- RMS
rhabdomyosarcoma
- HC
hydrocephalus
- PL
pancreatic lesions
- Hh
Hedgehog
- Sufu
Suppressor of fused
- Ptch1
Patched1
- Smo
Smoothened
- GCP
granule-cell precursor
- EGL
external granule layer
- wt
wild-type
- PFA
paraformaldehyde
- X-gal
X-galactoside 5-bromo-4-chloro-3-indlyl-beta-D-galactopyranoside
- β-gal
β-galactosidase
REFERENCES
- 1.Rohatgi R, Scott MP. Patching the gaps in Hedgehog signalling. Nat Cell Biol. 2007;9:1005–1009. doi: 10.1038/ncb435. [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW, Placzek M. Orchestrating ontogenesis: variations on a theme by sonic hedgehog. Nat Rev Genet. 2006;7:841–850. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- 3.Pasca di Magliano M, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer. 2003;3:903–911. doi: 10.1038/nrc1229. [DOI] [PubMed] [Google Scholar]
- 4.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 5.Kogerman P, Grimm T, Kogerman L, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–319. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- 6.Svard J, Heby-Henricson K, Persson-Lek M, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Cooper AF, Yu KP, Brueckner M, et al. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132:4407–4417. doi: 10.1242/dev.02021. [DOI] [PubMed] [Google Scholar]
- 8.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 9.Hahn H, Wojnowski L, Zimmer AM, Hall J, Miller G, Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat Med. 1998;4:619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- 10.Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 11.Gorlin RJ. Nevoid basal cell carcinoma (Gorlin) syndrome. Genet Med. 2004;6:530–539. doi: 10.1097/01.gim.0000144188.15902.c4. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RL, Rothman AL, Xie J, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 13.Polkinghorn WR, Tarbell NJ. Medulloblastoma: tumorigenesis, current clinical paradigm, and efforts to improve risk stratification. Nat Clin Pract Oncol. 2007;4:295–304. doi: 10.1038/ncponc0794. [DOI] [PubMed] [Google Scholar]
- 14.Hahn H, Wojnowski L, Specht K, et al. Patched target Igf2 is indispensable for the formation of medulloblastoma and rhabdomyosarcoma. J Biol Chem. 2000;275:28341–28344. doi: 10.1074/jbc.C000352200. [DOI] [PubMed] [Google Scholar]
- 15.Wetmore C, Eberhart DE, Curran T. Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res. 2001;61:513–516. [PubMed] [Google Scholar]
- 16.Lee Y, Kawagoe R, Sasai K, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007 doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 17.Aszterbaum M, Epstein J, Oro A, et al. Ultraviolet and ionizing radiation enhance the growth of BCCs and trichoblastomas in patched heterozygous knockout mice. Nat Med. 1999;5:1285–1291. doi: 10.1038/15242. [DOI] [PubMed] [Google Scholar]
- 18.Milenkovic L, Goodrich LV, Higgins KM, Scott MP. Mouse patched1 controls body size determination and limb patterning. Development. 1999;126:4431–4440. doi: 10.1242/dev.126.20.4431. [DOI] [PubMed] [Google Scholar]
- 19.Wetmore C, Eberhart DE, Curran T. The normal patched allele is expressed in medulloblastomas from mice with heterozygous germ-line mutation of patched. Cancer Res. 2000;60:2239–2246. [PubMed] [Google Scholar]
- 20.Berman DM, Karhadkar SS, Hallahan AR, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 21.Oliver TG, Read TA, Kessler JD, et al. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 22.Romer JT, Kimura H, Magdaleno S, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/−)p53(−/−) mice. Cancer Cell. 2004;6:229–240. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y, Miller HL, Russell HR, Boyd K, Curran T, McKinnon PJ. Patched2 modulates tumorigenesis in patched1 heterozygous mice. Cancer Res. 2006;66:6964–6971. doi: 10.1158/0008-5472.CAN-06-0505. [DOI] [PubMed] [Google Scholar]
- 24.Pazzaglia S, Mancuso M, Atkinson MJ, et al. High incidence of medulloblastoma following X-ray-irradiation of newborn Ptc1 heterozygous mice. Oncogene. 2002;21:7580–7584. doi: 10.1038/sj.onc.1205973. [DOI] [PubMed] [Google Scholar]
- 25.Pazzaglia S, Tanori M, Mancuso M, et al. Two-hit model for progression of medulloblastoma preneoplasia in Patched heterozygous mice. Oncogene. 2006;25:5575–5580. doi: 10.1038/sj.onc.1209544. [DOI] [PubMed] [Google Scholar]
- 26.Magdaleno S, Jensen P, Brumwell CL, et al. BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor MD, Liu L, Raffel C, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 28.Koch A, Waha A, Hartmann W, et al. No evidence for mutations or altered expression of the Suppressor of Fused gene (SUFU) in primitive neuroectodermal tumours. Neuropathol Appl Neurobiol. 2004;30:532–539. doi: 10.1111/j.1365-2990.2004.00560.x. [DOI] [PubMed] [Google Scholar]
- 29.Adolphe C, Hetherington R, Ellis T, Wainwright B. Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res. 2006;66:2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 30.Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208:17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 31.Mao J, Ligon KL, Rakhlin EY, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dagg C. In: Biology of the Laboratory Mouse. Green E, editor. McGraw-Hill; New York: 1966. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.