Abstract
TNF-α is a potent proinflammatory cytokine that induces endothelial cell (EC) adhesion molecules. In addition, TNF promotes angiogenesis by inducing an EC tip cell phenotype and the expression of jagged-1, a ligand for the notch pathway. Notch signaling is critical for vascular patterning and helps to restrict the proliferation of tip cells. Here we demonstrate that TNF induction of jagged-1 in human EC is rapid and dependent upon signaling through TNFR1, but not TNFR2. A luciferase reporter construct carrying 3.7 kb of 5′ promoter sequence from the human gene was responsive to both TNF and overexpression of NFκB pathway components. TNF-induced promoter activation was blocked by treatment with an NFκB inhibitor or co-expression of dominant-negative IKKβ. Mutations in a putative NFκB-binding site at −3.0 kb, which is conserved across multiple species, resulted in a loss of responsiveness to TNF and NFκB. Electromobility shift and chromatin immunoprecipitation assays revealed binding of both p50 and p65 to the promoter in response to TNF treatment. Full promoter activity also depends on an AP-1 site at −2.0 kb. These results indicate that canonical NFκB signaling is required for TNF induction of the notch ligand jagged-1 in EC.
Keywords: Angiogenesis, transcription, endothelial, TNF, NFκB, Notch
1. INTRODUCTION
The demands on endothelial cells (EC) vary under different physiological states. EC are non-thrombogenic, express blood components, regulate transfer of nutrients and waste between blood and tissues, regulate immune cell activation and recruitment, and under conditions of growth or tissue repair, undergo angiogenic sprouting to generate new vessels. How EC switch from the quiescent, homeostatic maintenance phenotype to the proliferative, migratory, pro-angiogenic phenotype is currently the focus of intense study as the regulation of this switch has implications for development, wound healing, diabetic retinopathy and tumor growth.
Recently, we identified the inflammatory mediator TNF as a key effector in wound healing that coordinates the onset of angiogenesis with the resolution of inflammation (Sainson et al., 2008). In an inflammatory setting TNF, which is largely derived from infiltrating monocyte/macrophages, blocks EC proliferation and migration mediated by VEGF, while concomitantly inducing an EC tip cell phenotype, characterized by distinct morphology and gene expression. Tip cells lead developing sprouts and are highly motile, non-proliferative and do not form a lumen (Gerhardt et al., 2003; Sainson et al., 2005). TNF induces a number of genes that are enriched in these tip cells and that are necessary for their function, including VEGFR2 and PDGFB (Sainson et al., 2008). In addition, we showed that TNF also induces expression of the notch ligand jagged-1. Notch signaling is critical for vascular development (Krebs et al., 2000; Duarte et al., 2004; Gale et al., 2004) and in particular it regulates EC proliferation and limits tip cell sprouting. Indeed, in the absence of a notch signal an excess of sprouting is seen and vessels are enlarged as a result of EC hyperproliferation (Limbourg et al., 2005; Sainson et al., 2005; Hellstrom et al., 2007; Leslie et al., 2007; Lobov et al., 2007; Siekmann and Lawson, 2007; Suchting et al., 2007).
There is considerable interest in the regulation of gene expression during angiogenesis and much of the focus has been on the role of VEGF. Very little is known about coordination of angiogenic gene expression with inflammation and so the role of TNF in this process is of particular interest We have, therefore, sought the mechanistic basis for regulation of jagged-1 expression by TNF and show that NFκB and AP-1 regulatory elements in the 5′ promoter region of this gene are critical, and that p50/p65 heterodimers are the key effector of the TNF-TNFR1-IKK pathway in EC upstream of the jagged-1 promoter.
2. MATERIALS AND METHODS
2.1 Cell culture conditions
Primary human umbilical vein endothelial cells (HUVEC) were isolated from neonatal umbilical cords as described previously (Hughes et al., 1990). Cells were routinely maintained at 37°C, 5% CO2 in EGM-2 medium (Clonetics, Walkersville, MD) and used between passages 3 and 6. Before addition of TNF cells were rested overnight in low (1%) serum. Recombinant human TNF-α (Invitrogen, Carlsbad, CA) was added to cultures for the times indicated to a final concentration of 10 ng/ml. For NFκB inhibitor studies, cells were treated with 10 or 40 μM BAY 11-7082 ((E)-3-(4-Methylphenylsulfonyl)-2-propenenitrile, CalBiochem, San Diego, CA) or an equivalent concentration of DMSO carrier as control for the times indicated. For jnk inhibition we used 10 or 40 μM SP-600125 (Calbiochem). Blocking antibodies to TNF-R1 and TNF-R2 were from R&D Systems and were used at 5x their reported median neutralizing dose (ND50).
2.2 Cloning, and analysis of jagged1 promoter sequence
Genomic DNA was isolated from HUVEC by standard protocol. Using the GenBank sequence for human chromosome 20 as a template, we designed PCR primers to amplify 2.6 kb or 3.8 kb of sequence upstream of the jagged1 transcriptional start site. Primers contained restriction enzyme site linkers as follows (restriction sites underlined): KpnJP2.6 fwd: 5′-CGCGGTACCCACCAGCCTTTTTCAGC-3′, KpnJP3.8 fwd: 5′-CGCGGTACCCACCCACCCTCAAAATCA-3′, and BamJP rev: 5′-CCGCGGGATCCGGGACGCCGCCGCTGCT-3′. PCR was performed on a PTC-200 thermocycler (MJ Research, MA) using genomic DNA and Phusion HS DNA polymerase (Finnzymes, Finland) with the following parameters: 98°C for 1.5 min (1 cycle); 98°C for 15 s, 62°C for 1 min, 72°C for 3 min (25 cycles); and 72°C for 5 min (1 cycle). PCR products were electrophoretically separated on 1.25% agarose gels and the appropriate sized band cut out and purified using the PerfectPrep Gel Cleanup kit (Eppendorf, Westbury, NY). Purified PCR products were digested with KpnI and BamHI restriction enzymes (Invitrogen) and ligated into KpnI and BglII-digested pGL3enhancer Luciferase Reporter vector (Promega, Madison, WI). The putative NFκB binding site in the jagged-1 promoter was mutated from (mutated bases underlined) GGGAGTCCC to TCTAGTCCC, and the AP-1 site was mutated from TGTTTCA to TATTAAC (lower strand sequence) using the QuickChange XL Site-Directed Mutagenesis kit (Stratagene, Cedar Creek, TX). All ligation reactions were transformed into E. coli DH5α (Invitrogen), amplified and purified by MaxiPrep (Qiagen, Valencia, CA). All constructs were verified by sequencing (Laguna Scientific, Laguna Hills, CA) and subsequent analysis using Lasergene software (DNAStar Inc, Madison, WI). We identified putative transcription factor binding sites using the TRANSFAC Database (www.gene-regulation.com).
