Abstract
Comparisons of complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera to six published grass chloroplast genomes reveal that gene content and order are similar but two microstructural changes have occurred. First, the expansion of the IR at the SSC/IRa boundary that duplicates a portion of the 5′ end of ndhH is restricted to the three genera of the subfamily Pooideae (Agrostis, Hordeum and Triticum). Second, a 6 bp deletion in ndhK is shared by Agrostis, Hordeum, Oryza and Triticum, and this event supports the sister relationship between the subfamilies Erhartoideae and Pooideae. Repeat analysis identified 19–37 direct and inverted repeats 30 bp or longer with a sequence identity of at least 90%. Seventeen of the 26 shared repeats are found in all the grass chloroplast genomes examined and are located in the same genes or intergenic spacer (IGS) regions. Examination of simple sequence repeats (SSRs) identified 16–21 potential polymorphic SSRs. Five IGS regions have 100% sequence identity among Zea mays, Saccharum officinarum and Sorghum bicolor, whereas no spacer regions were identical among Oryza sativa, Triticum aestivum, H. vulgare and A. stolonifera despite their close phylogenetic relationship. Alignment of EST sequences and DNA coding sequences identified six C–U conversions in both Sorghum bicolor and H. vulgare but only one in A. stolonifera. Phylogenetic trees based on DNA sequences of 61 protein-coding genes of 38 taxa using both maximum parsimony and likelihood methods provide moderate support for a sister relationship between the subfamilies Erhartoideae and Pooideae.
Introduction
Chloroplasts are the most noticeable feature of green cells in leaves and, excluding the vacuole, probably constitute the largest compartment within mesophyll cells (Lopez-Juez and Pyke 2005). Plastids are multifunctional and are used by the plant for critical biochemical processes other than photosynthesis, including starch synthesis, nitrogen metabolism, sulfate reduction, fatty acid synthesis, DNA and RNA synthesis (Zeltz et al. 1993). The chloroplast genome generally has a highly conserved organization (Palmer 1991; Raubeson and Jansen 2005) with most land plant genomes composed of a single circular chromosome with a quadripartite structure that includes two copies of an inverted repeat (IR) that separate the large and small single copy regions (LSC and SSC). The size of this circular genome varies from 35 to 2,217 kb but among photosynthetic organisms the majority are between 115 and 165 kb (Jansen et al. 2005).
Our knowledge of the organization and evolution of chloroplast genomes has been expanding rapidly because of the large numbers of completely sequenced genomes published in the past decade. The use of information from chloroplast genomes is well established in the study of the evolutionary patterns and processes in plants (Avise 1994; Raubeson and Jansen 2005). Genetic markers derived from organelle genomes generally show simple, uniparental modes of inheritance, which makes them invaluable for the purposes of population genetic and phylogenetic studies (Bryan et al. 1999; Provan et al. 2001) and this feature also facilitates transgene containment (Daniell 2002).
Sorghum, with 25 species, is a member of the family Poaceae and tribe Andropogoneae (Garber 1950). Recent molecular phylogenetic analyses indicated that the genus may be paraphyletic (Spangler et al. 1999), and that it is comprised of three distinct lineages, Sorghum, Sarga and Vacoparis (Spangler 2003). The genus Sorghum was redefined to include three species, Sorghum bicolor, Sorghum halepense, and Sorghum nitidum. Sorghum bicolor, grain sorghum, is the third most important cereal crop in the United States and the fifth most important crop in the world (Crop Plant Resources 2000). Sorghum is well known for its capacity to tolerate conditions of limited moisture and to produce during periods of extended drought, in circumstances that would impede production in most other grains (Crop Plant Resources 2000). Sorghum is used for human nutrition and feed grain for livestock throughout the world (Carter et al. 1989). A more recent use of Sorghum is the production of ethanol, with one bushel producing the same amount of ethanol as one bushel of corn (National Sorghum Producers 2006). Some Sorghum varieties are rich in anti-oxidants and all varieties are gluten-free, an attractive alternative for those allergic to Triticum aestivum (US Grains Council 2006).
Of the various cereals, Hordeum vulgare L. (barley) is a major food, feed and malt crop. In 2005, H. vulgare ranked fourth in quantity produced and in area of cultivation of cereal crops in the world (http://faostat.fao.org/faostat/) demonstrating its broad consumption and wide adoption in a variety of climates, from sub-arctic to sub-tropical. According to the USDA/NASS, H. vulgare is the third major feed grain crop produced in the United States, after Zea mays (maize) and Sorghum bicolor. Production is concentrated in the Northern Plains and the Pacific Northwest. The United States is the eighth largest producer of H. vulgare in the world with current production estimated at 4.9 million acres. It is a short-season, early maturing crop grown on both irrigated and dry land production areas in the United States. Whole grain H. vulgare contains high levels of minerals and important vitamins, including calcium, magnesium, phosphorus, potassium, vitamin A, vitamin E, niacin and folate.
Among the non-food grasses, Agrostis stolonifera L. (creeping bentgrass) has attracted great attention in both academia and the biotech industry due to its social and economic importance. A. stolonifera is a wind-pollinated, highly outcrossing perennial grass used on golf courses worldwide. It can also enhance the natural beauty of the environment and increase the value of residential and commercial property, and provide many environmental benefits including preventing soil erosion, filtering water and trapping dust and pollutants (Bonos et al. 2006). It has been extensively used, covering millions of acres globally making it an economically valuable grass crop. Due to its aforementioned importance, transgenic A. stolonifera was produced conferring the herbicide resistance trait by engineering the CP4 EPSPS gene, which is one of the first transgenic, perennial, wind-pollinated crops intending to be grown outside of agricultural fields (i.e., on golf courses). Unfortunately, pollen-mediated transgene flow has been reported in several studies (Wipff and Fricker 2001; Watrud et al. 2004; Reichman et al. 2006) limiting its commercialization and demonstrating the requirement of effective containment strategies to protect the environment and to engineer this plant with environmentally friendly approaches like chloroplast engineering or cytoplasmic male sterility.
The agronomic, economic and/or social importance of H. vulgare, Sorghum bicolor and A. stolonifera has made them the focus of numerous studies attempting to improve these crop species. Much of this work has been restricted to investigations of nuclear genomes of these species (USDA 2006, Cheng et al. 2004). This has resulted in very limited information on the organization and evolution of chloroplast genomes of H. vulgare, Sorghum bicolor and A. stolonifera. Therefore, the current study could enhance our understanding of the chloroplast genome organization of grasses facilitating the improvement of those crops by chloroplast genetic engineering. The plastid transformation approach has been shown to have a number of advantages, most notably with regard to its high transgene expression levels (De Cosa et al. 2001), capacity for multi-gene engineering in a single transformation event (De Cosa et al. 2001; Lossl et al. 2003; Ruiz et al. 2003; Quesada-Vargas et al. 2005; Daniell and Dhingra 2002), and ability to accomplish transgene containment via maternal inheritance (Daniell 2002). Moreover, chloroplasts appear to be an ideal compartment for the accumulation of certain proteins, or their biosynthetic products, which would be harmful if they accumulated in the cytoplasm (Daniell et al. 2001; Lee et al. 2003; Leelavathi and Reddy 2003; Ruiz and Daniell 2005). In addition, no gene silencing has been observed in association with this technique, whether at the transcriptional or translational level (De Cosa et al. 2001; Lee et al. 2003; Dhingra et al. 2004). Because of these advantages, the chloroplast genome has been engineered to confer several useful agronomic traits, including herbicide resistance (Daniell et al. 1998), insect resistance (McBride et al. 1995; Kota et al. 1999), disease resistance (DeGray et al. 2001), drought tolerance (Lee et al. 2003), salt tolerance (Kumar et al. 2004a), and phytoremediation (Ruiz et al. 2003). The chloroplast genome has also been utilized in the field of molecular farming, for the expression of biomaterials, human therapeutic proteins, and vaccines for use in humans or other animals (Guda et al. 2000; Staub et al. 2000; Fernandez-San et al. 2003; Leelavathi et al. 2003; Molina et al. 2004; Vitanen et al. 2004; Watson et al. 2004; Koya et al. 2005; Grevich and Daniell 2005; Daniell et al. 2005a, b; Kamarajugadda and Daniell 2006; Chebolu and Daniell 2007; Arlen et al. 2007; Ruhlman et al. 2007; Daniell et al. 2004a, b).
In this article, we present the complete sequences of the chloroplast genomes of H. vulgare, Sorghum bicolor and A. stolonifera. One goal is to compare the genome organization of H. vulgare, Sorghum bicolor and A. stolonifera with six other completely sequenced grass chloroplast genomes; Oryza sativa, O. nivara, Saccharum hybrid, Saccharum officinarum, T. aestivum, and Z. mays. In addition to examining gene content and gene order, we determined the distribution and location of repeated sequences among these genomes, including potential microsatellite markers. A second goal is to compare levels of DNA sequence divergence of non-coding regions. Intergenic spacer (IGS) regions have been examined to identify ideal insertion sites for transgene integration, and to assess the utility of these regions for resolving phylogenetic relationships among closely related species (Kelchner 2002; Shaw et al. 2005, 2007; Saski et al. 2005; Daniell et al. 2006; Timme et al. 2007). A third goal of this paper is to examine the extent of RNA editing in the H. vulgare, Sorghum bicolor and A. stolonifera chloroplast genomes by comparing the DNA sequences with available expressed sequence tag (EST) sequences. RNA editing is a co- or post-transcriptional process that occurs in organelles and changes the coding information in mRNAs (Kugita et al. 2003; Wolf et al. 2004; Peeters and Hanson 2002). Most of our knowledge about the frequency of this process in crop plants comes from studies in Z. mays (Maier et al. 1995) and Nicotiana tabacum (Hirose et al. 1999), and additional comparative studies are needed in other plant species to understand the extent of RNA editing in chloroplast genomes. A final goal is to assess phylogenetic relationships between H. vulgare, Sorghum bicolor, A. stolonifera and other completely sequenced angiosperm chloroplast genomes.
Materials and methods
DNA sources
Bacterial artificial chromosome (BAC) libraries of H. vulgare cv Morex and Sorghum bicolor cv BTX623 were constructed by ligating size fractionated partial HindIII digests of total cellular, high molecular weight DNA with the pINDIGOBAC536 vector. The average insert size of H. vulgare (HV_MBa) and Sorghum bicolor (SB_BBc) libraries was 106 and 120 kb, respectively. BAC related resources for these public libraries can be obtained from the Clemson University Genomics Institute BAC/EST Resource Center (www.genome.clemson.edu).
