Abstract
At least 11 complementation groups (CGs) have been identified for the peroxisome biogenesis disorders (PBDs) such as Zellweger syndrome, for which seven pathogenic genes have been elucidated. We have isolated a human PEX19 cDNA (HsPEX19) by functional complementation of peroxisome deficiency of a mutant Chinese hamster ovary cell line, ZP119, defective in import of both matrix and membrane proteins. This cDNA encodes a hydrophilic protein (Pex19p) comprising 299 amino acids, with a prenylation motif, CAAX box, at the C terminus. Farnesylated Pex19p is partly, if not all, anchored in the peroxisomal membrane, exposing its N-terminal part to the cytosol. A stable transformant of ZP119 with HsPEX19 was morphologically and biochemically restored for peroxisome biogenesis. HsPEX19 expression also restored peroxisomal protein import in fibroblasts from a patient (PBDJ-01) with Zellweger syndrome of CG-J. This patient (PBDJ-01) possessed a homozygous, inactivating mutation: a 1-base insertion, A764, in a codon for Met255, resulted in a frameshift, inducing a 24-aa sequence entirely distinct from normal Pex19p. These results demonstrate that PEX19 is the causative gene for CG-J PBD and suggest that the C-terminal part, including the CAAX homology box, is required for the biological function of Pex19p. Moreover, Pex19p is apparently involved at the initial stage in peroxisome membrane assembly, before the import of matrix protein.
Peroxisomal proteins, including membrane proteins, are encoded by nuclear genes, translated on free polyribosomes in the cytosol (1). Peroxisomes are thought to be formed by division of preexisting peroxisomes after import of newly synthesized proteins (1). It is noteworthy that recent evidence suggests involvement of the endoplasmic reticulum in peroxisomal membrane biogenesis in yeast (2). Peroxisomal functions are highlighted by the existence of fatal human genetic peroxisomal biogenesis disorders (PBDs), such as Zellweger syndrome (ZS), neonatal adrenoleukodystrophy, and infantile Refsum disease (3). In addition, rhizomelic chondrodysplasia punctata manifesting the defect in import of peroxisome-targeting signal type 2 (PTS2) proteins (3) is identified as a group of PBDs. In mammals, including humans, genetic heterogeneity comprising 13 complementation groups (CGs) has been found in peroxisome deficiency (3–9). This finding implies that more than 13 genes are involved in mammalian peroxisome assembly. Much progress has been made in the identification of a number of protein factors, called peroxins, that are essential for peroxisome assembly by genetic analysis of peroxisome-deficient mutants of yeast and mammalian cells (3, 10–12). To date, seven peroxin cDNAs have been cloned in mammals by genetic phenotype-complementation assay of Chinese hamster ovary (CHO) cell mutants and by the expressed sequence tag search of the human database by using the yeast PEX genes.
To investigate molecular mechanisms involved in peroxisome biogenesis and the genetic cause of PBD, we have so far isolated seven CGs of peroxisome-deficient CHO cell mutants (4–7, 13) (see Table 2). Very recently, we identified CG-J (8). Two CHO cell mutants, ZP119 and ZP165, were also found to belong to this group (8). In no CG-J mutant cell were peroxisomal ghosts found (8). We isolated PEX1, PEX2 (formerly PAF-1), PEX5, PEX6, and PEX12 by genetic phenotype-complementation assay of CHO cell mutants ZP107, Z65, ZP105/ZP139, ZP92, and ZP109, respectively (13–18), and demonstrated that these PEX genes are defective in the various patients with PBD (13, 14, 17–20). Thus, peroxisome biogenesis-defective CHO cell mutants are a useful mammalian somatic cell system for the investigation of peroxisome assembly at the molecular and cellular level, as well as for delineation of the genetic basis of PBD (3). However, until now no peroxins evidently required for the biogenesis of peroxisomal membranes have been elucidated in mammals.
Table 2.
Complementation of CHO cell mutants and patient fibroblasts by HsPEX19
| CHO mutant | Peroxisomepositive clone | Patient fibroblasts from CG | Peroxisomepositive | Gene |
|---|---|---|---|---|
| ZP119 | 24/30 | J (PBDJ-01) | + | PEX19 |
| ZP165 | 21/30 | |||
| ZP107 | − | E (I) | − | PEX1 |
| ZP139 | − | II | − | PEX5 |
| ZP109 | − | III | − | PEX12 |
| ZP92 | − | C (IV) | − | PEX6 |
| (VI) | ND | |||
| B (VII) | − | PEX10 | ||
| A (VIII) | − | |||
| D (IX) | − | PEX16 | ||
| Z65 | − | F (X) | − | PEX2 |
| G | − | |||
| ZP110 | − | |||
| ZP114 | − |
Peroxisome-deficient CHO mutants of 8 CGs (4–8, 13–18) and fibroblasts from 10 CG PBD patients (3, 4, 8, 9) were transfected with pUcD2Hyg⋅HsPEX19 and examined for peroxisome assembly by immunostaining with antisera to rat and human catalase, respectively, at 3 days after transfection. Parentheses indicate fibroblasts of CG not used in this experiment. In CG-J ZP119 and ZP165, peroxisome-positive colonies were counted in 30 colonies; in other cells: +, complemented; −, not complemented; ND, not determined.