2.3 Quantitative RT-PCR
Total RNA was isolated from confluent HUVEC grown in 6 well plates (Falcon) using the Aurum Total RNA Mini kit (Bio-Rad, Hercules, CA) according to manufacturer’s instructions. 1 μg of total RNA from triplicate samples was used for cDNA synthesis using the iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer’s instructions. Quantitative RT-PCR was performed using SYBR Green ER (Invitrogen) and HotStarTaq DNA Polymerase (Qiagen) on a Bio-Rad iCycler. Data were analyzed using iQ5 software (Bio-Rad). All samples were run in triplicate and normalized to a GAPDH standard curve. Primer sequences available on request.
2.4 Transfections and luciferase reporter assays
Confluent HUVEC grown in 6-well or 10 cm plates were transfected according to manufacturer’s instructions, with modifications, using Lipofectamine 2000 (Invitrogen). Briefly, 70–80% confluent HUVEC in 6-well plates were washed 3X with M199 medium (Gibco/Invitrogen) before incubation with 3 ml transfection cocktail containing 1–1.5 μg total DNA per well. After 3 hours, the transfection cocktail was replaced with fresh M199 supplemented with 10% fetal bovine serum. Transfected cells were incubated overnight in low (1%) serum before treatment or lysis as indicated. Transfection efficiencies were determined by analyzing pEGFP-transfected cells by flow cytometry. Fluorescence intensities were collected in the FL1 (GFP+) and FL2 (control) channels and dot plots were generated. The number of GFP-positive cells was determined by counting the number of cells that fall “off axis”. This method identifies cells with low fluorescence which may be masked in single histogram plots. Transfection efficiencies were routinely > 80% GFP-positive. For cotransfection experiments equivalent concentrations of DNA were transfected per condition, with EGFP serving as balancer and/or negative control DNA. Luciferase assays were performed as previously described (Nakatsu et al., 2003). Notch signaling was assayed by measuring induction of RBP-luciferase, a gift of Dr. Zimber-Strobl (Munich, Germany). Expression plasmids for NFκB components p50, p65, and cRel were gifts of Dr. Nigel Mackman (Scripps Research Institute, CA), constitutively active (CA) IKKβ and dominant-negative (DN) IKKβ were gifts of Dr. Craig Walsh (UC Irvine). The c-jun expression plasmid was from Dr. Al Bothwell Yale). The c-fos plasmid was from Open Biosystems.
2.5 Chromatin immunoprecipitation and gel shift assays
Chromatin immunoprecipitation (ChIP) was performed according to manufacturer’s instructions (Millipore, Danvers, MA) using antibodies directed against p50 and p65 (Santa Cruz Biotechnologies, Santa Cruz, CA). PCR amplification of specific and control sequences used the following primers. Jagged promoter flanking the NFκB site at −3034: Fwd – CTC TCG GCA GCA GTT CCT CAT; Rev – TAG GTG AAG CCA GGT GGA GAT CT (product 457bp); VCAM promoter flanking the tandem NFκB sites: Fwd – CCA CCC CCT TAA CCC ACA TT; Rev – TAA AAT GCC TGC GAA GAT GGT C (product 456bp); β-actin promoter: Fwd – GGC CCC ACC TCA CCA CTC TTC CTA; Rev – AGA CAT ACA ACG GAC GGT GGG CCC (product 423bp). Electrophoretic mobility shift assays (EMSA) were performed using the LightShift Chemilluminescent EMSA kit (Pierce Biotechnology, Rockford, IL) according to manufacturer’s instructions. Briefly, 5 μg HUVEC nuclear protein extracts were combined with 20 fmol biotinylated duplex DNA probe (IDT, Coralville, IA), 50 ng/ml poly dI:dC and 1X binding buffer in a 20 μl volume and incubated for 20 minutes at room temperature. For competition reactions, a 50-fold excess of unlabeled duplex probe (IDT) was added to each reaction. For supershift studies, reaction mixtures were first incubated on ice for 20 minutes with 2 μg antibodies directed against p50, p65, c-rel, or control (Santa Cruz Biotechnologies) before addition of biotinylated probe and incubation at room temperature for 20 min. Reaction mixtures were electrophoresed in 5% polyacrylamide gels (Bio-Rad) in 0.5X TBE buffer at 100V for 60 minutes before electrophoretic transfer to a positively-charged nylon membrane (Ambion, Austin, TX) at 100V for 45 minutes. Membranes were UV-cross-linked for 60 seconds at 120 mJ/cm2, before LightShift detection according to manufacturer’s instructions.
2.6 Flow Cytometry
Surface expression of jagged1 protein was determined by immunostaining cells with polyclonal goat anti-jagged1 (R&D Systems, Minneapolis, MN) or isotype control primary antibody followed by FITC-conjugated anti-goat secondary antibody (Jackson ImmunoResearch, West Grove, PA). Cells were subsequently characterized using a FACSCalibur flow cytometer and CellQuest software (Becton Dickinson, NJ).
2.7 Statistical analyses
The differences between experimental groups of equal variance were analyzed using Student’s t-test with p<0.05 being considered significant. All experiments were performed at least three times with similar results, except where indicated.
3. RESULTS
3.1 TNF-α induces expression of jagged1 in EC via TNFR1
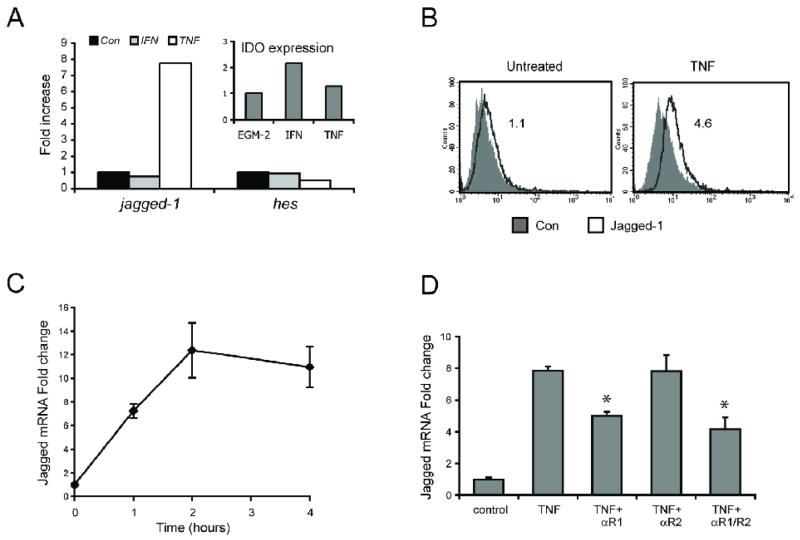
Jagged-1 mRNA expression in EC was assessed by qRT-PCR in cells treated with the pro-inflammatory cytokines TNF or IFN-γ for 4 hours. IFN-γ treatment had no effect on Jagged-1 expression or on expression of the notch target gene HES1 (Fig. 1A), although it did induce indolamine 2,3-dioxygenase (Fig. 1A inset). In sharp contrast TNF strongly induced jagged-1 expression, and had a mildly inhibitory effect on HES1. TNF also induced a 4-fold increase in surface expression of jagged-1 (Fig. 1B). Induction of mRNA was rapid, reaching peak expression by 2 hours (Fig. 1C). Notch1 expression was decreased by TNF treatment (data not shown) consistent with the decrease in expression of the notch target HES1. EC express two distinct TNF receptors and each triggers a different signaling cascade, with TNFR1 triggering the canonical NFκB pathway in EC (Zhou et al., 2007). To determine which is involved in the induction of jagged-1 mRNA we used blocking antibodies specific to each receptor. Only the antibody to TNFR1 blocked jagged-1 induction and there was no additive effect when both were used together (Fig. 1D). The failure to see complete blocking is likely due to the relatively low affinity of antibodies for the receptors relative to the pM binding affinity of the cytokine. These data suggest that in EC TNF signals through TNFR1, and likely through the NFκB pathway, to induce jagged-1 expression.