Bacterial artificial chromosome clones containing chloroplast genome inserts were isolated by screening the library with a soybean chloroplast DNA probe. The first 96 positive clones from screening were pulled from the library, arrayed in a 96 well microtitre plate, copied and archived. Selected clones were then subjected to HindIII fingerprinting and NotI digests. End-sequences were determined and localized on the chloroplast genome of Arabidopsis thaliana to deduce the relative positions of the clones; then clones that covered the entire chloroplast genomes of H. vulgare and Sorghum bicolor were chosen for sequencing.
Preparation of intact chloroplasts and rolling circle amplification
The A. stolonifera L. cultivar Penn A-4 was supplied by HybriGene, Inc. (Hubbard, OR, USA). Prior to chloroplast isolation, plants were kept in dark for 2 days to reduce levels of starch. Chloroplasts from young leaves were isolated using the sucrose step gradient method of Palmer (1986) as modified by Jansen et al. (2005). About 10 g of leaf tissue was homogenized in Sandbrink isolation buffer using pre-chilled tissue blender bursts at high speed for 5 s to get sufficient quantities of chloroplasts. The homogenate was filtered using four layers of cheesecloth and one layer of miracloth (Calbiochem, catalog number 474855) without squeezing. The filtrate was transferred to pre-chilled centrifuge tubes and centrifuged at 1,000 g for 15 min at 4°C. Pellets were resuspended in 7 ml of ice-cold wash buffer and gently loaded over the step gradient consisting of 18 ml of 52% sucrose, over-layered with 7 ml of 30% sucrose. The sucrose step gradient was centrifuged at 25,000 rpm for 30–60 min at 4°C in a SW-27 rotor (Beckman). The chloroplast band from the 30–52% interface was removed using a wide bore pipette, diluted with ten volumes wash buffer, and centrifuged at 1,500 g for 15 min at 4°C. Purified chloroplast pellets were resuspended in a final volume of 2 ml. The entire chloroplast genome was amplified by Rolling Circle Amplification (RCA) using the Repli-g RCA kit (Qiagen, Inc.) following the methods described in (Jansen et al. 2005). RCA was performed at 30°C for 16 h; the reaction was terminated with final incubation at 65°C for 10 min. Digestion of the RCA product with the restriction enzymes BstXI, EcoRI and HindIII verified successful genome amplification, as well as DNA quality for sequencing.
DNA sequencing and genome assembly
The nucleotide sequences of the BAC clones and RCA product were determined by the bridging shotgun method. The purified BAC DNA or RCA product was subjected to hydroshearing, end repair and then size-fractionated by agarose gel electrophoresis. Fractions of approximately 3.0–5.0 kb were eluted and ligated into the vector pBLUE-SCRIPT IIKS+. The libraries were plated and arrayed into 40 96-well microtitre plates for the sequencing reactions.
Sequencing was performed using the Dye-terminator cycle sequencing kit (Perkin Elmer Applied Biosystems, USA). Sequence data from the forward and reverse priming sites of the shotgun clones were accumulated. Sequence data equivalent to eight times the size of the genome was assembled using Phred-Phrap programs (Ewing et al. 1998).
Gene annotation
Annotation of the Sorghum bicolor, H. vulgare and A. stolonifera chloroplast genomes was performed using DOGMA (Dual Organellar GenoMe Annotator, Wyman et al. 2004, http://bugmaster.jgi-psf.org/dogma/). This program uses a FASTA-formatted input file of the complete genomic sequences and identifies putative protein-coding genes by performing BLASTX searches against a custom database of previously published chloroplast genomes. The user must select putative start and stop codons for each protein-coding gene and intron and exon boundaries for intron-containing genes. Both tRNAs and rRNAs are identified by BLASTN searches against the same database of chloroplast genomes.
Molecular evolutionary comparisons
Comparisons of gene content and gene order
Gene content comparisons were performed with Multipipmaker (Schwartz et al. 2003). Comparisons included nine genomes: O. sativa (NC_001320, Hiratsuka et al. 1989), O. nivara (NC_005973, Shahid-Masood et al. 2004), Saccharum officinarum (NC_006084, Asano et al. 2004), Saccharum hybrid (NC_005878, Calsa et al. unpublished), T. aestivum (NC_002762, Ogihara et al. 2000), Z. mays (NC_001400, Maier et al. 1995), H. vulgare (NC_008590, current study), Sorghum bicolor (NC_008602, current study) and A. stolonifera (NC_008591, current study) using O. sativa as the reference genome. Gene orders were examined by pair-wise comparisons between the above genomes using PipMaker (Elnitski et al. 2002).
Examination of repeat structure
Shared and unique repeats were identified for H. vulgare, Sorghum bicolor and A. stolonifera genomes and compared to other grass genomes using Comparative Repeat Analysis (CRA, N. Holtshulte and S. K. Wyman, unpublished, http://bugmaster.jgi-psf.org/repeats/). This program filters the redundant output of REPuter (Kurtz et al. 2001) and identifies shared repeats among the input genomes. For repeat identification, the following constraints were set in CRA: a minimum repeat size of 30 bp and a Hamming distance of 3 (i.e., a sequence identity of ≥90%). Oryza sativa was used as the reference genome. Blast hits 30 bp and longer with a sequence identity of ≥90% were identified to determine the shared repeats among the seven genomes examined. To detect SSRs we used a modified version of the Perl script SSRIT (Temnykh et al. 2001). The modified script, CUGISSR (Jung et al. 2005), was used to search for SSRs ranging from di-to penta-nucleotide repeats.
Comparison of intergenic spacer regions
Intergenic spacer regions from seven grass chloroplast genomes were compared using MultiPipMaker (Schwartz et al. 2003, http://pipmaker.bx.psu.edu/pipmaker/tools.html). MultiPipMaker has a suite of software tools to analyze relationships among more than two sequences. We used a program known as ‘all_bz’ that iteratively compares a pair of nucleotide sequences at a time until all possible pairs from all species have been examined. However, this program processes only one set of IGS regions at a time. For genome-wide comparisons of corresponding intergenic regions from all species, we developed two programs written in PERL. The first iteration creates a set of input files containing corresponding intergenic regions from each species and compares them using ‘all_bz’ program, until all the intergenic regions in the chloroplast genome are processed. The second program parses the output from the above comparisons, calculates percent identity by using the number of identities over the length of the longer sequence, and generates results in tab-delimited tabular format.
Variation between coding sequences and cDNAs
Each of the genes from the H. vulgare, Sorghum bicolor and A. stolonifera chloroplast genomes were used to perform a BLAST search of expressed sequence tags (ESTs) from the NCBI Genbank. The retrieved EST sequences from A. stolonifera, H. vulgare and Sorghum bicolor were then aligned with the corresponding annotated gene for each species separately, using Clustal X. The aligned sequences were then screened and nucleotide and amino acid changes were detected using the Megalign software and the plastid/bacterial genetic code. Due to variation in length between an EST and the corresponding gene, the length of the analyzed sequence was recorded.
Phylogenetic analyses
The 61 genes included in the analyses of Goremykin et al. (2003a, 2004a, 2005), Leebens-Mack et al. (2005), Chang et al. (2006), Lee et al. (2006a, b), Jansen et al. (2006) and Ruhlman et al. (2006) were extracted from the chloroplast genome sequence of A. stolonifera, H. vulgare and Sorghum bicolor using DOGMA (Wyman et al. 2004). The same set of 61 genes was extracted from chloroplast genome sequences of 35 other sequenced genomes (see Table 1 for complete list). All 61 protein-coding genes of the 38 taxa were translated into amino acid sequences, aligned using MUSCLE (Edgar 2004) followed by manual adjustments, and then nucleotide sequences of these genes were aligned by constraining them to the aligned amino acid sequences. A Nexus file with character sets for phylogenetic analyses was generated after nucleotide sequence alignment was completed. The complete nucleotide alignment is available online at Chloroplast Genome Database (Cui et al. 2006, http://chloroplast.cbio.psu.edu).
Table 1.
Taxa included in phylogenetic analyses with GenBank accession numbers and references
Phylogenetic analyses using maximum parsimony (MP) and maximum likelihood (ML) were performed with PAUP* version 4.10b10 (Swofford 2003) and GARLI version 0.942 (Zwickl 2006, http://www.bio.utexas.edu/grad/zwickl/web/garli.html), respectively. Phylogenetic analyses excluded gap regions to avoid alignment ambiguities in regions with variation in sequence lengths. All MP searches included 100 random addition replicates and TBR branch swapping with the Multrees option. Non-parametric bootstrap analyses (Felsenstein 1985) were performed for MP analyses with 1,000 replicates with TBR branch swapping, one random addition replicate, and the Multrees option. Modeltest 3.7 (Posada and Crandall 1998) was used to determine the most appropriate model of DNA sequence evolution for the combined 61-gene dataset. Hierarchical likelihood ratio tests and the Akaike information criterion were used to assess which of the 56 models best fit the data, which was determined to be GTR + I + Γ by both criteria. For ML analyses in GARLI two independent runs were performed using the default settings (see Garli manual at http://www.bio.utexas.edu/grad/zwickl/web/garli.html). Non-parametric bootstrap analyses (Felsenstein 1985) were performed in GARLI for ML analyses using default settings.
Results
Size, gene content and organization of the H. vulgare, S. bicolor and A. stolonifera chloroplast genomes
The complete sizes of the H. vulgare, Sorghum bicolor and A. stolonifera chloroplast genomes are 136,462, 140,754 bp and 136,584 bp, respectively (Fig. 1). The genomes include a pair of IRs of 21,579 bp (H. vulgare), 22,782 bp (Sorghum bicolor) and 21,649 bp (A. stolonifera) separated by a small single copy region of 12,704 bp (H. vulgare), 12,502 bp (Sorghum bicolor) and 12,740 bp (A. stolonifera) and a large single copy region of 80,600 bp (H. vulgare), 82,688 bp (Sorghum bicolor) and 80,546 bp (A. stolonifera).
Fig. 1.
