The influenza vaccine failed this winter. Steven Salzberg suggests that future success relies on sharing data ore widely and making the virus strain selection process more transparent.
During the past 50 years, the scientific community has studied the influenza virus in great detail, and has developed an effective vaccine that is administered widely each year. The vaccine contains isolates from each of the three strains commonly circulating in humans: H3N2, H1N1 and influenza B. H3N2 has been the dominant strain of influenza A in most years, since it first emerged in 1968, and it is responsible for the most serious infections. The milder H1N1 has been co-circulating since it reappeared in Russia in 1977, and influenza B is milder yet. Because the virus mutates rapidly, the vaccine strains — especially H3N2 — need to be changed almost every year in order to remain effective. In some respects, the influenza-vaccine programme is a remarkable success: every year a new vaccine is developed and distributed, and most of the time it works.
This year, however, the vaccine was a failure: the strain of H3N2 that was used provided very little protection from infection. After a mild start dominated by H1N1 a new type of H3N2 emerged in mid-winter and quickly dominated, soaring to 71% of cases in the first 8 weeks of 2008 and overwhelming medical clinics in many places. A study in Wisconsin found the vaccine to be only 44% effective compared with the 70–90% effectiveness expected1, and a Harris Poll of more than 2,500 people revealed that for the first time in at least four winters, people who were vaccinated seemed no less likely to become infected2. The harm was thus twofold; people fell ill and their trust in the vaccine system was undermined. This failure could have been predicted, if not prevented, through a more open system of vaccine design, a stronger culture of sharing in the influenza research community and a serious commitment to new technologies for production. The habits of the vaccine community must change for the sake of public health.
What happened?
Twice each year, the World Health Organization (WHO) organizes a meeting with the directors of the four WHO Collaborating Centres and three Reference Laboratories to make recommendations for the composition of the influenza vaccine. The experts convene in February to determine the vaccine for the Northern Hemisphere, and in September for the Southern Hemisphere. The WHO publishes its recommendations immediately afterwards. In February 2007, the recommended strains for the 2007–08 season were: for H3N2, a 2005 strain from Wisconsin; for H1N1, a 2006 strain for the Solomon Islands; and for influenza B, a 2004 strain from Malaysia.
H1N1 dominated the 2006–07 season, and the WHO panel decided — correctly, it turned out — that the 1999 strain in that season’s vaccine was no longer effective. They also looked at surveillance data on H3N2, which showed that a new strain of H3N2 was emerging. According to a WHO report, “an increasing proportion of recent viruses showed antigenic differences from the vaccine virus”3. However, there was some uncertainty about what the new H3N2 strain would be, and furthermore “the lack of egg isolates precluded the selection of a new vaccine candidate”. In other words, the strains that seemed to match the most recent H3N2 cases could not be adapted to grow well in eggs. The committee therefore chose to take no action: they recommended that the vaccine continue to use the 2005 Wisconsin strain.
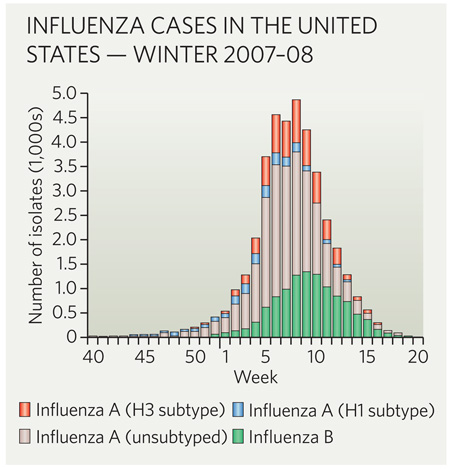
This strategy was likely to fail, given that the WHO already had evidence that H3N2 was changing. Had the season’s influenza A infections been caused mainly by H1N1, few would have noticed the H3N2 mismatch outside the influenza research community. But from January 2008, the profile changed, and H3N2 isolates dominated for the rest of the flu season (see graph).
The choice of the wrong strain for the vaccine had dire consequences: in addition to a rapid increase in H3N2 in early 2008 (a 17-fold increase in the first eight weeks of the year), pneumonia and influenza mortality exceeded epidemic levels — defined as 6.3% of all deaths reported to the Centers for Disease Control and Prevention (CDC) — for 19 consecutive weeks starting with the second week of 2008, according to CDC data from 122 US cities.
A closed process
The WHO met on 11–13 February this year to decide on the strains to be included in the vaccine for the 2008–09 season. As usual, the meeting was closed to all but invited participants, who this year included members of the WHO influenza surveillance network, representatives of national drug regulatory agencies, and influenza vaccine manufacturers4. The experts involved chose to replace the H3N2 strain with a more recent isolate, from 2007, which should be a better match to the circulating viruses next season. Neither the WHO nor the CDC publishes the evidence used to support their decision. That evidence includes hemagglutinin inhibition tests of hundreds of isolates, genome sequences of some isolates and data on the ease with which the isolates can be grown in eggs.