Figure 1. TNF induces jagged-1 expression through TNFR1.
A. EC were treated with either IFN-γ (100ng/ml) or TNF (10ng/ml) for 4 hours and mRNA was harvested for qRT-PCR analysis. Jagged-1 and HES-1 expression were normalized to GAPDH and fold-increase is plotted. Inset – Indolamine 2,3-dioxygenase (IDO) mRNA expression was normalized to GAPDH and fold-increase is plotted. B. EC were rested or treated with TNF (10ng/ml) for 4 hours and harvested for FACS analysis using either an isotype control or a jagged-1-specific primary antibody and a FITC-conjugated secondary. C. EC were treated with TNF (10ng/ml) for the indicated times and mRNA was harvested for qRT-PCR analysis. Jagged-1 expression was normalized to GAPDH and fold-increase (mean and SD) is plotted. D. EC were treated with TNF (10ng/ml) for 4 hours in the presence of control antibody (TNF alone), a blocking antibody to TNF-R1, a blocking antibody to TNF-R2, or a combination of both antibodies, and mRNA was harvested for qRT-PCR analysis. Jagged-1 expression was normalized to GAPDH and fold-increase (mean and SD) is plotted. * p<0.005 by t-test, compared to TNF + control antibody.
3.2 The jagged-1 promoter contains a distal TNF response element
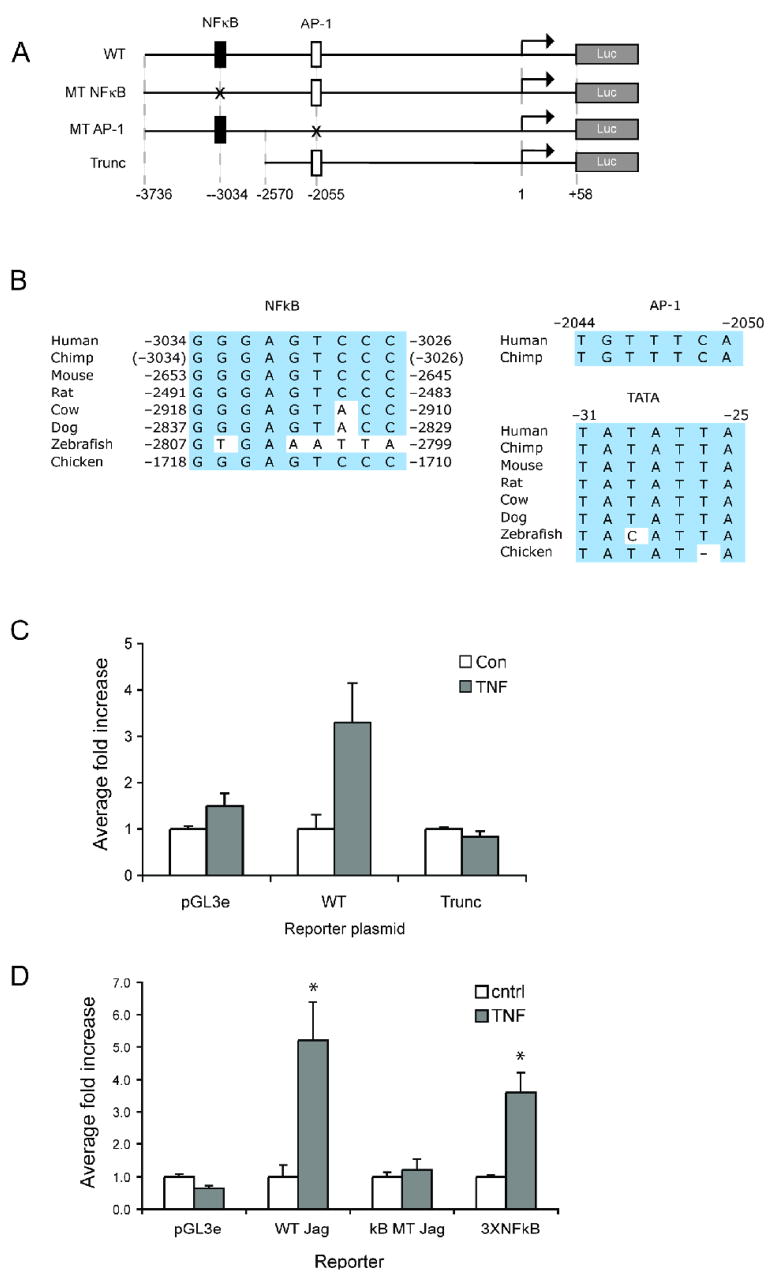
We analyzed the human jagged-1 promoter using the TRANSFAC database and identified numerous potential binding sites for known TNF-inducible transcription factors. Of particular interest was a perfect consensus NFκB site at −3034/−3026 upstream of the transcription start site (TSS) (Fig. 2A). Also of interest was a variant AP-1 site at −2055/−2050. Important sites are often conserved across species and this is the case for the NFκB site (Fig. 2B). The identical sequence is present in human, chimp, mouse, rat and chicken. The same C to A variant is present in both dog and cow. We could not identify a similar sequence in the zebrafish promoter. Interestingly, the AP-1 site seems to be specific for human and chimp as a similar sequence was not found in the promoters of the other species. The TATA box at −31 is also conserved across all species, except for zebrafish where a TACA variant is present upstream of the putative TSS. We PCR-amplified a 3.8 kb fragment of the human promoter region and cloned this into the pGL3e-Luc reporter vector. This construct, which we refer to as wild type (WT), contains 3736 bp upstream of the TSS and 58 bp downstream (Fig. 2A).
Figure 2. The jagged-1 promoter contains an NFκB element and responds to TNF.