In the study reported here, we isolated human PEX19 cDNA (HsPEX19) encoding a farnesylated protein that restored peroxisome assembly in a CHO cell mutant ZP119 devoid of peroxisomal membrane vesicles (8). HsPEX19 expression also complemented impaired peroxisome biogenesis in fibroblasts from a patient with CG-J PBD. In a patient with CG-J, we identified a mutation site that inactivated Pex19p. The biological function of Pex19p is also discussed.
MATERIALS AND METHODS
Cell Lines.
Patient skin fibroblast cell lines and CHO cell mutants were cultured as described (4, 5). ZP119EG1, ZP119 stably expressing “enhanced” green fluorescent protein (EGFP) tagged with PTS1 (EGFP-PTS1), was isolated by transfecting pUcD2Hyg⋅EGFP-PTS1, as described (14).
Screening of a cDNA Library.
A human liver cDNA library in pCMVSPORT vector divided into small pools, each containing about 4,000 clones (14), was transfected to ZP119 cells with Lipofectamine (GIBCO/BRL), as described (14, 18). Among the cDNA pools examined, a positive one (F6–19) that restored peroxisomes in ZP119 was further divided into subpools and screened. A single positive clone, F6-19-30, was isolated. The nucleotide sequence of ≈900 bp on the 5′ side of F-6–19-30 showed a sequence to be identical to human HK-33 (21) but shorter by nine nucleotides downstream of the initiation codon in HK-33. To construct a full-length cDNA, the lacking nine nucleotides were inserted as follows. PCR was done by using the F6-19-30 as a template, with primers, 5′-CGCGGATCCAAGATGGCCGCCGCTGAG-3′ (initiation codon, italic) and 5′-TGTGGATCCTCACATGATCAGACACTG-3′ (termination codon, italic). The BamHI fragment (nucleotide residues −8–905) of the PCR product was inserted into the BamHI site of the pUcD2SRαMCSHyg vector (18). The resulting plasmid was termed pUcD2Hyg⋅HsPEX19.
Transfection of PEX19.
CHO cell mutants were transfected with pUcD2Hyg⋅HsPEX19, as described above. A stable clone of the HsPEX19 transformant of ZP119, 119P19, was isolated by transfection of pUcD2Hyg⋅HsPEX19 and then by selection with hygromycin B and limiting dilution. Patient fibroblasts were transfected with pUcD2Hyg⋅HsPEX19 by electroporation, as described (14).
Expression of Epitope-Tagged Pex19p.
An epitope flag tagging to the N terminus of Pex19p was done by a PCR-based technique. HsPEX19 (nucleotide residues 4–900) was amplified using a forward primer HsPEX19.FLAG.F (5′-GCGAACTAGTGGATCCAGGCCGCCGCTGAGGAAGGCTG-3′) and a reverse primer HsPEX19.FLAG.R (5′-CTGCCTCGAGGTACCTCACATGATCAGACACTGTTC-3′). The PCR products were digested with BamHI and KpnI and then ligated into the BamHI–KpnI site of a vector fragment containing a flag tag sequence, originated from pUcD2Hyg⋅flag-RnPEX12 (18). Flag-tagged Pex19p was detected by using anti-flag antibody, as described (18).
Morphological Analysis.
Peroxisomes in CHO cells and human fibroblasts were visualized by indirect immunofluorescence light microscopy, as described (4, 14), with rabbit antibodies to rat liver catalase (5), human catalase (4), PTS1 (13), and a 70-kDa peroxisomal integral membrane protein (PMP70) (5, 8).
Mutation Analysis.
Reverse transcription (RT)–PCR, with poly(A)+ RNA prepared as described (14), was done with a pair of human PEX19-specific PCR primers: sense RT1, 5′-AAGATGGCCGCCGCTGAGGAAG-3′ (initiation codon, italic), and antisense RT2, 5′-CGTGTTGTGTTTCAC-3′ (termination codon, italic), to cover the full length of the PEX19 ORF. The nucleotide sequence of the PCR products cloned into pGEM-T Easy Vector (Promega) was determined. Patient PEX19 cDNA was cloned into the pUcD2Hyg vector (18) by inserting its NotI fragment (residues at −3–911) in pGEM-T Easy Vector. Genomic DNA was prepared from cultured fibroblasts (20) with a QIAamp tissue kit (Qiagen, Chatsworth, CA). To investigate the zygosity of the PEX19A764ins allele in patient PBDJ-01, PCR amplification of the sequence between residues 691 and 808 was done with the genomic DNA and a pair of PEX19-specific primers: sense 19GF, 5′-CAGTTTGAGGCAGAGACC-3′, and antisense 19GR, 5′-CAGCCAGCTCTTTTGGAG-3′. PCR products were extracted and sequenced directly.
In Vitro Translation.
Anti-Pex19p antibody was raised in rabbits by immunizing them with synthetic peptide comprising the C-terminal, 19-aa sequence (see Fig. 2, underline) supplemented with cysteine at the N terminus that had been linked to key-hole limpet hemocyanin (15). In vitro transcription/translation was done essentially as described (22). Translation in a rabbit reticulocyte cell-free protein-synthesizing system was performed with 1.2 mCi/ml [35S]methionine and [35S]cysteine (Amersham Pharmacia) or 80 μCi/ml [3H]farnesyl pyrophosphate (New England Nuclear). Immunoprecipitation of Pex19p was done as described (22).
Figure 2.
Amino acid sequence alignment of PEX19 protein from two mammalian species and S. cerevisiae Pex19p. Deduced amino acid sequence of human (Hs) PEX19 was compared with those of Pex19p from Chinese hamster (Cl) and S. cerevisiae (Sc). -, a space. Identical amino acids between species, including two mammalian ones, are shaded, and the CAAX motif is boxed. The sequence used for chemical synthesis of Pex19p peptide is underlined. The solid arrowhead indicates the position of mutation in CG-J PBD patient (see Fig. 4). The database accession numbers for the human PEX19 cDNA is AB018541.