Gene map of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera chloroplast genomes. The thick lines indicate the extent of the inverted repeats (IRa and IRb), which separate the genome into small (SSC) and large (LSC) single copy regions. Genes on the outside of the map are transcribed in the clockwise direction and genes on the inside of the map are transcribed in the counterclockwise direction
The H. vulgare, Sorghum bicolor and A. stolonifera chloroplast genomes contain 113 different genes, and 18 of these are duplicated in the IR, giving a total of 131 genes (Fig. 1). There are 30 distinct tRNAs, and 7 of these are duplicated in the IR. Sixteen genes contain one or two introns, and six of these are in tRNAs. The H. vulgare chloroplast genome consists of 56.7% coding regions that includes 48% protein coding genes, 8.7% RNA genes and 43.3% non-coding regions, containing both IGS regions and introns. The Sorghum bicolor chloroplast genome is composed of 52.1% coding regions that includes 43.4% protein coding genes, 8.7% RNA genes and 47.9% non-coding regions. The A. stolonifera chloroplast genome is composed of 53.6% coding regions that includes 44.7% protein coding genes, 8.9% RNA genes and 46.4% non-coding regions. The overall GC and AT content of the H. vulgare, Sorghum bicolor and A. stolonifera chloroplast genomes are 38.31% (H. vulgare), 38.50% (Sorghum bicolor), 38.45% (A. stolonifera) and 61.69% (H. vulgare), 61.50% (Sorghum bicolor) and 61.55% (A. stolonifera), respectively.
Gene content and gene order
Gene content and order of the H. vulgare, Sorghum bicolor and A. stolonifera chloroplast genomes are similar to the other six sequenced grass chloroplast genomes (O. sativa, O. nivara, Saccharum hybrid, Saccharum officinarum, T. aestivum, and Z. mays). Like other grass chloroplast genomes, the IR in H. vulgare, Sorghum bicolor and A. stolonifera has expanded to include rps19. However, the extent of the IR at the SSC/IRa boundary differs between two of the genomes with the IR of H. vulgare and A. stolonifera expanded to duplicate a portion of ndhH, a feature that is shared with the T. aestivum chloroplast genome (Ogihara et al. 2000). This expansion includes 207 bp (69 amino acids) in H. vulgare, 174 bp (58 amino acids) in A. stolonifera, and 96 bp (32 amino acids) in T. aestivum. The H. vulgare, Sorghum bicolor and A. stolonifera genomes also share the loss of introns in clpP and rpoC1 with other grasses. There are insertions and deletions (indels) of nucleotides within several coding sequences. For example, CAAAAC is uniquely present within matK of Sorghum bicolor, but absent in the rest of the grasses examined (Supplementary Figure 1). There is also a 6 bp deletion in the ndhK gene in H. vulgare, A. stolonifera, T. aestivum and both species of Oryza (Supplementary Figure 1).
Repeat structure
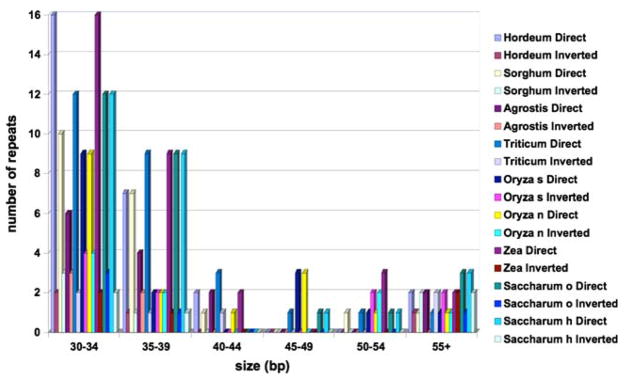
Repeat analyses identified 19–37 direct and IRs 30 bp or longer with a sequence identity of at least 90% among the nine chloroplast genomes examined (Fig. 2). With one exception of a 91 bp repeat, all other repeats range in size between 30 and 60 bp, and 78.4% are in the direct orientation while 21.6% are inverted. The longest repeats other than the IRs found in H. vulgare and Sorghum bicolor are 540 and 524 bp, respectively. BlastN comparisons of the O. sativa repeats against the chloroplast genomes of the eight other grasses identified 26 shared repeats ≥30 bp with a sequence identity ≥90% (Table 2). H. vulgare and T. aestivum share four repeats (31, 32, 36, and 38 bp) not found in any other genomes. Both Oryza species share 41 and 59 bp repeats. Zea mays has the most repeats with 37 and A. stolonifera has the fewest with 19. Seventeen of the 26 repeats are found in all eight chloroplast genomes and all of these are located in the same genes or IGS regions.
Fig. 2.
Histogram showing the number of repeated sequences ≥30 bp long with a sequence identity ≥90% in nine grass chloroplast genomes
Table 2.
Oryza sativa repeats blasted against all eight chloroplast genomes
| Repeat number | Size (bp) | Number of hits | Orientation | Location | Genomes |
|---|---|---|---|---|---|
| 1 | 30 | 2 | Direct | IGS—(trnN-GUU-rps15) | Sb, So, Sh, On, Zm |
| 2 | 30 | 2 | Direct | rps3 | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 3 | 30 | 2 | Direct | IGS—(trnM-CAU-trnG-UCC), trnM-CAU | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 4 | 30 | 2 | Direct | Intron—(ndhB) | Sb, On, Hv, Sh, So, Zm, As |
| 5 | 31 | 3 | Direct | IGS—(trnG-GCC—trnM-CAU), IGS—(trnM-CAU—rps14) | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 6 | 31 | 2 | Direct | rpoC2 | Sb, On, Sh, So, Zm, As |
| 7 | 32 | 2 | Inverted | trnS-UGA | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 8 | 32 | 3 | Inverted | rpl23 | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 9 | 32 | 3 | Inverted | rpl23 | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 10 | 33 | 2 | Inverted | trnT-GGU | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 11 | 34 | 2 | Direct | psaB, psaA | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 12 | 34 | 2 | Direct | rpoC2 | Sb, On, Ta, Hv, Sh, So |
| 13 | 34 | 2 | Direct | trnfM-CAU | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 14 | 36 | 3 | Inverted | Intron—(ycf3 Exon1—ycf3 Exon2), IGS—(trnV-GAC—rps12_3end) | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 15 | 36 | 3 | Direct | rpoC2 | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 16 | 36 | 2 | Inverted | trnS-GCU | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 17 | 37 | 2 | Direct | rpoC2 | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 18 | 45 | 3 | Direct | rps8 | Sb, On, Ta, Hv, Sh, Zm, As |
| 19 | 45 | 2 | Direct | rpoC2 | Sb, On, Ta, Sh, So, Zm, As |
| 20 | 47 | 2 | Direct | IGS—(trnG-GCC—trnfM-CAU), Intron—(trnfM-CAU—trnG-UCC | On, Ta |
| 21 | 50 | 3 | Inverted | IGS—(psbE—petL), Intron—(rps12_3end—rps7) | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 22 | 52 | 2 | Direct | IGS—(trnN-GUU-rps15) | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 23 | 52 | 4 | Inverted | IGS—(ndhB-trnL-CAA) | Sb, On, Ta, Hv, Sh, So, Zm, As |
| 24 | 56 | 2 | Direct | rps18 | Sb, On, Sh, So, Zm, As |
| 25 | 59 | 2 | Inverted | IGS—(psaI-rpl23) | On |
| 26 | 91 | 3 | Inverted | rp123 (69 bp)—IGS (rp123—accD), rp123 (79 bp)—IGS (rp123—rp12) | Sb, On, Ta, Hv, Sh, So, Zm, As |
Includes blast hits at least 30 bp in size, a sequence identity ≥90%, and a bit-score of great than 40
Sb Sorghm bicolor, On Oryza nivara, Ta Triticum aestivum, Hv Hordeum vulgare, Sh Saccharum hybrid, So Saccharum officinarum, Zm Zea mays, As Agrostis stolonifera
Previous studies of grass chloroplast genomes identified three inversions relative to the established consensus chloroplast gene order identical to that found in tobacco (Hiratsuka et al. 1989, Doyle et al. 1992, Palmer and Stein 1986). Because inversions are often associated with repeated sequences (Palmer 1991) we examined inversion endpoint regions for repeats. We located shared repeats flanking the endpoints of the largest 28 kb inversion of grasses. Repeat analyses identified a 21 bp direct repeat in O. sativa that contains the motif GTGAGCTACCAAACTGCTCTA and flanks the inversion endpoints. This repeat has a Hamming distance of 2, and is shared by all the other grasses examined. Repeat analyses at the endpoints of the two other grass inversions failed to identify any shared repeats at the settings used in this analysis.
Our analyses identified 16–21 SSRs per genome and these are composed of di-to penta-nucleotide repeating units (Supplementary Table 3). Nearly 50% of all SSRs are tetra-nucleotide repeats with no common motif. The next most common SSR consists of di-nucleotide repeats and accounts for 30% of the SSRs with a predominant motif of TA or AT. The remaining 20% of the SSRs are composed of tri- and penta-nucleotide repeats. Of the SSRs identified, the same dinucleotide repeat (AT) is located within the coding region of the gene rpoC2 in all chloroplast genomes examined.
Intergenic spacer regions
We analyzed the similarity and divergence of IGS regions from seven grass chloroplast genomes including A. stolonifera, H. vulgare, Z. mays, O. sativa, Sorghum bicolor, Saccharum officinarum and T. aestivum. The results of these analyses are presented in Tables 3 and 4, Figs. 3 and 4, and in Supplementary Tables 1 and 2. These species were subdivided into two groups for comparative analyses based on their position in phylogenetic trees (Figs. 5, 6). The first group includes O. sativa, T. aestivum, H. vulgare and A. stolonifera and the second group contains Z. mays, Saccharum officinarum and Sorghum bicolor.
Table 3.
Analysis of intergenic spacer regions of O. sativa, T. aestivum, H. vulgare and A. stolonifera
| Intergenic region | A. stolonifera/H. vulgare | O. sativa/H. vulgare | T. aestivum/H. vulgare | A. stolonifera/O. sativa | A. stolonifera/T. aestivum | O. sativa/T. aestivum |
|---|---|---|---|---|---|---|
| trnA-UGC:trnA-UGC | 100 | 99 | 99 | 99 | 98 | 98 |
| trnH-GUG:rpl2 | 100 | 91 | 100 | 91 | 100 | 91 |
| trnA-UGC:trnI-GAU | 100 | 94 | 91 | 92 | 91 | 91 |
| rpl23:trnI-CAU | 97 | 97 | 100 | 97 | 97 | 97 |
| trnI-CAU:rpl23 | 97 | 97 | 100 | 97 | 97 | 97 |
| rrn4.5:rrn23 | 92 | 94 | 100 | 89 | 92 | 94 |
| rrn23:rrn4.5 | 91 | 94 | 100 | 88 | 92 | 94 |
| trnE-UUC:trnY-GUA | 89 | 92 | 100 | 90 | 89 | 92 |
| trnN-GUU:trnR-ACG | 88 | 85 | 100 | 94 | 88 | 85 |
| trnR-ACG:trnN-GUU | 88 | 85 | 100 | 94 | 88 | 85 |
| rps12_5end:clpP | 86 | 80 | 100 | 78 | 86 | 80 |
| ndhB:rps7 | 98 | 95 | 95 | 95 | 95 | 100 |
| rps7:ndhB | 98 | 94 | 94 | 94 | 94 | 100 |
| trnQ-UUG:psbK | 92 | 91 | 91 | 91 | 91 | 100 |
| rps16:trnQ-UUG | 40 | 36 | 36 | 56 | 56 | 100 |
Intergenic spacer regions that are 100% identical in at least two of the four species are shown
Table 4.