A. The human wild type (WT) jagged-1 promoter was cloned upstream of the luciferase gene in pGL3-Luc. The NFκB and AP-1 sites are indicated. A truncated version of the promoter was also generated that lacks the NFκB site. B. The NFκB site and the TATA box is conserved across species. Note that the lower (reverse) strand sequence of the AP-1 site is shown and numbered accordingly. C. EC were transfected with the control (pGL3e) vector or with the WT or truncated promoter reporters, rested overnight in low serum (1%), and then treated for 4 hours with TNF (10ng/ml) before lysis and luciferase assay. * p<0.02 by t-test compared to untreated cells. D. EC were transfected with the control (pGL3e) vector, with the WT or Mutant NFkB promoter reporter, or with an NFκB reporter containing multimerized canonical binding sites. Cells were rested overnight in low serum (1%) and then treated for 4 hours with TNF (10ng/ml) before lysis and luciferase assay. * p<0.005 by t-test compared to untreated cells.
Given our results implicating the NFκB pathway downstream of TNFR1 we chose to first focus our attention on the putative NFκB binding site at −3034. We transfected EC with the WT construct or a truncated version of the promoter (−2570 to + 58 bp), which lacks the NFκB site (Fig. 2A), and assayed for luciferase activity in control and TNF-treated cells (Fig. 2C). The WT promoter responded strongly to TNF, showing on average a 4–8 fold induction (Fig. 2C), whereas the truncated promoter was not TNF responsive. Interestingly, however, the truncated promoter consistently had a 3 to 5-fold higher basal activity than the WT promoter, suggesting that negative regulatory elements may lie upstream of −2570 (data not shown). We next generated a 3 bp mutation in the putative NFκB site of the WT promoter designed to block binding of NFκB proteins (GGGAGTCCC to TCTAGTCCC). When transfected into EC this promoter consistently failed to respond to TNF, whereas the WT promoter was strongly responsive (Fig. 2D). As an additional positive control we used an NFκB reporter (consisting of three canonical NFκB sites driving luciferase), and this also strongly responded to TNF (Fig. 2D). Thus the putative NFκB site at −3034 is a TNF-response element.
3.3 TNF induction of the jagged-1 promoter depends on NFκB signaling
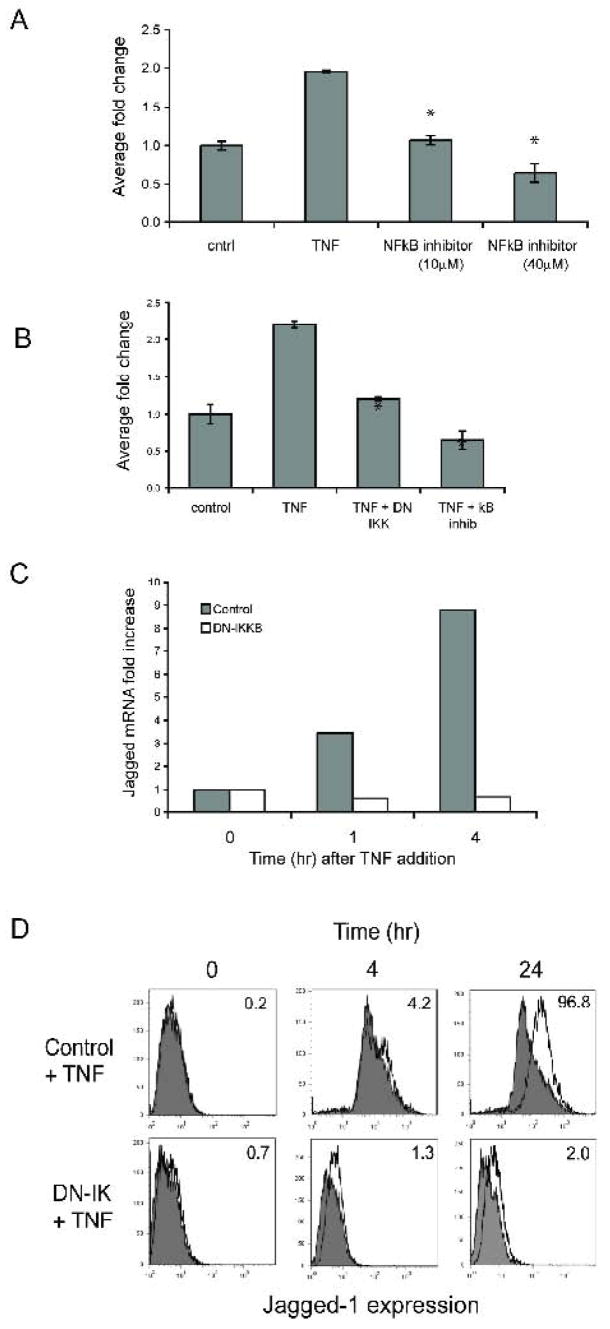
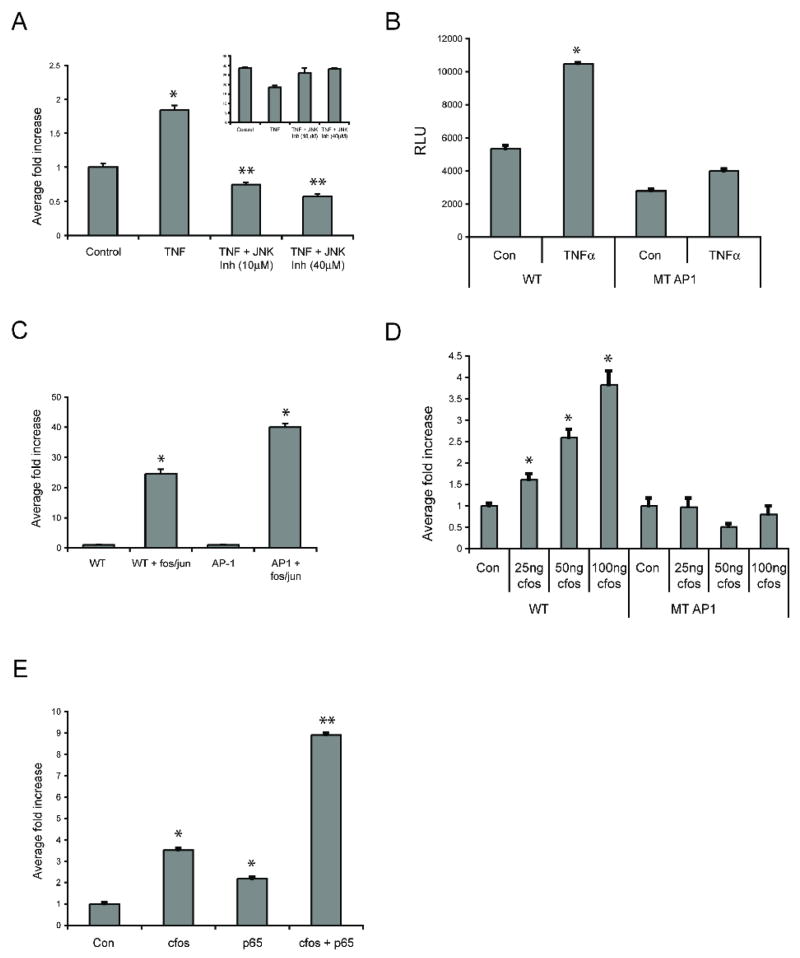
To test the role of the NFκB pathway downstream of TNF we first turned to a chemical inhibitor of NFκB signaling. BAY 11-7082 selectively inhibits TNF-induced phosphorylation of IκBα, thereby blocking release of NFκB to the nucleus. The inhibitor dose-dependently blocked TNF-induction of the jagged-1 promoter, completely blocking the response at 40μM (Fig. 3A), indicating that the TNF response is entirely dependent on the release of NFκB. As a further test of this hypothesis we co-transfected EC with the WT promoter and a dominant negative (DN) form of IKKβ, the enzyme that phosphorylates IκB. Again, the TNF induction of jagged-1 promoter activity was completely blocked by inhibition of the NFκB pathway (Fig. 3B). To confirm that the endogenous gene is similarly sensitive to blocking of the NFκB pathway we transfected EC with the DN-IKKβ, rested them for 4 hours and then treated with TNF before harvesting RNA for qRT-PCR analysis of jagged-1 expression. Jagged-1 mRNA was strongly induced by 1 hour and more so by 4 hours of TNF treatment, and in both cases this induction was completely blocked in cells expressing DN-IKKβ (Fig. 3C). To confirm that changes in mRNA levels correlate with changes in protein expression we transfected cells with the DN-IKKβ and treated these with TNF for 4 or 24 hours before examining jagged-1 expression by FACS. By 24 hours the control-transfected EC showed robust expression of jagged-1, whereas cells expressing DN-IKKβ showed no induction (Fig. 3D). Interestingly, the increased background staining, which is often seen with TNF treatment, was also suppressed. Thus, consistent with the promoter analysis, TNF induction of endogenous jagged-1 mRNA and protein expression is also dependent on NFκB signaling.