Other Methods.
The nucleotide sequence was determined by the dideoxy-chain termination method with a dye terminator DNA sequencing kit (Applied Biosystems). Alignment was done with the genetyx-mac program (Software Development, Tokyo). Western blot analysis was done with rabbit antibodies to rat acyl-CoA oxidase, 3-ketoacyl-CoA thiolase (thiolase), and Pex19p, as described (14, 18). The catalase latency assay with digitonin was done as described (5). P12/UV and P9OH/UV resistances were determined under conditions of 2 μM for 1.5 min and 6 μM for 2 min, respectively (4).
RESULTS
Cloning of a Human PEX19 cDNA.
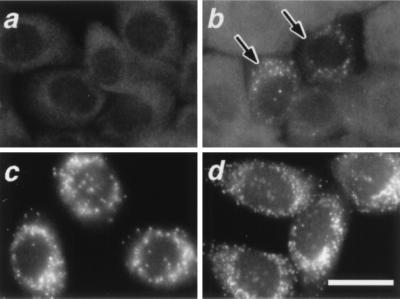
We used a transient expression assay (14, 18) to clone the PEX19 cDNA, making use of a human liver cDNA library. After transfection with a small pool of cDNA library, we searched for cells in which peroxisome biogenesis was restored, by staining ZP119 with anti-rat catalase antibody. In ZP119, catalase was localized in the cytosol (Fig. 1a), consistent with our earlier observation (8). Twenty-seven pools, which contained about 1.1 × 105 independent cDNA clones, were screened, and one pool (F6-19) yielded numerous catalase-positive particles, presumably peroxisomes, in several of ZP119 cells in a single dish (Fig. 1b, arrows). After a third round of screening, we isolated one positive clone, F6-19-30, that contained a 1.8-kb cDNA with an ORF encoding a 209-aa protein. A homology search suggested that this ORF was likely to be related to PEX19 from the yeast Saccharomyces cerevisiae (23) and identical to the amino acid sequence from residues at 91 to the C terminus of human HK33 (21), whereas the deduced sequence of isolated clone F6-19-30 was shorter by 90 aa compared with the sequence of HK33. F6-19-30 started the cDNA sequence nine nucleotides downstream of the initiation codon in the HK33, i.e., 261 nt upstream of a putative initiation codon in F6-19-30. Accordingly, we constructed a cDNA, termed HsPEX19, encoding the full-length, 299-aa PEX19 protein of 32,806 Da, Pex19p, by inserting a 9-mer oligonucleotide, ATGGCCGCC (see Materials and Methods). HsPEX19 complemented peroxisomal import of catalase more efficiently than F6-19-30 in both ZP119 and ZP165 (data not shown). The human Pex19p was shorter by 51 aa than S. cerevisiae Pex19p (23), with 20% amino acid identity, showing 92% identity to the same-length Chinese hamster (Cl) Pex19p (24) (Fig. 2). Hydropathy analysis suggested that Pex19p is a hydrophilic protein with no apparent membrane-spanning region (data not shown).
Figure 1.
Restoration of peroxisomes in CG-J CHO mutant cells. (a) Peroxisome-deficient mutant ZP119 cells. (b) Peroxisome-restored ZP119, after lipofection with a combined pool (F6-19) of human cDNA library. Arrows indicate the complemented cells. Cytosolic appearance of catalase was apparent in the other cells. (c and d) 119P19 cells, stable HsPEX19-transformants of ZP119 cells. Cells were stained with antisera to catalase (a and b), PTS1 peptide (c), and PMP70 (d), respectively. (Magnification: ×630; bar = 20 μm.)
Restoration of Peroxisome Biogenesis in ZP119 by PEX19.
In 119P19, a stable HsPEX19 transformant of ZP119, numerous peroxisomes were detected by immunofluorescent staining of PTS1 and PMP70 (Fig. 1 c and d), which were seen in a diffuse staining in ZP119 (data not shown; ref. 8). Import of peroxisomal thiolase, a PTS2 protein, was also restored (data not shown). These results demonstrated that 119P19 cells had morphologically normal peroxisomes, as seen in the wild-type CHO-K1 cells.
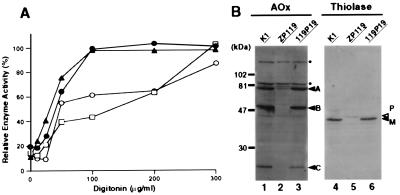
In peroxisome-deficient cells, peroxisomal proteins are mislocalized to the cytosol, rapidly degraded, or not converted to mature forms, despite normal synthesis (4–8, 13, 14). According to results of the digitonin titration assay, nearly 60% of catalase activity was latent at 100 μg/ml digitonin in the wild-type cells, consistent with earlier observations (5–8, 13, 14) (Fig. 3A; Table 1). In ZP119 cells, full activity of catalase was detected at 100 μg/ml digitonin, which was also the case for a cytosolic enzyme, lactate dehydrogenase (Fig. 3A), suggesting that catalase is present in the cytosol, in agreement with the morphological observation (Fig. 1a; ref. 8). In 119P19 cells, catalase latency was comparable to that of the wild-type CHO-K1 cells (Fig. 3A). Moreover, 119P19 was resistant to P12/UV treatment and sensitive to P9OH/UV, similar to CHO-K1 (Table 1). In contrast, mutant ZP119 was resistant to P9OH/UV treatment and sensitive to P12/UV (8).