Analysis of intergenic spacer regions of Z. mays, S. officinarum and S. bicolor
| Intergenic spacer region | Z. mays/S. officinarum | Z. mays/S. bicolor | S. officinarum/S. bicolor |
|---|---|---|---|
| ndhD:psaC | 100 | 100 | 100 |
| psbJ:psbL | 100 | 100 | 100 |
| psbN:psbH | 100 | 100 | 100 |
| rrn23:trnA-UGC | 100 | 100 | 100 |
| trnA-UGC:rrn23 | 100 | 100 | 100 |
| ndhB:trnL-CAA | 100 | 99 | 99 |
| trnL-CAA:ndhB | 100 | 99 | 99 |
| rps19:trnH-GUG | 100 | 96 | 96 |
| trnH-GUG:rps19 | 100 | 96 | 96 |
| ndhB:ndhB | 99 | 100 | 99 |
| rps12:trnV-GAC | 99 | 99 | 100 |
| trnA-UGC:trnA-UGC | 99 | 99 | 100 |
| trnV-GAC:rps12 | 99 | 99 | 100 |
| rrn16:trnV-GAC | 98 | 98 | 100 |
| trnN-GUU:trnR-ACG | 98 | 98 | 100 |
| trnR-ACG:trnN-GUU | 98 | 98 | 100 |
| trnV-GAC:rrn16 | 98 | 98 | 100 |
| rpl23:trnI-CAU | 97 | 97 | 100 |
| rps2:atpI | 97 | 97 | 100 |
| rps7:rps12 | 97 | 97 | 100 |
| rrn4.5:rrn5 | 97 | 97 | 100 |
| trnI-CAU:rpl23 | 97 | 97 | 100 |
| petG:trnW-CCA | 96 | 96 | 100 |
| ndhI:ndhA | 95 | 100 | 95 |
| psbC:trnS-UGA | 95 | 95 | 100 |
| rrn4.5:rrn23 | 95 | 95 | 100 |
| rpl22:rps19 | 94 | 94 | 100 |
| rpl36:infA | 94 | 94 | 100 |
| trnM-CAU:atpE | 93 | 93 | 100 |
| trnE-UUC:trnY-GUA | 92 | 92 | 100 |
| cemA:petA | 91 | 91 | 100 |
| ndhJ:ndhK | 90 | 90 | 100 |
| rps3:rpl22 | 89 | 89 | 100 |
| trnA-UGC:trnI-GAU | 86 | 86 | 100 |
| psbT:psbN | 69 | 69 | 100 |
| rps12:rps7 | 9 | 9 | 100 |
Intergenic spacer regions that are 100% identical in at least two of the three species are shown below
Fig. 3.
Histogram showing pairwise sequence divergence of the intergenic spacer regions of rice (Oryza sativa), wheat (Triticum aestivum) barley (Hordeum vulgare) and bentgrass (Agrostis stolonifera) chloroplast genomes. Comparisons of 19 most variable intergenic regions with less than 80% average sequence identity. The values plotted in this histogram come from Supplementary Table 1, which shows percent sequence identities for all intergenic spacer regions. The plotted values were converted from percent identity to sequence divergence on a scale from 0 to 1 and included on the Y-axis. Asterisk indicates regions that are in the top 25 most variable intergenic spacer regions in Solanaceae (adapted from Daniell et al. 2006), plus indicates regions that are in the top 25 most variable intergenic spacer regions in Asteraceae (adapted from Timme et al. 2007)
Fig. 4.
Histogram showing pairwise sequence divergence of the intergenic spacer regions of maize (Zea mays), sugarcane (Saccharum officinarum) and sorghum (Sorghum bicolor) chloroplast genomes. Comparisons of the nine most variable intergenic spacer regions with less than 80% average sequence identity. The values plotted in this histogram come from Supplementary Table 2, which shows percent sequence identities for all intergenic spacer regions. The plotted values were converted from percent identity to sequence divergence on a scale from 0 to 1 and included on the Y-axis. Asterisk indicates regions that are in the top 25 most variable intergenic spacer regions in Solanaceae (adapted from Daniell et al. 2006), plus indicates regions that are in the top 25 most variable intergenic spacer regions in Asteraceae (adapted from Timme et al. 2007)
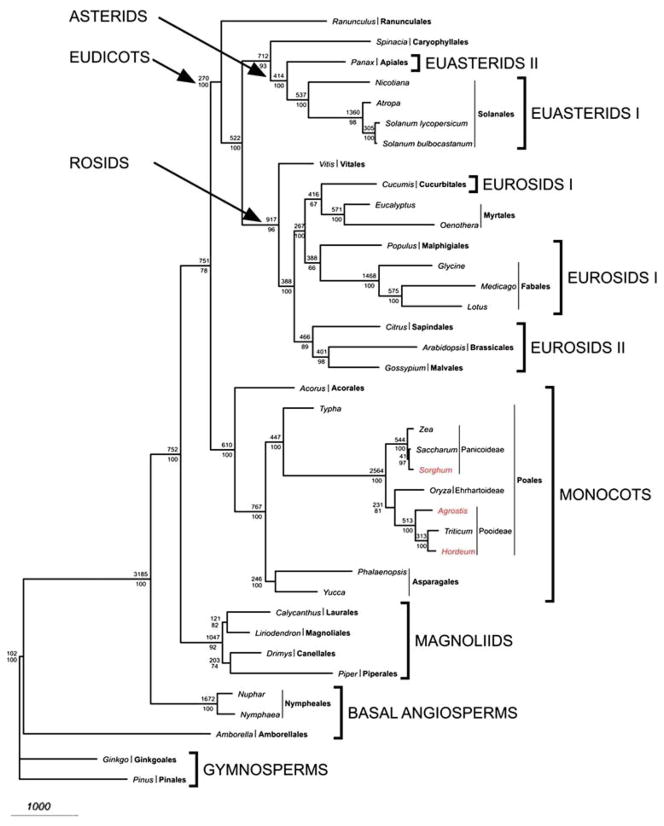
Fig. 5.
Phylogenetic tree of 38 taxa based on 61 plastid protein-coding genes using maximum parsimony. The tree has a length of 62,437, a consistency index of 0.407 (excluding uninformative characters) and a retention index of 0.627. Numbers above node indicate number of changes along each branch and numbers below nodes are bootstrap support values. Ordinal and higher level group names follow APG II (2003). Taxa in red are the new genomes reported in this paper
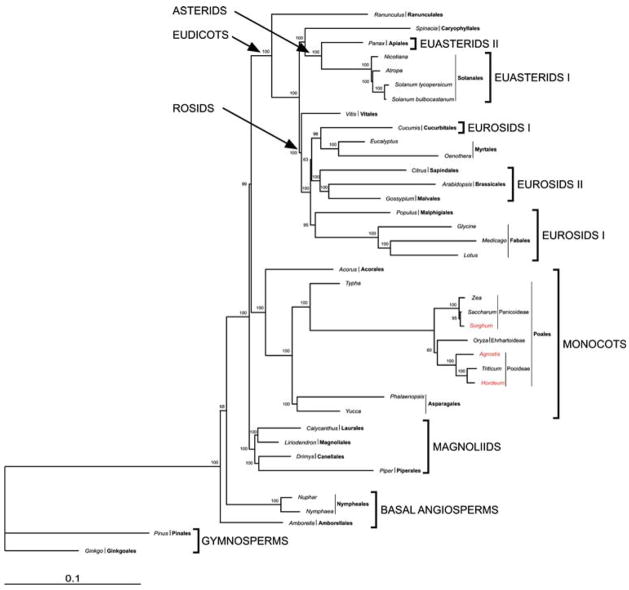
Fig. 6.
Phylogenetic tree of 38 taxa based on 61 plastid protein-coding genes using maximum likelihood. The tree has a ML value of −lnL = 348086.2268. Numbers at nodes are bootstrap support values 50%. Ordinal and higher level group names follow APG II (2003). Taxa in red are the new genomes reported in this paper
Five IGS regions (ndhD:psaC, psbJ:psbL, psbN:psbH, rrn23:trnA-UGC, trnA-UGC:rrn23) have 100% sequence identity among Z. mays, Saccharum officinarum and Sorghum bicolor, whereas no spacer regions are identical among O. sativa, T. aestivum, H. vulgare and A. stolonifera despite of their close phylogenetic relationship. Divergence among Z. mays, Sorghum bicolor and Saccharum officinarum chloroplast genomes is much less because there are only nine IGS regions with less than 80% average sequence identity versus 19 among O. sativa, T. aestivum, H. vulgare and A. stolonifera (Figs. 3, 4). Only three of the intergenic regions in the two sets of comparisons have more than 80% average sequence divergence (rpl16:rps3, psbH:petB, and rps12_3end:rps7; compare Figs. 3, 4). Some spacer regions have indels resulting in extremely low sequence identity. For example, in Z. mays, deletion of a 558 bp intergenic region between rps12 3′ end and rps7 IGS has resulted in only 9% sequence identity between Z. mays:Sorghum bicolor and Z. mays:Saccharum officinarum comparisons. Nevertheless, this region shows 100% identity between Sorghum bicolor and Saccharum officinarum (see Supplementary Table 2). Regions marked with asterisks or plus signs in Figs. 3 and 4 are in the top 25 most variable IGSs in Solanaceae (Daniell et al. 2006) and Asteraceae (Timme et al. 2007), respectively.