Figure 3. TNF regulates the jagged-1 promoter through NFκB.
A. EC were transfected with the WT jagged-1 promoter reporter, rested overnight in low serum (1%), and then treated for 4 hours with TNF (10ng/ml) in the presence or absence of the NFκB inhibitor BAY 11-7082 or DMSO, as indicated. Cells were then lysed for luciferase assay. * p<0.005 by t-test compared to cells treated with TNF and DMSO. B. EC were transfected with the WT jagged-1 promoter reporter along with an expression plasmid for DN-IKKβ or a control (GFP) plasmid, rested overnight in low serum (1%), and then treated for 4 hours with TNF (10ng/ml) in the presence or absence of the NFκB inhibitor BAY 11-7082 (40μM), as indicated. Cells were then lysed for luciferase assay. * p<0.005 by t-test compared to cells treated with TNF alone. C. EC were transfected with DN-IKKβ, rested overnight in low serum (1%), and then treated for the indicated times with TNF (10ng/ml). mRNA was harvested for qRT-PCR analysis and jagged-1 expression was normalized to GAPDH and fold-increase plotted. D. EC were transfected with control (GFP) or DN-IKKβ expression plasmids, rested overnight and then treated with TNF (10ng/ml) for 0, 4 or 24 hours before harvesting for FACS analysis using either an isotype control or a jagged-1-specific primary antibody and a FITC-conjugated secondary. Control staining is shown by the shaded plot, jagged staining by the open line. Corrected mean fluorescence intensity (jagged–control) is indicated.
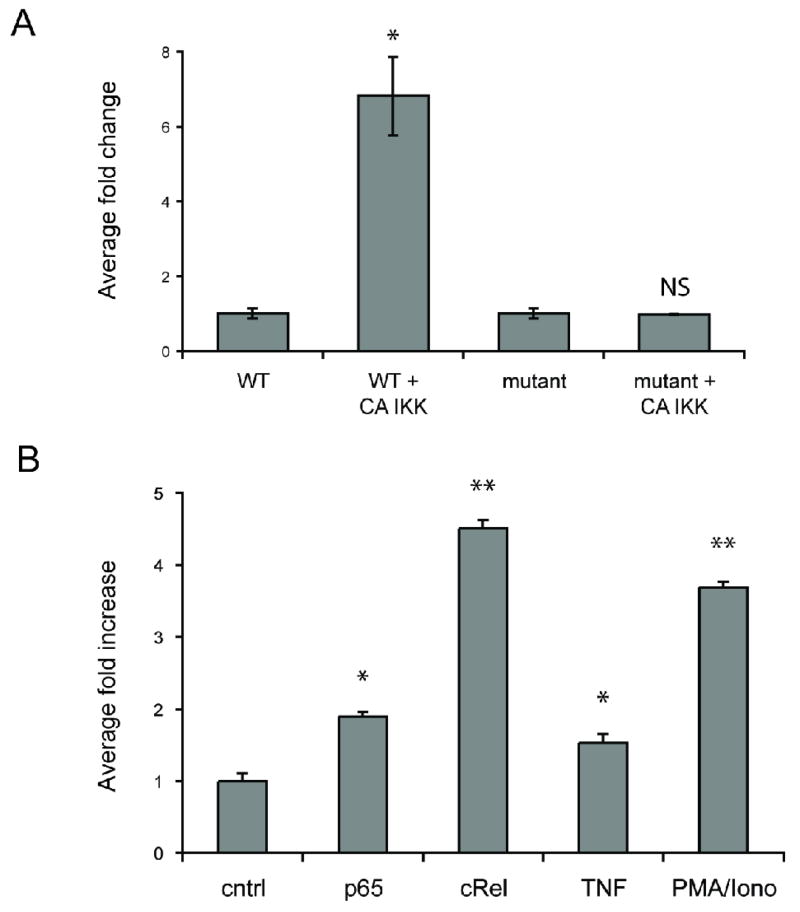
To test whether NFκB activation is sufficient to drive the jagged-1 promoter we cotransfected EC with the WT promoter-reporter and a constitutively active (CA) form of IKKβ that drives phosphorylation of IκB and thus NFκB activation. As shown in Fig. 4A CA-IKKβ induced the jagged-1 promoter by almost 7-fold compared to control (GFP-transfected) cells. Importantly, when we transfected EC with the reporter carrying a mutated NFκB site the promoter was not responsive to CA-IKKβ (Fig. 4A). Consistent with these findings, overexpression of the NFκB components p65 or c-rel also stimulated promoter activity – p65 by 2 to 3-fold and c-rel by 4 to 5-fold (Fig. 4B). PMA plus ionomycin served as a positive control. Taken together, these results suggest that NFκB signaling and the distal NFκB binding site are required for TNF-induced jagged-1 expression.
Figure 4. Components of the NFκB pathway activate the jagged-1 promoter.