Figure 3.
Complementation of biogenesis of peroxisomal enzymes. (A) Latency of catalase activity in CHO-K1, ZP119, and 119P19 cells. □, CHO-K1; ●, ZP119, ○, 119P19; ▴, lactate dehydrogenase in ZP119. Relative free enzyme activity is expressed as a percentage of the total activity measured in the presence of 1% Triton X-100 (5). The results represent a mean of duplicate assays. (B) Biogenesis of peroxisomal proteins. Cell lysates (≈1.8 × 105 cells) were subjected to SDS/PAGE and transferred to polyvinylidene difluoride membrane. Cell types are indicated at the top. Immunoblotting was done with antibodies to acyl-CoA oxidase and thiolase. Arrowheads show the positions of acyl-CoA oxidase components, A, B and C; open and solid arrowheads indicate a larger precursor (P) and mature protein (M) of thiolase, respectively. Dots indicate nonspecific bands (14).
Table 1.
Properties of wild-type CHO-K1, ZP119, and HsPEX19 transfected ZP119 (119P19) cells
| Cell | Peroxisome | Catalase latency, % | P12/UV, % | P9OH/UV, % |
|---|---|---|---|---|
| CHO-K1 | + | 57 | 83 | <0.03 |
| ZP119 | − | 0.6 | <0.01 | 98 |
| 119P19 | + | 39 | 73 | 12 |
Catalase latency represents peroxisomal catalase, calculated as described (5). For determination of P12/UV or P9OH/UV resistance, 200 or 1 × 105 cells were inoculated into 60-mm dishes and selected (4). The numbers of colonies were counted in duplicate experiments and expressed as percentages of that of unselected control.
Acyl-CoA oxidase is synthesized as a 75-kDa polypeptide (A component) and is proteolytically converted into 53- and 22-kDa polypeptides (B and C, respectively) in peroxisomes (22). In immunoblots, all three polypeptide components, A, B and C, were present in CHO-K1 as well as in 119P19 (Fig. 3B, lanes 1 and 3), but only the A component was seen in a smaller amount in the ZP119 (lane 2). Peroxisomal thiolase, a PTS2 protein, is synthesized as a larger 44-kDa precursor and undergoes maturation to the 41-kDa form in peroxisomes (14, 18). In wild-type CHO-K1 and 119P19 cells, only the 41-kDa mature thiolase was detected (Fig. 3B, lanes 4 and 6), whereas only the larger precursor was found in ZP119 (lane 5).
Taken together, these results indicate that HsPEX19 can fully complement the ZP119 mutation. HsPEX19 expression also complemented peroxisome assembly in ZP165 cells, the same CG as ZP119 (8), but did not restore peroxisome formation in six other CGs of peroxisome-deficient CHO cell mutants so far isolated (4–7), Z65, ZP92, ZP105, ZP109, ZP110, and ZP114. These data show that Pex19p is the peroxisome biogenesis factor for ZP119/ZP165 (Table 2).
PEX19 Specifically Complements Fibroblasts from CG-J Patient with ZS.
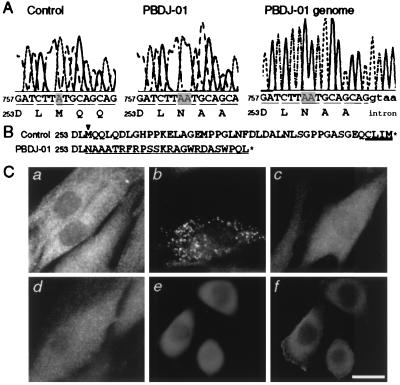
ZP119 and ZP165 belong to the same CG as human CG-J (8, 9). HsPEX19 was introduced into fibroblasts of 10 CGs of peroxisomal diseases, i.e., CGs A, B, C, D, E, F, G, and J of Gifu University, Japan, and groups II and III of the Kennedy-Krieger Institute, Baltimore. As expected, the fibroblasts derived from a CG-J patient (PBDJ-01) with ZS were morphologically restored for import of catalase (Fig. 4 Ca and Cb), but no restoration of peroxisome formation was seen in fibroblasts from the other, nine CGs (Table 2). Restoration of peroxisome assembly was also assessed by staining the transfectants with antibodies to PTS1 and PMP70 (data not shown). These results strongly suggest that HsPEX19 is the causal gene of CG-J PBD.
Figure 4.
Complementation of fibroblasts from a CG-J Zellweger patient. (A) Partial sequence and deduced amino acid sequence of PEX19 cDNA isolated from a normal control (Left) and a ZS patient PBDJ-01 (Center) are shown. PCR was also done for DNA from PBDJ-01 fibroblasts (Right). One-base insertion A764 (shaded), in a codon for Met255, causes a frameshift in PBDJ-01 PEX19 sequence. (B) Sequence comparison of the C-terminal part of Pex19p each from a control and a patient PBDJ-01. Amino acid sequence resulted from the frameshift by the 1-bp insertion in PBDJ-01 is underlined. The arrowhead indicates the position of frameshift mutation. The CAAX box is double-underlined. (C) Transfection of PEX19 from a normal control and a CG-J patient (PBDJ-01). (a) PBDJ-01 fibroblasts. (b) PBDJ-01 fibroblasts were transfected with pUcD2Hyg⋅HsPEX19. (c and d) PBDJ-01 fibroblasts were transfected with PBDJ-01-derived PEX19 cDNA, PEX19A764ins, and a mutant PEX19, PEX19C296S, respectively. (e and f) flag-tagged PEX19C296S, flag-PEX19C296S, was expressed in ZP119. Cells were stained with antibodies to human catalase (a–d), rat catalase (e), and flag (f). Note that peroxisomes were restored only in b, but not in c–f. (Bar = 20 μm.)