Variation between coding regions and cDNAs
Alignment of EST sequences and DNA coding sequences identified 15 nucleotide substitution differences in the Sorghum bicolor chloroplast genome (Table 5), 25 in the H. vulgare genome (Table 6) and 1 in A. stolonifera (not shown). Sorghum bicolor has six C–U conversions, five of which result in amino acid changes. H. vulgare also has six C–U conversions, all of which result in amino acid changes. Of these substitutions, 11 are non-synonymous and 4 are synonymous in Sorghum bicolor. In H. vulgare, 17 substitutions are non-synonymous and eight are synonymous. Sorghum bicolor experienced 1–2 substitutions per gene while H. vulgare has 1–5 variable sites per identified gene. H. vulgare and Sorghum bicolor share three variable positions in the rpoC2, psaA and atpB genes (Tables 5, 6). At the time of the analysis of A. stolonifera, there were only 9018 EST sequences available to analyze potential RNA editing sites. Comparing the coding regions of the A. stolonifera chloroplast genome to available ESTs reveals only one potential editing site. This site is located within the psbZ gene at position 54 and suggests a C–U change, which does not result in a change in the amino acid. There are 89 ESTs that show support for a C–U change, and five that don’t show the edit.
Table 5.
Differences observed by comparison of S. bicolor chloroplast genome sequences with EST sequences obtained by BLAST search of NCBI GenBank
| Gene | Gene size (bp) | Sequence analyzeda | Number of variable sites | Variation type | Position(s)b | Amino acid change |
|---|---|---|---|---|---|---|
| atpA | 1,523 | 1069–1523 | 1 | C–U | 1148 | S–L |
| ndhK | 746 | 1–297 | 1 | C–U | 128 | P–L |
| rpoC2 | 4,562 | 2728–3143 | 1 | C–U | 2753 | S–L |
| psaA | 2,284 | 893–1281 | 1 | T–G | 968 | L–W |
| atpB | 1,496 | 551–1488 | 2 | T–G | 535 | H–Q |
| A–G | 1466 | I–V | ||||
| psbJ | 122 | 1–122 | 2 | T–A | 35 | L–Q |
| T–C | 60 | L–L | ||||
| psbD | 1,061 | 306–1061 | 1 | G–A | 741 | M–I |
| psbC | 1,421 | 534–1065 | 1 | T–G | 1047 | G–G |
| psaB | 2,204 | 95–587 | 1 | T–G | 99 | S–R |
| ndhA | 1,089 | 1023–1089 | 1 | C–U | 1070 | S–F |
| rp12 | 843 | 1–511 | 2 | C–U | 14 | T–M |
| A–G | 405 | G–G | ||||
| ndhI | 543 | 1–543 | 1 | C–U | 513 | I–I |
Sequence based on the gene sequence, considering the first base of the initiation codon as 1
Variable position is given in reference to the first base of the initiation codon of the gene sequence
Table 6.
Differences observed by comparison of H. vulgare chloroplast genome sequences with EST sequences obtained by BLAST search of NCBI Gen-Bank
| Gene | Gene size | Sequence analyzeda | No of variable sites | Variation type | Nucleotide position(s)b | Amino acid change |
|---|---|---|---|---|---|---|
| rpoB | 3231 | 1–2150 | 4 | T–A | 241 | Y–N |
| G–C | 2,048 | S–T | ||||
| G–U | 2,050 | E–L | ||||
| A–U | 2,051 | E–L | ||||
| clpP | 651 | 265–651 | 5 | G–A | 337 | A–T |
| A–U | 417 | E–D | ||||
| T–C | 508 | S–P | ||||
| A–G | 598 | K–E | ||||
| G–A | 630 | P–P | ||||
| rpl2 | 390 | 1–390 | 1 | C–U | 2 | T–M |
| psaA | 2,253 | 117–894 | 3 | G–C | 81 | A–A |
| T–G | 138 | I–S | ||||
| C–A | 396 | F–L | ||||
| ycf4 | 558 | 38–376 | 3 | T–C | 319 | W–R |
| T–C | 342 | R–R | ||||
| T–C | 347 | V–A | ||||
| atpB | 1,497 | 1–670 | 3 | C–U | 490 | R–C |
| A–G | 663 | V–V | ||||
| T–C | 669 | N–N | ||||
| ycf3 | 228 | 1–228 | 1 | T–A | 23 | N–I |
| rpoC2 | 4,434 | 3640–4315 | 1 | C–U | 4,025 | S–L |
| psaJ | 129 | 1–129 | 1 | T–G | 72 | G–G |
| petA | 963 | 821–963 | 4 | T–C | 870 | P–P |
| C–U | 883 | R–C | ||||
| C–U | 917 | S–F | ||||
| C–U | 949 | V–I |
Sequence based on the gene sequence, considering the first base of the initiation codon as 1
Variable position is given in reference to the first base of the initiation codon of the gene sequence
Phylogenetic analyses
The data matrix comprises 61 protein-coding genes for 38 taxa, including 36 angiosperms and two gymnosperm out-groups (Pinus and Ginkgo, Table 1). The aligned sequences include 46,188 nucleotide positions but when the gaps are excluded to avoid ambiguities due to insertion/deletions there are 39,574 characters. MP analyses resulted in a single most-parsimonious tree with a length of 62,437, a consistency index of 0.407 (excluding uninformative characters) and a retention index of 0.627 (Fig. 5). Bootstrap analyses indicate that 26 of the 35 nodes have bootstrap values ≥95%, five nodes have 80–94%, and four nodes have 50–79%. ML analysis results in a single tree with a ML value of −lnL = 348,086.2268 (Fig. 6). Support is very strong for most clades in the ML tree with 32 of the 35 nodes with ≥95% bootstrap values and 3 with 60–69% support. The ML and MP trees only differ in the relationships among the rosids (compare Figs. 5, 6), although this difference is not strongly supported in the ML tree (63% bootstrap value). In the MP tree the eurosid II clade is sister to a clade that includes both members of eurosid I and Myrtales, whereas in the ML tree the eurosid II clade is sister to a clade that includes the Myrtales and one member of the eurosid I (Cucurbitales).
Discussion
Significance of transgene integration into grass chloroplast genomes
Although plastid transformation has been accomplished via organogenesis in a number of eudicots, two major obstacles have been encountered to extend plastid transformation technology to crop plants that regenerate via somatic embryogenesis: (1) the expression of transgenes in non-green plastids, in which gene expression and gene regulation systems are quite distinct from those of mature green chloroplasts, and (2) our current inability to generate homoplastomic plants via subsequent rounds of regeneration, using leaves as explants. Despite these limitations, plastid transformation has recently been accomplished via somatic embryogenesis in several eudicot crops, including Glycine max L. Merr. (soybean), Daucus carota L. (carrot) and Gossypium hirsutum L. (cotton, Dufourmantel et al. 2004, 2005; Kumar et al. 2004a, b) and foreign genes have been expressed in high levels in non-green plastids, including proplastids and chromoplasts (Kumar et al. 2004a). Breakthroughs in plastid transformation of recalcitrant crops, such as G. hirsutum and G. max, have raised the possibility of engineering plastid genomes of other major crops via somatic embryogenesis. To date, only fragmentary data were reported for O. sativa plastid transformation (Khan and Maliga 1999). However, a promising step toward stable plastid transformation in O. sativa has been reported recently (Lee et al. 2006b). Transplastomic O. sativa plants generated in this study exhibited stable integration and expression of the aadA and sgfp transgenes in their plastids. Moreover, the transplastomic O. sativa plants generated viable seeds, which were confirmed to transmit the transgenes to the T1 progeny. Unfortunately, conversion of the transplastomic O. sativa plants to homoplasmy was not successful, even after two generations of continuous selection. Thus, tissue culture and selection of transformed events continues to be a major challenge.
The success of chloroplast genetic engineering of crop plants is dependent, at least in part, on access to conserved spacer regions for inserting transgenes. The availability of sequences of complete chloroplast genomes for multiple crop plants in the grass family should facilitate plastid genetic engineering. Several studies have demonstrated that the use of IGS regions that have low sequence identities between the target genome and the flanking sequences in the chloroplast transformation vectors can result in substantially lower frequencies of transformants (Nguyen et al. 2005; Ruf et al. 2001; Sidorov et al. 1999). Given the low number of intergenic sequences that have high sequence identities among the seven sequenced chloroplast genomes (Tables 3, 4) it is unlikely that a single, highly conserved IGS region will be appropriate throughout the grass family. Among Solanaceae chloroplast genomes, only four spacer regions have 100% sequence identity among all sequenced genomes and three of these regions are within the IR region (Daniell et al. 2006). Five IGS regions have 100% sequence identity among Z. mays, Saccharum officinarum and Sorghum bicolor chloroplast genomes. Thus the variation in the IGS region is quite similar between solanaceae and grass chloroplast genomes. However, not a single IGS region is identical among O. sativa, T. aestivum and H. vulgare chloroplast genomes. Thus, conservation of IGS regions is not uniform even within the same family. However, it is noteworthy that the same IGS regions have very low sequence identity within Poaceae, Solanaceae and Asteraceae, as discussed below.
Genome organization and evolutionary implications
Organization and evolution of grass chloroplast genomes
The organization of chloroplast genomes is highly conserved in most land plants but alterations in gene content and order have been identified in several lineages (Raubeson and Jansen 2005). Notable rearrangements are known in two families with many crop species, a single 51-kb inversion common to most papilionoid legumes (Palmer et al. 1988; Doyle et al. 1996; Saski et al. 2005) and three inversions in the grasses (Quigley and Weil 1985; Howe et al. 1988; Hiratsuka et al. 1989; Doyle et al. 1992; Katayama and Ogihara 1996). The H. vulgare, Sorghum bicolor and A. stolonifera chloroplast genomes contain all three of the inversions present in grasses.
Gene order and content of the sequenced grass chloroplast genomes are similar. However, two microstructural changes have occurred. First, the expansion of the IR at the SSC/IR boundary that duplicates a portion of the 5′ end of ndhH is restricted to the three genera of the subfamily Pooideae (Agrostis, Hordeum and Triticum). These three genera form a monophyletic group in the phylogenetic trees based on DNA sequences of protein-coding genes (Figs. 5, 6) but the extent of the IR expansion differs in each of the three genera (32, 69 and 58 amino acids in wheat, barley and bentgrass, respectively). Thus, it is not possible to determine if there have been three independent expansions or a single expansion followed by two subsequent contractions. Second, a 6 bp deletion in ndhK (Supplementary Figure 1) is shared by Agrostis, Hordeum, Oryza and Triticum, and this event supports the sister relationship between the subfamilies Erhartoideae and Pooideae (Figs. 5, 6).