A. EC were transfected with the WT or NFκB mutant jagged-1 promoter reporter along with an expression plasmid for CA-IKKβ or a control (GFP) plasmid. Cells were then lysed after 4 hours for luciferase assay. * p<0.002 by t-test compared to cells not expressing CA-IKKβ. NS – not significant compared to mutant promoter in the absence of CA-IKKβ. B. EC were transfected with the WT jagged-1 promoter reporter along with expression plasmids for p65, c-rel or control (GFP). EC transfected with WT reporter alone were treated with TNF (10ng/ml) or PMA/ionomycin (25nM/1μM, respectively). Cells were then lysed after 4 hours for luciferase assay. * p<0.01 by t-test compared to control cells. ** p<0.005 by t-test compared to control cells.
3.4 NFκB binds to the jagged-1 promoter
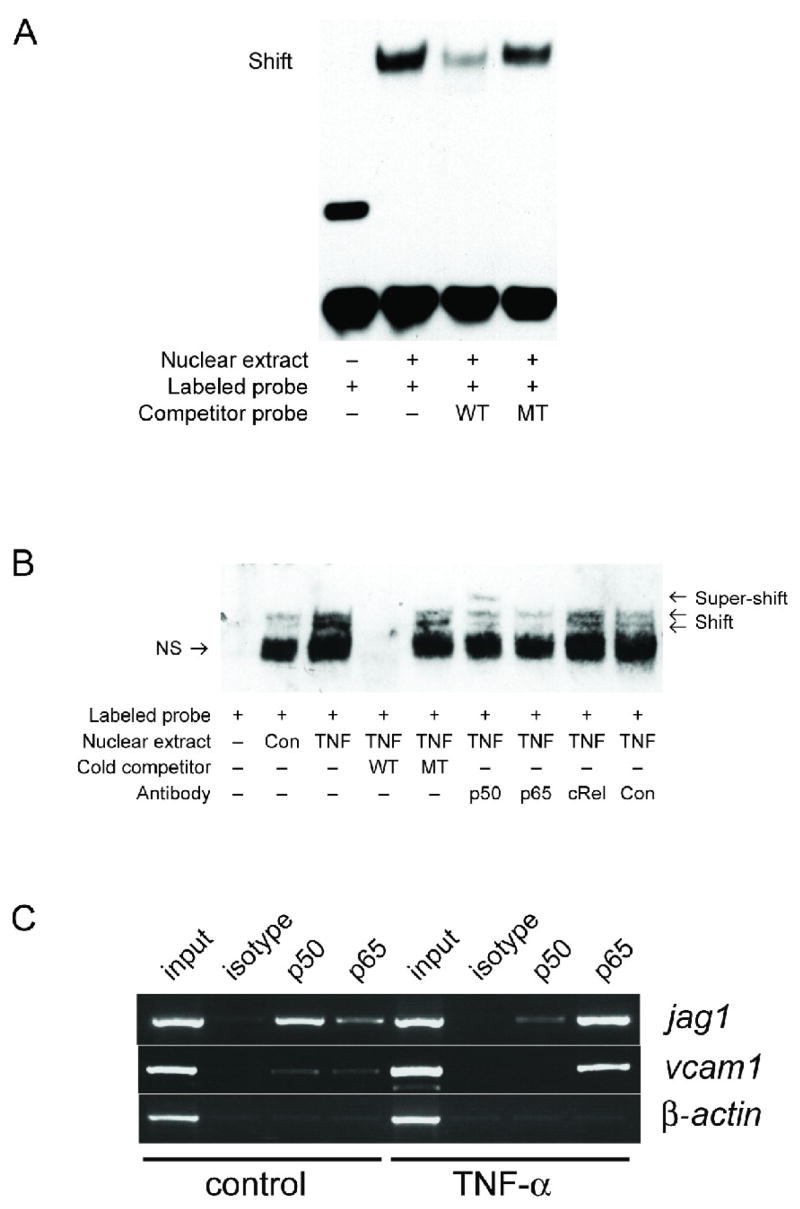
To determine whether NFκB proteins can bind to sequences in the jagged-1 promoter we first turned to electrophoretic mobility shift assays (EMSA). Probes were designed that covered the NFκB-response sequence identified above, as well as the mutant sequence and these were incubated with extracts from TNF-treated EC. These extracts strongly shifted the WT probe (Fig. 5A), and this binding was inhibited by an excess of WT but not mutant probe, indicating the presence of nuclear proteins in TNF-treated EC capable of binding specifically to the NFκB consensus sequence. To investigate further the nature of these proteins we used subunit-specific antibodies to either generate supershifts, or to block binding. As shown in Fig. 5B, nuclear extracts from TNF-treated EC contained considerably more NFκB binding activity than extracts from resting cells and again this was competed by an excess of WT but not mutant probe. An antibody to p50 generated a supershift, whereas an antibody to p65 inhibited protein binding (Fig. 5B). In contrast, an antibody to c-rel had no effect, indicating that the endogenous NFκB binding activity is composed of p50 and p65, but not c-rel, proteins, although as shown above, overexpressed c-rel can drive the promoter.
Figure 5. The NFκB site in the jagged-1 promoter binds p50 and p65.
A. EC were treated with TNF (10ng/ml) for 4 hours and nuclear extracts prepared and analyzed by EMSA using a labeled probe covering the NFκB site from the jagged-1 promoter. Where indicated a 50-fold molar excess of unlabeled WT or NFκB mutant probe was added to demonstrate specificity. B. EC were untreated or treated with TNF (10ng/ml) for 4 hours and nuclear extracts prepared and analyzed by EMSA using a labeled probe covering the NFκB site from the jagged-1 promoter. Where indicated a 50-fold molar excess of unlabeled WT or NFκB mutant probe was added to demonstrate specificity. Antibodies to p50, p65, c-rel or control were added to extracts for 20 min. prior to the addition of labeled probe. NS – non-specific band. C. ChIP analysis was performed using control or TNF-treated (10ng/ml) EC and antibodies specific for p50 and p65, or an isotype control. After cross-linking and precipitation, PCR was performed using primers specific for the jagged-1, VCAM-1 or β-actin promoters. Input DNA was used as a positive control.
These data show that nuclear extracts contain NFκB proteins that can bind to isolated NFκB response elements, however, it is important to show that these proteins can also bind to the full-length, endogenous promoter. To confirm that the endogenous jagged-1 promoter does indeed bind NFκB proteins we turned to a chromatin immunoprecipitation assay (ChIP). Confluent EC monolayers, control and TNF-treated, were crosslinked to preserve protein: DNA interactions, and the chromatin was purified and immunoprecipitated with anti-NFκB and control antibodies. PCR was used to amplify a 400 bp fragment of the jagged-1 promoter that included the NFκB site at −3034. As a positive control, a fragment of the VCAM-1 promoter containing the previously-identified NFκB site was also amplified, and as a negative control we used a fragment of the β-actin gene. In control cells we found only a very weak VCAM-1 signal with either the p50 or p65 antibodies, whereas in TNF-treated cells we saw the expected strong p65 signal (Fig. 5C), which correlates with activation of the VCAM-1 promoter. The negative control, β-actin, was not detectable in either control or TNF-treated cells – the expected result as this gene is not regulated by NFκB. Untreated cells, which express only low amounts of jagged-1 on their surface, yielded a strong signal for p50 on the jagged-1 promoter but only a weak p65 signal (Fig. 5C). Generally, p50 homodimers are considered to be less transcriptionally active than p50:p65 heterodimers (Hoffmann et al., 2006). In sharp contrast to control cells, this ratio is reversed in TNF-treated cells where we found a weak p50 signal but a strong p65 signal, correlating with the higher transcriptional activity of the jagged-1 promoter in TNF-treated cells. Taken together the EMSA and ChIP data demonstrate that in resting cells the NFκB site is likely occupied mostly by p50 homodimers, whereas in TNF-treated cells there is a shift toward p65-containing complexes, which correlates with enhanced jagged-1 transcription.