CG-J Patient Analysis.
To determine dysfunction of PEX19 in the patient PBDJ-01, we isolated PEX19 cDNA from PBDJ-01 fibroblasts by RT-PCR. A point mutation was detected by subsequent sequencing: a 1-base insertion, A764, in a codon for Met255, named PEX19A764ins, resulted in a frameshift, inducing a 24-aa sequence distinct from that of normal Pex19p (Fig. 4A Left and Center and B). All of the 10 cDNA clones isolated showed the same site mutation, evidently implying that the patient was a homozygote for the mutation. To confirm the homozygosity of a PEX19A764ins allele, genomic PCR was done to amplify the sequence corresponding to nucleotide residues 691–808 in the PEX19 ORF. Only a single type of nucleotide sequence giving rise to the A insertion was identified in the PCR product (Fig. 4A Right), indicating that patient PBDJ-01 with ZS was a homozygote for the A764 insertion. This one-point mutation inactivated PEX19, as assessed by back-transfection of PEX19A764ins to both PBDJ-01 fibroblasts (Fig. 4Cc) and ZP119 cells (data not shown). Collectively, these data demonstrate that the dysfunction of PEX19 is responsible for CG-J peroxisome deficiency.
Pex19p Is Prenylated.
To investigate whether a conserved cysteine in the CAAX motif is essential for the function of Pex19p, Cys296 was mutated to Ser. Expression of C296S-mutated HsPEX19, termed HsPEX19C296S, could not rescue the impaired assembly of peroxisomes in PBDJ-01 fibroblasts (Fig. 4Cd) and ZP119 (data not shown). Flag-HsPEX19C296S was also inactive in complementing ZP119, where its expression was assessed (Fig. 4 Ce and Cf). Therefore, prenylation of Pex19p is most likely essential for its biological activity.
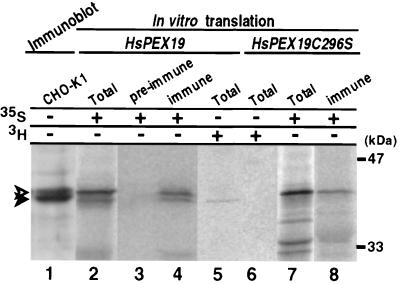
Rabbit antibody raised against the C-terminal peptide (residues 276–294) of Pex19p reacted with two protein bands with apparent masses of 40 kDa and 39 kDa, respectively, in immunoblots of CHO-K1 cells (Fig. 5, lane 1). The mobility of these two bands shown by SDS/PAGE was indistinguishable from that of Pex19p likewise showing two bands, synthesized in vitro by using [35S]methionine and [35S]cysteine by coupled transcription/translation of HsPEX19 (lane 2). Both 35S-Pex19p proteins were specifically immunoprecipitated by anti-Pex19p antibody but not by preimmune serum (lanes 3 and 4), thereby indicating that two bands were Pex19p and that a cloned HsPEX19 encoded bona fide Pex19p. In vitro synthesized Pex19p using [3H]farnesyl pyrophosphate as a prenyl-label showed a 3H-protein band (Fig. 5, lane 5), exactly with the same mobility as 35S-Pex19p as well as an immunoblotted band with a higher mobility (lanes 1, 2, and 4), thus demonstrating that the lower band corresponds to the farnesylated Pex19p. Moreover, the transcription/translation product of HsPEX19C296S, 35S-labeled Pex19pC296S, migrated as a single band with the same mobility as the larger 35S-Pex19p and was immunoprecipitated by anti-Pex19p antibody (Fig. 5, lanes 7 and 8). In contrast, transcription/translation of HsPEX19C296S with [3H]farnesyl pyrophosphate resulted in no 3H-labeled protein (Fig. 5, lane 6), indicating that Pex19pC296S was not prenylated. Together, the results demonstrate that Pex19p is partly farnesylated in vivo (see Fig. 5, lane 1).
Figure 5.
Farnesylation of Pex19p. Size comparison of in vitro transcription/translation product of normal and mutated PEX19 cDNA and Pex19p of CHO cells. In vitro transcription/translation product of HsPEX19 and CHO-K1 cell-lysates were subjected to SDS/PAGE. Immunodetection was done for lane 1, as in Fig. 3B, by using anti-Pex19p antibody; radioactive bands were detected by a FujiX BAS1500 Bio-Imaging Analyzer at exposures for 16 h (lanes 2–4, 7, and 8) and 72 h (lanes 5 and 6). Lanes: 1, 40 μg of CHO-K1 cell-lysates; 2, in vitro transcription/translation product (1 μl) of HsPEX19 in the presence of [35S]methionine and [35S]cysteine as label; 3 and 4, immunoprecipitation of 35S-Pex19p (3.5 μl) was done with preimmune and anti-Pex19p immune sera, respectively; 5 and 6, in vitro transcription/translation product (15 μl) of HsPEX19 and HsPEX19C296S, respectively, using [3H]farnesyl pyrophosphate as label; 7 and 8, total (1 μl) and immunoprecipitate (3.5 μl) from in vitro transcription/translation product of HsPEX19C296S using [35S]methionine/cysteine. Solid and open arrowheads indicate farnesylated and nonfarnesylated Pex19p, respectively (see text).