Other than the IR, repeated sequences are considered to be relatively uncommon in chloroplast genomes (Palmer 1991). The analysis of the repeated sequences of grass chloroplast genomes revealed 26 groups of repeats shared among various members of the family (Table 2, Fig. 2). Furthermore, 17 of the 26 repeats are shared among all eight of the chloroplast genomes examined suggesting a high level of conservation of repeat structure among grasses. Examination of the location of these repeats suggests that all of them occur in the same location, either in genes, introns or within IGS regions. This high level of conservation of both sequence identity and location suggests that these elements may play a functional role in the genome, although we cannot rule out the possibility that this conservation may simply be due to a common ancestry. Because organellar genomes are often uniparentally inherited, chloroplast DNA polymorphisms have become a marker of choice for investigating evolutionary issues such as sex-biased dispersal and the directionality of introgression (Willis et al. 2005). They are also invaluable for the purposes of population-genetic and phylogenetic studies (Bryan et al. 1999; Raubeson and Jansen 2005). Also, knowledge of mutation rates is important because they determine levels of variability within populations, and hence greatly influence estimates of population structure (Provan et al. 1999). Based on our mining for SSRs, we identified 16–18 SSRs within the nine genomes examined. These initial findings indicate a potential to test and utilize SSRs to rapidly analyze diversity in germplasm collections.
Previous studies of grass chloroplast genomes have identified three inversions in the family (Quigley and Weil 1985; Howe et al. 1988; Hiratsuka et al. 1989; Doyle et al. 1992; Katayama and Ogihara 1996). Our analysis of the inversion endpoints indicate that there are shared repeats flanking the endpoints of the largest 28 kb inversion. This first inversion has endpoints between trnG-UCC and trnR-UCU at one end and rps14 and trnfM-CAU at the other creating an intermediate form of the chloroplast genome prior to the second inversion when compared to N. tabacum (Hiratsuka et al. 1989; Doyle et al. 1992). Repeat analyses identified a 21 bp direct repeat in O. sativa that flanks the inversion endpoints, and this repeat is shared by all other grasses examined. It is likely that the shared repeat facilitated this large inversion by intramolecular recombination. Two additional inversions, one largely overlapping the 28 kb event, subsequently gave rise to the gene order observed in O. sativa and T. aestivum (Hiratsuka et al. 1989). The endpoints of the second inversion (ca 6 kb) occur between trnS and psbD on one end and trnG-UCC and trnT-GGU on the other (Doyle et al. 1992). The third inversion has endpoints between trnG-UCU and trntT-GGU and trnT-GGU and trnE-UUC. This inversion is quite small and accounts for the inverted orientation of trnT-GGU (Hiratsuka et al. 1989). Our repeat analyses found no shared repeats that may have played a role in these two inversions. Chloroplast genome organization is also known from other monocots based on both gene mapping and complete genome sequencing (de Heij et al. 1983; Chase and Palmer 1989; Chang et al. 2006). Four non-grass monocots Spirodela oligorhiza (Lemnaceae), two orchids (Oncidium excavatum and Phalaenopsis aphrodite), and members of the Alliaceae (Allium cepa), Asparagaceae (Asparagus sprengeri) and Amaryllidaceae (Narcissus × hybridus) have the same gene order as tobacco. Thus, the inversions in H. vulgare, Sorghum bicolor and A. stolonifera reported here are confined to the grass family as was previously suggested by Doyle et al. (1992).
Comparisons of DNA and EST sequences for H. vulgare, Sorghum bicolor and A. stolonifera identified many differences (Tables 5, 6), most of which are not likely due to RNA editing. Previous investigations of RNA editing in chloroplast genomes in the angiosperms N. tabacum (Hirose et al. 1999) and Atropa (Schmitz-Linneweber et al. 2002) and in the fern Adiantum (Wolf et al. 2004) indicated that RNA edits only result in C–U changes. In the case of H. vulgare, Sorghum bicolor and A. stolonifera, only seven differences in the DNA and EST sequences were C–U changes. Thus, these are the only changes that may be the result of RNA editing. The other 9 differences in Sorghum bicolor and 19 differences in H. vulgare are likely due to either polymorphisms resulting from the use of different plants or cultivars or sequencing errors. In the case of A. stolonifera, only one C–U change was found. This could be attributed to the lack of available expression information since only 9,018 EST sequences were available for A. stolonifera when the analysis was performed, suggesting a need for more comprehensive investigations into the chloroplast and nuclear transcriptomes.
Several recent comparisons of DNA and EST sequences for other crop species including G. hirsutum (Lee et al. 2006a), Vitis vinifera (Jansen et al. 2006), Citrus sinensis L. (Bausher et al. 2006), carrot (Ruhlman et al. 2006), Lactuca and Helianthus (Timme et al. 2007) and Solanum lycopersicum and Solanum bulboscastanum (Daniell et al. 2006) have identified both putative RNA editing sites and possible sequencing errors. The much greater depth of coverage in the chloroplast genome sequences (generally 4-20X coverage) suggests that most of the differences other than changes from C to U are likely due to errors in EST sequences.
Phylogenetic utility of intergenic spacer regions
Phylogenetic studies at the inter- and intraspecific levels in plants have relied extensively on IGS regions of chloroplast genomes because the coding regions are generally too highly conserved at these lower taxonomic levels (Kelchner 2002; Raubeson and Jansen 2005; Jansen et al. 2005; Shaw et al. 2005, 2007). There have been many efforts to identify the most divergent IGSs for phylogenetic comparisons at lower taxonomic levels with the hope that some universal regions could be found for angiosperms (Shaw et al. 2005, 2007, Daniell et al. 2006; Timme et al. 2007). Only two previous studies have performed genome-wide comparisons among multiple, sequenced genomes in the families Asteraceae (Timme et al. 2007) and Solanaceae (Daniell et al. 2006). Comparison of our results in the Poaceae with these earlier studies indicates that there are considerable differences regarding which IGS regions are most variable in these three families (see asterisks and plus signs in Figs. 3, 4). Only three (Fig. 4) to five (Fig. 3) of the 25 most variable regions of Solanaceae are among the most variable IGSs in grasses. The overlap in the regions with high sequence divergence between the Asteraceae and grasses is higher, with three (Fig. 4) to nine (Fig. 3) of the most variable IGS regions in the Poaceae among the 25 most variable regions in the Asteraceae. Overall, genome-wide comparisons among these three families indicate that there may be few universal IGS regions across angiosperms for phylogenetic studies at lower taxonomic levels. Thus, it will likely be necessary to identify variable IGS regions in chloroplast genomes for each family to locate the most appropriate markers for phylogenetic comparisons.
Phylogenetic relationships of angiosperms
During the past three years there has been a rapid increase in the number of studies using DNA sequences from completely sequenced chloroplast genomes for estimating phylogenetic relationships among angiosperms (Goremykin et al. 2003a, b, 2004, 2005; Leebens-Mack et al. 2005; Chang et al. 2005; Lee et al. 2006a; Jansen et al. 2006; Ruhlman et al. 2006; Bausher et al. 2006; Cai et al. 2006). These studies have resolved a number of issues regarding relationships among the major clades, including the identification of either Amborella alone or Amborella + Nymphaeales as the sister group to all other angiosperms, strong support for the monophyly of magnoliids, monocots and eudicots, the position of magnoliids as sister to a clade that includes both monocots and eudicots, the placement of Vitaceae as the earliest diverging lineage of rosids, and the sister group relationship between Caryophyllales and asterids. However, some issues remain unresolved, including the monophyly of the eurosid I clade and relationships among the major clades of rosids. The phylogenetic analyses reported here (Figs. 5, 6) with expanded taxon sampling are congruent with these earlier studies so our discussion will focus on relationships among grasses.
Our study has added complete chloroplast genome sequences for three genera of grasses representing two subfamilies (Pooideae and Erhartoideae, sensu Grass Phylogeny Working Group 2001). This expands the number sequenced grass genera to seven from three different subfamilies, Panicoideae, Pooideae and Erhartoideae. Our phylogenetic trees (Figs. 5, 6) indicate that the Erhartoideae is sister to the Pooideae with weak to moderate bootstrap support (60 or 81% in ML and MP trees, respectively). The sister relationship of these subfamilies is also supported by a 6 bp deletion in ndhK (Supplementary Figure 1). This result is congruent with phylogenetic trees based on sequences of six genes (four chloroplast and two nuclear, Grass Phylogeny Working Group 2001). This multigene tree, which included 68 genera of grasses, also provided only moderate bootstrap support (71%) for a close phylogenetic relationship between these two subfamilies. Furthermore, the clade including Pooideae and Erhartoideae also contained members of the Bambusioideae. Clearly, many additional chloroplast genome sequences are needed from the grasses to provide sufficient taxon sampling to generate a family-wide phylogeny based on whole genomes.
Supplementary Material
Acknowledgments
Investigations reported in this article were supported in part by grants from USDA 3611-21000-017-00D and NIH 2 R01 GM 063879 to Henry Daniell, from NSF DEB 0120709 to Robert K. Jansen, from USDA USDA-BRAG 2005-39454-16511, CREES SC-1700315 to Hong Luo and from the Research Council of Norway BILAT-174998/D15 to Jihong Liu Clarke.
Footnotes
Communicated by A. Paterson.
Electronic supplementary material The online version of this article (doi: 10.1007/s00122-007-0567-4) contains supplementary material, which is available to authorized users.
Contributor Information
Christopher Saski, Clemson University Genomics Institute, Clemson University, Biosystems Research Complex, 51 New Cherry Street, Clemson, SC 29634, USA.
Seung-Bum Lee, 4000 Central Florida Blvd, Department of Molecular Biology and Microbiology, Biomolecular Science, University of Central Florida, Building #20, Orlando, FL 32816-2364, USA.
Siri Fjellheim, Department of Plant and Environmental Sciences, Norwegian University of Life Sciences, 1432 Aas, Norway.
Chittibabu Guda, Gen*NY* Sis Center for Excellence in Cancer Genomics and Department of Epidemiology and Biostatistics, State University of New York at Albany, 1 Discovery Dr Rensselaer, New York, NY 12144, USA.
Robert K. Jansen, Section of Integrative Biology and Institute of Cellular and Molecular Biology, Biological Laboratories 404, University of Texas, Austin, TX 78712, USA
Hong Luo, Department of Genetics and Biochemistry, Clemson University, 51 New Cherry Street, Clemson, SC 29634, USA.
Jeffrey Tomkins, Clemson University Genomics Institute, Clemson University, Biosystems Research Complex, 51 New Cherry Street, Clemson, SC 29634, USA.
Odd Arne Rognli, Department of Plant and Environmental Sciences, Norwegian University of Life Sciences, 1432 Aas, Norway.