3.5 An AP-1 site also contributes to jagged-1 transcriptional induction
In addition to its effects on the NFκB pathway TNF is also known to activate c-jun N-terminal kinase (jnk), thereby affecting AP-1-dependent transcription. To determine whether jnk has a role in jagged induction downstream of TNF we transfected cells with the WT promoter reporter and stimulated them in the presence of increasing concentrations of the jnk inhibitor SP-600125. Jnk inhibition strongly reduced TNF-induced jagged-1 expression, as well as basal level expression (Fig. 6A), indicating that along with NFκB, jnk activity is also necessary for TNF-induced jagged-1 up regulation. The inhibitor was not toxic to the cells over this time course as protein levels were not affected (indeed, the inhibitor prevented the TNF-induced downregulation of protein synthesis (Fig. 6A inset)), and in addition, activity of the minimal TK promoter was also not affected (data not shown). The human and chimp promoters contain a putative AP-1 site at −2055 (TGTTTCA on the lower strand, compared to the consensus TGACTCA). This variant is also present and functional in the IL-2 promoter (Macian et al., 2001). We made a 4 bp mutation in this site (to TATTAAC) and tested responsiveness of the promoter to TNF. Loss of this site almost completely blocked TNF induction, indicating that both the AP-1 and NFκB sites are necessary downstream of TNF (Fig. 6B).
Figure 6. AP-1 also contributes to TNF-driven jagged-1 expression.
A. EC were transfected with the WT jagged-1 promoter, rested overnight in low serum (1%), and then treated for 4 hours with TNF (10ng/ml) in the presence or absence of the jnk inhibitor SP-600125 or DMSO, as indicated. Cells were then lysed for luciferase assay. * p<0.005 by t-test compared to control cells. ** p<0.005 by t-test compared to TNF-treated cells in the absence of inhibitor. Aliquots of cells were also used for protein assay (inset). B. EC were transfected with the WT or AP-1 mutant jagged-1 promoter, rested overnight in low serum (1%), and then treated for 4 hours with TNF (10ng/ml). Cells were then lysed for luciferase assay. * p<0.005 by t-test compared to control cells. C. EC were transfected with the WT jagged-1 promoter or an AP-1 reporter containing multimerized canonical AP-1 sites, along with expression plasmids for c-fos and c-jun, or control (GFP). Cells were incubated for 4 hours and then lysed for luciferase assay. * p<0.001 by t-test compared to control cells in the absence of fos/jun. D. EC were transfected with the WT or AP-1 mutant jagged-1 promoter along with increasing amounts of an expression plasmid for c-fos or control (GFP). Cells were incubated for 4 hours and then lysed for luciferase assay. * p<0.01 by t-test compared to control cells in the absence of c-fos. E. EC were transfected with the WT jagged-1 promoter along with suboptimal concentrations of expression plasmids for c-fos (100ng) and/or p65 (10ng), or control (GFP). Cells were incubated for 4 hours and then lysed for luciferase assay. * p<0.01 and ** p<0.001 by t-test compared to control (GFP) cells.
To confirm the responsiveness of the jagged-1 promoter to AP-1 we co-transfected EC with the WT promoter and c-jun and c-fos expression constructs. The promoter was strongly induced, by greater than 20-fold, as was a positive control AP-1 reporter (Fig. 6C). As expected, the mutant AP-1 promoter did not respond to increasing doses of a c-fos expression plasmid, whereas the WT promoter was strongly induced (Fig. 6D). Finally, as a direct test of cooperativity between NFκB and AP-1 we cotransfected EC with the WT promoter and sub-optimal amounts of expression plasmids for p65 and c-fos. While both transcription factors were able to induce modest induction alone (4-fold for c-fos and 2-fold for p65), they were strongly synergistic, inducing a 9.5-fold induction of luciferase when expressed together (Fig. 6E).
4. DISCUSSION
In previous studies we and others have shown that the notch pathway is a critical regulator of EC function during angiogenesis (Krebs et al., 2000; Limbourg et al., 2005; Sainson et al., 2005), and that the inflammatory mediator TNF induces expression of the notch ligand jagged-1 on the tip cells of developing sprouts (Sainson et al., 2008). TNF is an important regulator of the inflammatory response and acts to coordinate the onset of angiogenic sprouting with the resolution of inflammation, likely through targeting of the NFκB transcription factor family (Sainson et al., 2008). Here we have explored the mechanism underlying TNF regulation of jagged-1 expression in EC and show that this is dependent upon both NFκB and AP-1.
The NFκB pathway is a major effector of gene expression downstream of TNF signaling. The Rel or NFκB family of transcription factors is comprised of homo- and heterodimeric molecules made up from 5 subunits, p50/p105 (NFκB1), p52/p100 (NFκB2), p65 (RelA), RelB, and c-rel, related via their Rel homology domain, which mediates DNA binding (Hayden and Ghosh, 2008). The best characterized NFκB pathway involves the activation and nuclear translocation of a p50/p65 heterodimer, which can interact with a number of other different transcription factors and co-activators to form an array of regulatory complexes with varying effects on expression. Cytokine-inducible NFκB-responsive promoter elements have been identified in numerous cell types, including immune system cells and EC (Madge and Pober, 2001).
Our studies indicate that the endogenous jagged-1 promoter constitutively binds p50/p50 homodimers, which are replaced by p50/p65 heterodimers in TNF-stimulated cells. Several studies have indicated that p50/p50 homodimers can bind to an NFκB consensus site and repress transcription (Plaksin et al., 1993; Grundstrom et al., 2004; Guan et al., 2005), most likely by competing for the NFkB binding sequence site. Homodimers of p50 are not retained in the cytoplasm by IκB and are therefore free to enter the nucleus. Also, unlike p65, which has a transactivation domain and can be activated via phosphorylation or acetylation, p50 molecules have no transactivation domain. Our results are therefore in agreement with a model of NFκB-mediated transcriptional regulation in which p50/p50 homodimers are bound to the jagged-1 promoter and repress transcription in resting cells, and are then displaced by activated p50/p65 heterodimers that drive transcription in response to TNF signaling. Although we found no binding of c-rel to the endogenous promoter we did find that overexpression of c-rel drove jagged-1 transcription, a finding consistent with a previous report in HeLa cells (Bash et al., 1999). We have identified a perfect consensus NFκB site at −3034 that is absolutely required for TNF and NFκB-mediated jagged-1 transcription.