Intracellular Location of Pex19p.
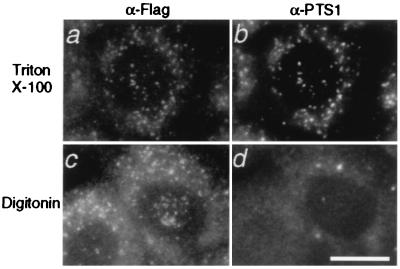
Subcellular localization of Pex19p was determined by immunofluorescent microscopy of flag-Pex19p ectopically expressed in CHO-K1 cells. Flag-Pex19p was detected in both punctate structures and a diffuse staining pattern in the cytoplasm (Fig. 6a). The punctate pattern was superimposable on that obtained with anti-PTS1 antibody (Fig. 6b), thereby suggesting that flag-Pex19p was localized partly to peroxisomes. Punctate staining was hardly discernible, however, in flag-Pex19pC296S-expressing CHO-K1 as expressed in ZP119 (Fig. 4Cf), suggesting that prenylation is required for peroxisomal localization (data not shown). Membrane topology of Pex19p was determined by a differential permeabilization procedure. Upon cell permeabilization with 25 μg/ml digitonin (17, 18), flag-Pex19p was likewise observed in a punctate staining pattern, whereas there was hardly any staining of cells with anti-PTS1 antibody (Fig. 6 c and d), thus strongly suggesting that the N-terminal part of peroxisomal Pex19p was exposed to the cytosol, presumably anchored by the farnesylated C terminus (see Figs. 2 and 5). A diffuse flag-staining was also seen, suggesting the cytosolic localization of a part of Pex19p (Fig. 6c). It is noteworthy that ScPex19p (23) and ClPex19p, originally named PxF (24), were also shown to be localized to peroxisomes.
Figure 6.
Intracellular localization and topology of Pex19p. N-terminally flag-tagged human Pex19p was expressed in CHO-K1 cells. Cells were treated with 0.1% Triton X-100 (a and b), or with 25 μg/ml of digitonin, under which the plasma membrane was permeabilized (17, 18) (c and d). Cells were stained with antibodies to flag (a and c) and PTS1 (b and d). Note that punctate structures, peroxisomes, are superimposable in a and b and that flag-Pex19p was detected after both types of treatments (a and c). A diffuse staining pattern was partly detected in a and c (see text). (Bar = 20 μm.)
Kinetics of Peroxisome Biogenesis.
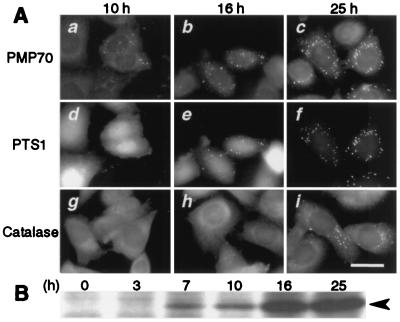
We investigated the kinetics of peroxisome assembly with respect to membrane vesicle formation and matrix protein import. ZP119EG1, ZP119 stably expressing EGFP-PTS1, was transfected with HsPEX19 and monitored under a fluorescent microscope. HsPex19p was detectable by immunoblot analysis at 7 h after the transfection and increased with time (Fig. 7B). At 10 h, PMP70 became visible in punctate structures in a part of transfected cells, possibly representing assembled peroxisomal membranes, whereas EGFP-PTS1 showed a diffuse fluorescence pattern implying localization in the cytoplasm and nucleus (Fig. 7 Aa and Ad) as seen in the untransfected cells (8). At 16 h, PMP70-positive vesicles increased in number, which were then partly colocalized with EGFP-PTS1 (Fig. 7 Ab and Ae), although EGFP-PTS1 still showed a predominantly diffuse pattern. This was interpreted to mean that a part of the assembled peroxisomal membrane vesicles imported EGFP-PTS1. At 25 h, numerous PMP70-positive punctates were superimposable with those of EGFP-PTS1, demonstrating re-establishment of membrane assembly and matrix protein import of peroxisomes (Fig. 7 Ac and Af). In contrast, catalase was noticeably imported only at 25 h after the HsPEX19 transfection (Fig. 7 Ag–Ai), into PTS1-positive vesicles (data not shown), suggesting the import of catalase at a slower rate as compared with PTS1. Collectively, peroxisomal membrane vesicles containing PMP70 are likely to form before the import of matrix proteins. The import kinetics of matrix proteins appears to be variable.
Figure 7.
Kinetics of peroxisome biogenesis. (A) ZP119EG1 cells expressing EGFP-PTS1 were transfected with pUcD2Hyg⋅HsPEX19, then monitored by fluorescent microscope. (a–c) PMP70 was visualized by using rabbit anti-PMP70 antibody and Texas Red-labeled goat anti-rabbit IgG antibody. (d–f) EGFP-PTS1. (g–i) Catalase in other cells detected by anti-catalase antibody, as for PMP70. (a, d, and g) 10 h after transfection, (b, e, and h) 16 h. (c, f, and i), 25 h. Note that PMP70, but not EGFP-PTS1 and catalase, is already in numerous vesicular structures at 10 h (see text). (Bar = 20 μm.) (B) Expression of human Pex19p in ZP119. Pex19p was detected by immunoblotting HsPEX19-transfected ZP119 lysates (1.3 × 105 cells at 0 h), at indicated time, where cell-doubling time was 22 h. Arrowhead, Pex19p.