The process of choosing flu-vaccine strains needs to be much more open. Other scientists, such as those in evolutionary biology with expertise in sequence analysis, could meaningfully contribute to the selection. At present, external scientists cannot review the data that went into the decision, nor can they suggest other types of data that might improve it.
The selection of a vaccine strain has tremendous public-health consequences. Such a decision should be described in at least as much detail as a scientific publication. Although time constraints make formal peer review impractical, at a minimum all the data used to justify the selection should be immediately made public. The WHO and the CDC should encourage other scientists to re-analyse the data themselves and to critique the decision.
The leaders of the influenza community, especially the WHO and the CDC, should create policies — for sharing data and isolates — that are more open, and should insist that their own scientists follow those policies. When these leading organizations set an example, the rest of the community will follow.
Of course, preparing vaccine in cell culture could reduce some of the pressures put on that dark room of vaccine predictors. The current system, in which most of the world’svaccine supply is grown in chicken eggs, is an antiquated, inefficient method requiring six months or more to ramp up production, which in turn means that the vaccine strains must be chosen far in advance of each flu season. More crucially it sometimes prevents the use of the optimal strain, as it did in 2007. And, if the next pandemic is an avian-influenza strain such as H5N1, then it could easily sweep through the chicken farms that we rely on to produce eggs for vaccines. Cell-culture methods for influenza vaccines are under development in Europe and the United States, with strong government support — the National Institutes of Health (NIH) awarded a contract to pharmaceutical company Sanofi Pasteur in 2005 to work on these — but the investment required to gain approval remains a significant hurdle. These efforts need to be accelerated if we are to be ready for the next pandemic.
Share sequences and isolates
Genome sequences of currently circulating isolates, both human and avian, are among the most important sources of information about influenza. Despite growing calls for the rapid release of influenza sequence data, the practices of most major influenza research groups — including the WHO and the CDC’s own influenza centres, and most large, NIH-funded influenza centres in the United States — have been slow to change. Scientists at these centres hold onto biological samples and sequences for as long as possible, usually until a scientific article is published, and sometimes indefinitely.
This practice stands in stark contrast to the genome-sequencing community, which realized at least ten years ago that society reaps the greatest benefit when scientific data are shared rapidly and openly. Rapid sharing was a major tenet of the Human Genome Project, and it has become a requisite for publicly funded research in the United States and United Kingdom. In 2004, in an effort to establish a similar model for influenza, David Lipman and I started the Influenza Genome Sequencing Project, an NIH-funded effort dedicated to sequencing as many influenza genomes as possible, with the goal of dramatically expanding our knowledge of influenza epidemiology and evolution5. The project has just finished its 3,000th genome, and all data have been deposited in GenBank, the publicly available NIH genetic sequence database.
The WHO and the CDC have stated publicly that they support placing sequence data in the public domain6,7. Unfortunately, the WHO’s own centres do not release all their influenza sequences, and when they do, they often use the Los Alamos National Laboratory influenza database. This database is, as reported on its own website, “a private database for collaborators” — access is restricted to a private group of subscribers. A closed database limits the free exchange that is so important to scientific research, and it sets the wrong example.
The database created recently by the Global Initiative on Sharing Avian Influenza organization (GISAID, www.gisaid.org) might represent a new model for influenza data sharing. GISAID allows registered users to have access to all its data with a few restrictions, designed to address legitimate concerns that some scientists and governments have expressed: users must agree to acknowledge the data providers, and they are expected not to make intellectualproperty claims that might tie up the data or restrict use for others. The database is still too new to know if it will succeed in its goal of encouraging greater data sharing, but if it does, it could be an important step forward.
We can’t control how the influenza virus mutates from year to year. But we can do much more to track those mutations, and by sharing what we learn, we can increase our odds of beating the flu.
References
- 1. www.cdc.gov/mmwr/preview/mmwrhtml/mm5715a1.htm.
- 2. www.harrisinteractive.com/harris_poll/index.asp?PID=906.
- 3. www.who.int/csr/disease/influenza/20078anorthreport.pdf.
- 4. www.who.int/csr/disease/influenza/vaccine_north2008_9/en/index.html.
- 5.Ghedin E, et al. Nature. 2005;437:1162–1116. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 6. www.who.int/csr/disease/avian_influenza/guidelines/h5n1sequences2006_08_23/en/index.html.
- 7. www.cdc.gov/od/oc/media/pressrel/r060822.htm.