Henry Daniell, 4000 Central Florida Blvd, Department of Molecular Biology and Microbiology, Biomolecular Science, University of Central Florida, Building #20, Orlando, FL 32816-2364, USA, e-mail: daniell@mail.ucf.edu.
Jihong Liu Clarke, Department of Genetics and Biotechnology, Norwegian Institute for Agricultural and Environmental Sciences, 1432 Aas, Norway.
References
- APG II An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- Arlen PA, Falconer R, Cherukumilli S, Cole A, Cole AM, Oishi K, Daniell H. Field production and functional evaluation of chloroplast-derived interferon alzpha 2b. Plant Biotechnol J. 2007 doi: 10.1111/j.1467-7652.2007.00258.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K. Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res. 2004;11:93–99. doi: 10.1093/dnares/11.2.93. [DOI] [PubMed] [Google Scholar]
- Avise JC. Molecular markers, natural history, and evolution. Chapman & Hall; New York: 1994. [Google Scholar]
- Bausher MG, Singh ND, Lee S-B, Jansen RK, Daniell H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 2006;6:21. doi: 10.1186/1471-2229-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonos SA, Clarke BB, Meyer WA. Breeding for disease resistance in the major cool-season turfgrass. Annu Rev Phytopathol. 2006;44:213–234. doi: 10.1146/annurev.phyto.44.070505.143338. [DOI] [PubMed] [Google Scholar]
- Bryan GJ, McNicoll J, Ramsey G, Meyer RC, De Jong WS. Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theor Appl Genet. 1999;99:859–867. [Google Scholar]
- Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson J, dePamphilis CW, Jansen RK. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogeny of magnoliids. BMC Evol Biol. 2006;6:77. doi: 10.1186/1471-2148-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PR, Hicks DR, Oplinger ES, Doll JD, Bundy LG, Schuler RT, Holmes BJ. Alternative field crops manual. University of Wisconsin-Extension; 1989. Grain Sorghum (Milo) Cooperative Extension. http://www.hort.Perdue.edu/newcrop/afcm/sorghum.html. [Google Scholar]
- Chang C-C, Lin H-C, Lin I-P, Chow T-Y, Chen H-H, Chen W-H, Cheng C-H, Lin C-Y, Liu S-M, Chang C-C, Chaw S-M. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- Chase MW, Palmer JD. Chloroplast DNA systematics of lilioid monocots: resources, feasibility, and an example from the Orchidaceae. Am J Bot. 1989;76:1720–1730. [Google Scholar]
- Chebolu S, Daniell H. Stable expression of GAL/GALNAc lectin of Entamoeba histolytica in transgenic chloroplast and immunogenicity in mice towards vaccine development for amebiasis. Plant Biotechnol J. 2007;2:230–239. doi: 10.1111/j.1467-7652.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Lowe BA, Spencer MT, Ye X, Armstrong CL. Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. In Vitro Cell Dev Biol. 2004;40:31–45. [Google Scholar]
- Crop Plant Resources. [Accessed May 18, 2006];Sorghum: Sorghum bicolor. 2000 http://darwin.nmsu.edu/~molbio/plant/sorghum.html.
- Cui L, Veeraraghavan N, Richer A, Wall K, Jansen RK, Leebens-Mack J, Makalowska I, dePamphillis CW. ChloroplastDB: the chloroplast genome database. Nucleic Acids Res. 2006;34:D692–D696. doi: 10.1093/nar/gkj055. [ http://chloroplast.cbio.psu.edu/] [DOI] [PMC free article] [PubMed]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nat Biotechnol. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A. Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol. 2002;13:136–141. doi: 10.1016/s0958-1669(02)00297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Pahchal T, Wiebe P. Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol. 2001;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Camrmona-Sanchez O, Burns B. Chaper 8, Chloroplast derived antibodies, biopharmaceuticals and edible vaccines. In: Rischer R, Schillberg S, editors. Molecular Farming. Wiley-VCH; Weinheim: 2004a. pp. 113–133. [Google Scholar]
- Daniell H, Cohill P, Kumar S, Dufourmantel N, Dubald M. Chloroplast genetic engineering. In: Daniell H, Chase C, editors. Molecular biology and biotechnology of plant organelles. Springer; Dordrecht: 2004b. pp. 423–468. [Google Scholar]
- Daniell H, Chebolu S, Kumar S, Singleton M, Falconer R. Chloroplast-derived vaccine antigens and other therapeutic proteins. Vaccine. 2005a;23:1779–1783. doi: 10.1016/j.vaccine.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Daniell H, Kumar S, Dufourmantel N. Breakthroughs in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 2005b;23:238–245. doi: 10.1016/j.tibtech.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Grevich J, Saski C, Guda C, Tomkins J, Jansen RK. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet. 2006;112:1503–1518. doi: 10.1007/s00122-006-0254-x. [DOI] [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Saford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- de Heij HT, Lustig H, Moeskops DM, Bovenberg WA, Bisanz C, Groot GSP. Chloroplast DNAs of Spinacia, Petunia, and Spirodela have similar gene organization. Curr Genet. 1983;7:1–6. doi: 10.1007/BF00365673. [DOI] [PubMed] [Google Scholar]
- Dhingra A, Portis A, Jr, Daniell H. Enhanced translation of a chloroplast-expressed rbcS gene restores small subunit levels and photosynthesis in nuclear rbcS antisense plants. Proc Natl Acad Sci USA. 2004;101:6315–6320. doi: 10.1073/pnas.0400981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Davis JI, Soreng RJ, Garvin D, Anderson MJ. Chloroplast DNA inversions and the origin of the grass family (Poaceae) Proc Natl Acad Sci USA. 1992;89:7723–7726. doi: 10.1073/pnas.89.16.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Ballenger JA, Palmer JD. The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol Phylogenet Evol. 1996;5:429–438. doi: 10.1006/mpev.1996.0038. [DOI] [PubMed] [Google Scholar]
- Dufourmantel N, Pelissier B, Garcon F, Peltier G, Ferullo J-M, Tissot G. Generation of fertile transplastomic soybean. Plant Mol Biol. 2004;55:479–489. doi: 10.1007/s11103-004-0192-4. [DOI] [PubMed] [Google Scholar]
- Dufourmantel N, Tissot G, Goutorbe F, Garcon F, Jansens S, Pelissier B, Peltier G, Dubald M. Generation and analysis of soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Plant Mol Biol. 2005;58:659. doi: 10.1007/s11103-005-7405-3. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, Riemer C, Petrykowska H, et al. PipTools: a computational toolkit to annotate and analyze pairwise comparisons of genomic sequences. Genomics. 2002;80:681–690. doi: 10.1006/geno.2002.7018. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl M, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-San MA, Mingeo-Castel AM, Miller M, Daniell H. A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J. 2003;1:71–79. doi: 10.1046/j.1467-7652.2003.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ED. Cytotaxonomic studies in the genus Sorghum. Univ Calif Publ Bot. 1950;23:283–361. [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol. 2003a;20:1499–1505. doi: 10.1093/molbev/msg159. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. The chloroplast genome of the “basal” angiosperm Calycanthus fertilis— structural and phylogenetic analyses. Plant Syst Evol. 2003b;242:119–135. [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. The chloroplast genome of Nymphaea alba: whole-genome analyses and the problem of identifying the most basal angiosperm. Mol Biol Evol. 2004;21:1445–1454. doi: 10.1093/molbev/msh147. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH. Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol. 2005;22:1813–1822. doi: 10.1093/molbev/msi173. [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group. Phylogeny and subfamilial classification of the grasses (Poaceae) Ann Missouri Bot Gard. 2001;88:373–457. [Google Scholar]
- Grevich JJ, Daniell H. Chloroplast genetic engineering: recent advances and future perspectives. Crit Rev Plant Sci. 2005;24:83–108. [Google Scholar]
- Guda C, Lee SB, Daniell H. Stable expression of biodegradable protein based polymer in tobacco chloroplasts. Plant Cell Rep. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- Hiratsuka J, Shimada H, Whittier R, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Hirose T, Kusumegi T, Tsudzuki T, Sugiura M. RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol Gen Genet. 1999;262:462–467. doi: 10.1007/s004380051106. [DOI] [PubMed] [Google Scholar]
- Howe CJ, Barker RF, Bowman CM, Dyer TA. Common features of three inversions in wheat chloroplast DNA. Curr Genet. 1988;13:343–349. doi: 10.1007/BF00424430. [DOI] [PubMed] [Google Scholar]
- Hupfer H, Swaitek M, Hornung S, et al. Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome 1 of the five distinguishable Euoenthera plastomes. Mol Gen Genet. 2000;263:581–585. doi: 10.1007/pl00008686. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Raubeson LA, Boore JL, et al. Methods for obtaining and analyzing chloroplast genome sequences. Methods Enzymol. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Kaittanis C, Saski C, Lee S-B, Tompkins J, Alverson AJ, Daniell H. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 2006;6:32. doi: 10.1186/1471-2148-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Abbott A, Jesudurai C, Tomkins J, Main D. Frequency, type, distribution, and annotation of simple sequence repeats in Rosaceae ESTs. Funct Integr Genomics. 2005;5:136–143. doi: 10.1007/s10142-005-0139-0. [DOI] [PubMed] [Google Scholar]
- Kamarajugadda S, Daniell H. Choroplast derived anthrax and other vaccine antigens: their immunogenic and immunoprotective properties. Expert Rev Vaccines. 2006;5:839–849. doi: 10.1586/14760584.5.6.839. [DOI] [PubMed] [Google Scholar]
- Katayama H, Ogihara Y. Phylogenetic affinities of the grasses to other monocots as revealed by molecular analysis of chloroplast DNA. Curr Genet. 1996;29:572–581. doi: 10.1007/BF02426962. [DOI] [PubMed] [Google Scholar]
- Kato T, Kaneko T, Sato S, Nakamura Y, Tabata S. Complete structure of the chloroplast genome of a legume, Lotus japonicus. DNA Res. 2000;7:323–330. doi: 10.1093/dnares/7.6.323. [DOI] [PubMed] [Google Scholar]
- Kelchner SA. The evolution of non-coding chloroplast DNA and its application in plant systematics. Ann Missouri Bot Gard. 2002;87:482–498. [Google Scholar]
- Khan M, Maliga P. Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat Biotechnol. 1999;17:910–915. doi: 10.1038/12907. [DOI] [PubMed] [Google Scholar]
- Kim K-J, Lee H-L. Complete chloroplast genome sequence from Korean Ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski S, Gould F, William M. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya V, Moayeri M, Leppla SH, Daniell H. Plant based vaccine: mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect Immun. 2005;73:8266–8274. doi: 10.1128/IAI.73.12.8266-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res. 2003;31:2417–2423. doi: 10.1093/nar/gkg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots and leaves confers enhanced salt tolerance. Plant Physiol. 2004a;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol Biol. 2004b;56:203–216. doi: 10.1007/s11103-004-2907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kwon H, Kwon S, et al. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed. 2003;11:1–13. [Google Scholar]
- Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics. 2006a;7:61. doi: 10.1186/1471-2164-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Kang K, Chung H, Yoo SH, Xu XM, Lee SB, Cheong JJ, Daniell H, Kim M. Plastid transformation in the monocotyledonous cereal crop, rice (Oryza sativa) and transmission of transgenes to their progeny. Mol Cells. 2006b;21:401–410. [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack J, Raubeson LA, Cui L, Kuehl J, Fourcade M, Chumley T, Boore JL, Jansen RK, dePamphilis CW. Identifying the basal angiosperms in chloroplast genome phylogenies: sampling one’s way out of the Felsenstein zone. Mol Biol Evol. 2005;22:1948–1963. doi: 10.1093/molbev/msi191. [DOI] [PubMed] [Google Scholar]
- Leelavathi S, Reddy V. Chloroplast expression of His-tagged GUS fusions: a general strategy to overproduce and purify foreign proteins using transplastomic plants as bioreactors. Mol Breed. 2003;11:49–58. [Google Scholar]
- Leelavathi S, Gupta N, Maiti S, Ghosh A, Reddy VS. Overproduction of an alkali- and thermo-stable xylanase in tobacco chloroplasts and efficient recovery of the enzyme. Mol Breed. 2003;11:59–67. [Google Scholar]
- Lopez-Juez E, Pyke KA. Plastids unleashed: their development and their integration in plant development. Int J Dev Biol. 2005;49:557–577. doi: 10.1387/ijdb.051997el. [DOI] [PubMed] [Google Scholar]
- Lossl A, Eibl C, Harloff HJ, Jung C, Koop H-U. Polyester synthesis in transplastomic tobacco (Nicotiana tabacum L.): significant contents of polyhydroxybutyrate are associated with growth reduction. Plant Cell Rep. 2003;21:891–899. doi: 10.1007/s00299-003-0610-0. [DOI] [PubMed] [Google Scholar]
- Maier RM, Neckermann K, lgloi GL, Kossel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- McBride K, Svab Z, Schaaf D, Hogan P, Stalker D, Maliga P. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- Molina A, Herva-Stubbs S, Daniell H, Mingo-Castel AM, Veramendi J. High yield expression of a viral peptide animal vaccine in transgenic tobacco chloroplasts. Plant Biotechnol J. 2004;2:141–153. doi: 10.1046/j.1467-7652.2004.00057.x. [DOI] [PubMed] [Google Scholar]
- National Sorghum Producers. What is Sorghum? 2006 www.sorghum.growers.com/Sorghum-101. Cited 06 Nov 2006.