DISCUSSION
Our earlier work showed that CHO cell mutants, ZP119 and ZP165, were defective in import of both matrix proteins and membrane polypeptides, a phenotype distinct from other CGs cells (8). Peroxisomal remnants were seen in the other 7 CGs of CHO cell mutants (4, 6, 7, 13) and 7 CGs of fibroblasts from PBD patients (25, 26). In the present work, we isolated a human Pex19p cDNA by functional complementation of peroxisome-deficient CG-J CHO cell mutants, although the initially isolated PEX19 cDNA clone was shorter by 9 nucleotides. The result implies that Pex19p translated at the second potential initiator methionine, comprising the primary sequence from residues 91–299 of the full-length Pex19p, is functional in complementing the impaired assembly of peroxisomes in CG-J cells. The entire N-terminal region of 90 amino acids appears not be required. Expression of the full-length HsPEX19 fully restored peroxisome biogenesis, including membrane assembly, in ZP119 and ZP165. Restoration of peroxisome-deficiency in fibroblasts from a CG-J patient indicated that the dysfunction of PEX19 is causal in this group. We delineated the homozygous mutant PEX19 allele from a CG-J patient: one-base insertion, A764, in a codon for Met255, resulted in a frameshift, inducing a 24-amino acid sequence entirely different from the normal sequence. Accordingly, PEX19 is the eighth gene identified to date responsible for the peroxisome-deficient diseases (Table 2). Moreover, given the fact that a mutation C296S abolished the complementation of peroxisomes in ZP119 and CG-J patient fibroblasts, we conclude that prenylation is essential for the biological activity of Pex19p. Very recently, Goette et al. (23) reported similar findings, including farnesylation of S. cerevisiae Pex19p being required for peroxisome biogenesis and growth in oleic acid.
Upon transfection of HsPEX19 into ZP119 devoid of peroxisomal “ghosts,” most striking was the formation of peroxisomal membranes, apparently followed by import of matrix proteins such as PTS1 proteins and catalase. To our knowledge, this is the first observation of the membrane assembly process that is temporally differentiated from and precedes the import of soluble proteins during peroxisome biogenesis. Therefore, it is possible that Pex19p functions as a factor in membrane protein-translocation process and/or membrane vesicle assembly. This result may also suggest that peroxisome can form de novo and does not have to arise from pre-existing, morphologically recognizable peroxisomes. It is noteworthy that Pex3p expression complemented peroxisome biogenesis in two yeast mutants, Hansenula polymorpha pex3 and Pichia pastoris pex3, apparently absent from peroxisomal structures (27, 28). Pex3p is more likely to be involved in peroxisomal membrane biogenesis. Moreover, import of PTS1 proteins and catalase at a different rate in PEX19-transfected ZP119 implies temporally differential translocation of some matrix proteins, if not all, into peroxisomal membrane vesicles. Interestingly, a similar differential import has been observed in rat liver, where several matrix proteins, including catalase and urate oxidase, a PTS1 protein, were translocated from the site of synthesis, the cytosol, to peroxisomes at various half times (29). It is plausible that Pex19p interacts with other PEX proteins (3, 10, 12, 30, 31), including both cytoplasmic and membrane peroxins. It is noteworthy that S. cerevisiae Pex19p binds Pex3p (23).
Further investigation, with ZP119 and ZP165 together with PEX19, should shed light on the molecular mechanisms involved in peroxisome biogenesis, and will allow dissection of the mechanism of membrane vesicle formation and import of matrix enzymes at the molecular level.
Acknowledgments
We thank T. Utsumi for helpful advice concerning the in vitro prenylation assay and the members of the Fujiki laboratory for discussion. This work was supported in part by a Core Research for Evolutional Science and Technology (CREST) grant (to Y.F.) from the Science and Technology Corporation of Japan and Grants-in-Aid for Scientific Research (08557011 to Y.F.) from The Ministry of Education, Science, Sports and Culture.
ABBREVIATIONS
- CG
complementation group
- CHO
Chinese hamster ovary
- EGFP
“enhanced” green fluorescent protein
- PBD
peroxisome biogenesis disorders
- PEX19
cDNA encoding the peroxin Pex19p
- PMP70
70-kDa peroxisomal integral membrane protein
- PTS1 and PTS2
peroxisome targeting signal types 1 and 2
- RT
reverse transcription
- ZS
Zellweger syndrome
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB018541).
References
- 1.Lazarow P B, Fujiki Y. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 2.Titorenko V I, Rachubinski R A. Trends Biochem Sci. 1998;23:231–233. doi: 10.1016/s0968-0004(98)01226-2. [DOI] [PubMed] [Google Scholar]
- 3.Fujiki Y. Biochim Biophys Acta. 1997;1361:235–250. doi: 10.1016/s0925-4439(97)00051-3. [DOI] [PubMed] [Google Scholar]
- 4.Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Fujiki Y. J Clin Invest. 1992;90:1864–1870. doi: 10.1172/JCI116063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukamoto T, Yokota S, Fujiki Y. J Cell Biol. 1990;110:651–660. doi: 10.1083/jcb.110.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okumoto K, Bogaki A, Tateishi K, Tsukamoto T, Osumi T, Shimozawa N, Suzuki Y, Orii T, Fujiki Y. Exp Cell Res. 1997;233:11–20. doi: 10.1006/excr.1997.3552. [DOI] [PubMed] [Google Scholar]
- 7.Tateishi K, Okumoto K, Shimozawa N, Tsukamoto T, Osumi T, Suzuki Y, Kondo N, Okano I, Fujiki Y. Eur J Cell Biol. 1997;73:352–359. [PubMed] [Google Scholar]
- 8.Kinoshita N, Ghaedi K, Shimozawa N, Wanders R J A, Matsuzono Y, Imanaka T, Okumoto K, Suzuki Y, Kondo N, Fujiki Y. J Biol Chem. 1998;273:24122–24130. doi: 10.1074/jbc.273.37.24122. [DOI] [PubMed] [Google Scholar]
- 9.Shimozawa N, Suzuki Y, Zhang Z, Imamura A, Kondo N, Kinoshita N, Fujiki Y, Tsukamoto T, Osumi T, Imanaka T, et al. Am J Hum Genet. 1998;63:1898–1903. doi: 10.1086/302142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Distel B, Erdmann R, Gould S J, Blobel G, Crane D I, Cregg J M, Dodt G, Fujiki Y, Goodman J M, Just W W, et al. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erdmann R, Veenhuis M, Kunau W-H. Trends Cell Biol. 1997;7:400–407. doi: 10.1016/S0962-8924(97)01126-4. [DOI] [PubMed] [Google Scholar]
- 12.Honsho M, Tamura S, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y. Am J Hum Genet. 1998;63:1622–1630. doi: 10.1086/302161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otera H, Tateishi K, Okumoto K, Ikoma Y, Matsuda E, Nishimura M, Tsukamoto T, Osumi T, Ohashi K, Higuchi O, Fujiki Y. Mol Cell Biol. 1998;18:388–399. doi: 10.1128/mcb.18.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura S, Okumoto K, Toyama R, Shimozawa N, Tsukamoto T, Suzuki Y, Osumi T, Kondo N, Fujiki Y. Proc Natl Acad Sci USA. 1998;95:4350–4355. doi: 10.1073/pnas.95.8.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsukamoto T, Miura S, Fujiki Y. Nature (London) 1991;350:77–81. doi: 10.1038/350077a0. [DOI] [PubMed] [Google Scholar]
- 16.Tsukamoto T, Miura S, Nakai T, Yokota S, Shimozawa N, Suzuki Y, Orii T, Fujiki Y, Sakai F, Bogaki A, et al. Nat Genet. 1995;11:395–401. doi: 10.1038/ng1295-395. [DOI] [PubMed] [Google Scholar]
- 17.Okumoto K, Fujiki Y. Nat Genet. 1997;17:265–266. doi: 10.1038/ng1197-265. [DOI] [PubMed] [Google Scholar]
- 18.Okumoto K, Shimozawa N, Kawai A, Tamura S, Tsukamoto T, Osumi T, Moser H, Wanders R J A, Suzuki Y, Kondo N, et al. Mol Cell Biol. 1998;18:4324–4336. doi: 10.1128/mcb.18.7.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Shirayoshi Y, Mori T, Fujiki Y. Science. 1992;255:1132–1134. doi: 10.1126/science.1546315. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda S, Shimozawa N, Suzuki Y, Zhang Z, Tomatsu S, Tsukamoto T, Hashiguchi N, Osumi T, Masuno M, Imaizumi K, et al. Am J Hum Genet. 1996;59:1210–1220. [PMC free article] [PubMed] [Google Scholar]
- 21.Braun A, Kammerer S, Weissenhorn W, Weiss E H, Cleve H. Gene. 1994;146:291–295. doi: 10.1016/0378-1119(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 22.Miyazawa S, Osumi T, Hashimoto T, Ohno K, Miura S, Fujiki Y. Mol Cell Biol. 1989;9:83–91. doi: 10.1128/mcb.9.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goette K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau W-H, Erdmann R. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James G L, Goldstein J L, Pathak R K, Anderson R G W, Brown M S. J Biol Chem. 1994;269:14182–14190. [PubMed] [Google Scholar]
- 25.Santos M J, Hoefler S, Moser A B, Moser H W, Lazarow P B. J Cell Physiol. 1992;151:103–112. doi: 10.1002/jcp.1041510115. [DOI] [PubMed] [Google Scholar]
- 26.Wendland M, Subramani S. J Clin Invest. 1993;92:2462–2468. doi: 10.1172/JCI116854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baerends R J S, Rasmussen S W, Hilbrands R E, van der Heide M, Faber K N, Reuvekamp P T W, Klei J A K W, Cregg J M, van der Klei I J, Veenhuis M. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- 28.Wiemer E A C, Luers G H, Faber K N, Wenzel T, Veenhuis M, Subramani S. J Biol Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- 29.Lazarow P B, Robbi M, Fujiki Y, Wong L. Ann NY Acad Sci. 1982;386:285–300. doi: 10.1111/j.1749-6632.1982.tb21423.x. [DOI] [PubMed] [Google Scholar]
- 30.Okumoto K, Itoh R, Shimozawa N, Suzuki Y, Tamura S, Kondo N, Fujiki Y. Hum Mol Genet. 1998;7:1399–1405. doi: 10.1093/hmg/7.9.1399. [DOI] [PubMed] [Google Scholar]
- 31.Warren D S, Morrell J C, Moser H W, Valle D, Gould S J. Am J Hum Genet. 1998;63:347–359. doi: 10.1086/301963. [DOI] [PMC free article] [PubMed] [Google Scholar]