- Nguyen TT, Nugent G, Cardi T, Dix PJ. Generation of homoplasmic plastid transformants of a commercial cultivar of potato (Solanum tuberosum L) Plant Sci. 2005;168:1495–1500. [Google Scholar]
- Ogihara Y, Isono K, Kojima T, et al. Chinese spring wheat (Triticum aestivum L.) chloroplast genome: complete sequence and contig clones. Plant Mol Biol Rep. 2000;18:243–253. [Google Scholar]
- Palmer JD. Isolation and structural analysis of chloroplast DNA. Methods Enzymol. 1986;118:167–186. [Google Scholar]
- Palmer JD. Plastid chromosomes: structure and evolution. In: Hermann RG, editor. The molecular biology of plastids. Cell culture and somatic cell genetics of plants. 7A. Springer; Vienna: 1991. pp. 5–53. [Google Scholar]
- Palmer JD, Stein DB. Conservation of chloroplast genome structure among vascular plants. Curr Genet. 1986;10:823–833. [Google Scholar]
- Palmer JD, Osorio B, Thompson WF. Evolutionary significance of inversions in legume chloroplast DNAs. Curr Genet. 1988;14:65–74. [Google Scholar]
- Peeters NM, Hanson MR. Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA. 2002;8:497–511. doi: 10.1017/s1355838202029424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Provan J, Soranzo N, Wilson N, Goldstein D, Powell W. A low mutation rate for chloroplast microsatellites. Genetics. 1999;153:943–947. doi: 10.1093/genetics/153.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Powell W, Hollingsworth PM. Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol. 2001;16:142–147. doi: 10.1016/s0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- Quesada-Vargas T, Ruiz ON, Daniell H. Characterization of heterologous multigene operons in transgenic chloroplasts: transcription, processing, translation. Plant Physiol. 2005;128:1746–1762. doi: 10.1104/pp.105.063040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley F, Weil JH. Organization and sequence of five tRNA genes and of an unidentified reading frame in the wheat chloroplast genome: evidence for gene rearrangements during the evolution of chloroplast genomes. Curr Genet. 1985;9:495–503. doi: 10.1007/BF00434054. [DOI] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast genomes of plants. In: Henry R, editor. Diversity and evolution of plants-genotypic and phenotypic variation in higher plants. CABI Publishing; Wallingford: 2005. pp. 45–68. [Google Scholar]
- Reichman JR, Watrud LS, Lee EH, Burdick C, Bollman M, Storm M, King G, Mallory-Smith C. Establishment of transgenic herbicide-resistant creeping bentgrass (Agrostis stolonifera L.) in nonagronomic habitats. Mol Ecol. 2006;15:4243–4255. doi: 10.1111/j.1365-294X.2006.03072.x. [DOI] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger I, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell D. Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC Genomics. 2006;7:224. doi: 10.1186/1471-2164-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H. Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts––oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotechnol J. 2007 doi: 10.1111/j.1467-7652.2007.00259.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz ON, Daniell H. Engineering cytoplasmic male sterility via the chloroplast genome by expression of β-ketothiolase. Plant Physiol. 2005;138:2–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz O, Hussein S, Terry N, Daniell H. Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol. 2003;132:1344–1352. doi: 10.1104/pp.103.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saski C, Lee S-B, Daniell H, Wood TC, Tomkins J, Kim H-G, Jansen RK. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Maier RM, Alcaraz JP, Cottet A, Herrman RG, Mache R. The plastid chromosome of spinach (Spinacia oleracea) complete nucleotide sequence and gene organization. Plant Mol Biol. 2001;45:307–315. doi: 10.1023/a:1006478403810. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Regel R, Du TG, Hupfer H, Herrmann RG, Maier RM. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol Biol Evol. 2002;19:1602–1612. doi: 10.1093/oxfordjournals.molbev.a004222. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Program NCS, Green ED, Hardison RC, Miller W. MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid-Masood M, Nishikawa T, Fukuoka S, Njenga PK, Tsudzuki T, Kadowaki K. The complete nucleotide sequence of wild rice (Oryza nivara) chloroplast genome: first genome wide comparative sequence analysis of wild and cultivated rice. Gene. 2004;340:133–139. doi: 10.1016/j.gene.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, et al. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analyses. Am J Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PT, Staub JM, Nehra NS. Technical advance: stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Spangler RE. Taxonomy of Sarga, Sorghum and Vacoparis (Poaceae: Andropogoneae) Aust Syst Bot. 2003;16:279–299. [Google Scholar]
- Spangler RE, Zaitchik B, Russo E, Kellogg E. Andropogoneae evolution and generic limits in Sorghum (Poaceae) using ndhF sequences. Syst Bot. 1999;24:267–281. [Google Scholar]
- Staub JM, Garcia B, Graves J, et al. High yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- Steane DA. Complete nucleotide sequence of the chloroplast genome from the Tasmanian Blue Gum, Eucalyptus globulus (Myrtaceae) DNA Res. 2005;12:215–220. doi: 10.1093/dnares/dsi006. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), ver. 4.0. Sinauer Associates; Sunderland: 2003. [Google Scholar]
- Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, Mc-Couch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am J Bot. 2007;94:302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- US Grains Council. 2006 http://www.grains.org/page.ww?section=Barley%2C+Corn+%26+Sorghum&name=Sorghum. Cited 06 Nov 2006.
- USDA. 2006 http://www.ars.usda.gov/research/projects/projects.htm?accn_no=408935. Cited 08 Nov 2006.
- Vitanen PV, Devine AL, Kahn S, Deuel DL, Van-Dyk DE, Daniell H. Metabolic engineering of the chloroplast genome using the E. coli ubiC gene reveals that corismate is a readily abundant precursor for 4-hydroxybenzoic acid synthesis in plants. Plant Physiol. 2004;136:4048–4060. doi: 10.1104/pp.104.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T, Sugiura M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc Natl Acad Sci USA. 1994;91:9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrud LS, Lee EH, Fairbrother A, Burdick C, Reichman JR, Bollman M, Storm M, King G, Van de Water PK. Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass with CP4 EPSPS as a marker. Proc Natl Acad Sci USA. 2004;101:14533–14538. doi: 10.1073/pnas.0405154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J, Koya V, Leppla SH, Daniell H. Expression of Bacillus anthracis protective antigen in transgenic chloroplasts of tobacco, a non-food/feed crop. Vaccine. 2004;22:4374–4384. doi: 10.1016/j.vaccine.2004.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D, Hester M, Liu A, Burke J. Chloroplast SSR polymorphisms in the compositae and the mode of organellar inheritance in Helianthus annuus. Theor Appl Genet. 2005;110:941–947. doi: 10.1007/s00122-004-1914-3. [DOI] [PubMed] [Google Scholar]
- Wipff JK, Fricker C. Gene flow from transgenic creeping bentgrass (Agrostis stolonifera L.) in the Willamette valley, Oregon. Int Turfgrass Soc Res J. 2001;9:224–242. [Google Scholar]
- Wolf PG, Rowe CA, Hasebe M. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene. 2004;339:89–97. doi: 10.1016/j.gene.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Boore JL, Jansen RK. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Zeltz P, Hess WR, Neckermann K, Borner T, Kossel H. Editing of the chloroplast rpoB transcript is independent of chloroplast translation and shows different patterns in barley and maize. EMBO J. 1993;12:4291–4296. doi: 10.1002/j.1460-2075.1993.tb06113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. PhD dissertation. The University of Texas; Austin: 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [ www.bio.utexas.edu/faculty/antisense/garli/Garli.html] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.