The jnk pathway has also been implicated in TNF signaling and previous studies have shown this pathway to be active in EC (Min and Pober, 1997; Zhang et al., 2007). Our studies show an absolute requirement for jnk activity in the TNF-mediated induction of jagged-1, and we have identified a variant AP-1 site at −2055 that mediates this response. Interestingly, the same sequence – TGTTTCA – is also present and functional in the IL-2 promoter (Macian et al., 2001). Our data further indicate that these sites cooperate to drive transcription of the jagged-1 gene as suboptimal doses of p65 and c-fos strongly synergize to activate the jagged-1 promoter-reporter, and loss of either site almost completely abolishes promoter activity. However, it should be emphasized that our data do not currently address AP-1 activity at the level of the endogenous promoter, rather, we infer that AP-1 and NFκB work in synergy based on the extensive reporter analysis we have performed for both activators, and our ChIP data for NFκB.
Important transcription factor binding sites are often conserved across multiple species and we find that to be the case with the NFκB site. The sequence is perfectly conserved between human, chimp, mouse, rat, and chicken, and is only one base pair different in cow and dog. Although these promoters vary somewhat in length, the position of the NFκB element is approximately the same in all – between 2.5 and 3.0 kb upstream of the transcription start site, with the exception of the chicken, where it is at −1.7kb. We were not able to identify a similar NFκB element in the putative zebrafish proximal promoter, although TNF and NFκB genes have been identified in this organism (Correa et al., 2004; Savan et al., 2005). There were weak NFκB sites in the more proximal promoter, however as a promoter fragment containing these, but not the site at −3034, did not respond to TNF we did not investigate these further (data not shown).
Interestingly, the AP-1 site does not appear to be conserved across species – being present in human and chimp but absent in all other sequences examined, suggesting that regulation of jagged-1 expression may be subtly different in rodents, for example, than in primates. Such differences have been noted before between human and mouse with regard to immune function and suggest caution in translating findings in mouse models to the human arena (Mestas and Hughes, 2004). In contrast, the TATA box at −31 is completely conserved between all species examined except zebrafish, where a TACA sequence is found at this location. There is also a variant AP-1 site at −1405 in the human promoter that we have not investigated, however it does not seem to compensate for the loss of the AP-1 site at −2055.
Our results demonstrate how jagged-1 expression can be regulated by the inflammatory mediator TNF, however this is not the only mechanism for regulation of this gene. A previous study found that jagged-1 was induced by wnt signaling in hair follicles (Estrach et al., 2006) and we have identified several LEF/TCF consensus sequences in the proximal promoter, including a perfect match across multiple species at position −116 (C.C.W. Hughes, unpublished observations). Our previous study demonstrated upregulation of jagged-1 but not dll4 on angiogenic EC under inflammatory conditions (Sainson et al., 2008), and jagged-1 is expressed at the leading edge of repairing vessels (Lindner et al., 2001). Jagged-1 may play a similar role in inflammatory angiogenesis as dll4 does during development, regulating vessel branching, and potentially recruitment and activation of pericytes and vascular smooth muscle cells (High et al., 2008). Understanding the regulation of jagged-1 expression thus has implications for multiple steps in the angiogenic process, and these studies further expand our knowledge of the role of TNF in the co-regulation of angiogenesis and inflammation.
Acknowledgments
This work was supported, in part, by NIH RO1 HL60067 and HC086959.
Abbreviations
- EC
endothelial cell(s)
- TNF
tumor necrosis factor
- VEGF
vascular endothelial growth factor
- PDGF
platelet-derived growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bash J, Zong WX, Banga S, Rivera A, Ballard DW, Ron Y, Gelinas C. Rel/NF-kappaB can trigger the Notch signaling pathway by inducing the expression of Jagged1, a ligand for Notch receptors. EMBO J. 1999;18:2803–11. doi: 10.1093/emboj/18.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa RG, Tergaonkar V, Ng JK, Dubova I, Izpisua-Belmonte JC, Verma IM. Characterization of NF-kappa B/I kappa B proteins in zebra fish and their involvement in notochord development. Mol Cell Biol. 2004;24:5257–68. doi: 10.1128/MCB.24.12.5257-5268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A, Hirashima M, Benedito R, Trindade A, Diniz P, Bekman E, Costa L, Henrique D, Rossant J. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18:2474–8. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;133:4427–38. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- Gale NW, Dominguez MG, Noguera I, Pan L, Hughes V, Valenzuela DM, Murphy AJ, Adams NC, Lin HC, Holash J, Thurston G, Yancopoulos GD. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci U S A. 2004;101:15949–54. doi: 10.1073/pnas.0407290101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–77. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundstrom S, Anderson P, Scheipers P, Sundstedt A. Bcl-3 and NFkappaB p50-p50 homodimers act as transcriptional repressors in tolerant CD4+ T cells. J Biol Chem. 2004;279:8460–8. doi: 10.1074/jbc.M312398200. [DOI] [PubMed] [Google Scholar]
- Guan H, Hou S, Ricciardi RP. DNA binding of repressor nuclear factor-kappaB p50/p50 depends on phosphorylation of Ser337 by the protein kinase A catalytic subunit. J Biol Chem. 2005;280:9957–62. doi: 10.1074/jbc.M412180200. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalen M, Gerhardt H, Betsholtz C. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–80. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- High FA, Lu MM, Pear WS, Loomes KM, Kaestner KH, Epstein JA. Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2008;105:1955–9. doi: 10.1073/pnas.0709663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–16. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- Hughes CC, Savage CO, Pober JS. Endothelial cells augment T cell interleukin 2 production by a contact-dependent mechanism involving CD2/LFA-3 interaction. J Exp Med. 1990;171:1453–67. doi: 10.1084/jem.171.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, Smith GH, Stark KL, Gridley T. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–52. [PMC free article] [PubMed] [Google Scholar]
- Leslie JD, Ariza-McNaughton L, Bermange AL, McAdow R, Johnson SL, Lewis J. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–44. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- Limbourg FP, Takeshita K, Radtke F, Bronson RT, Chin MT, Liao JK. Essential role of endothelial Notch1 in angiogenesis. Circulation. 2005;111:1826–32. doi: 10.1161/01.CIR.0000160870.93058.DD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner V, Booth C, Prudovsky I, Small D, Maciag T, Liaw L. Members of the Jagged/Notch gene families are expressed in injured arteries and regulate cell phenotype via alterations in cell matrix and cell-cell interaction. Am J Pathol. 2001;159:875–83. doi: 10.1016/S0002-9440(10)61763-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobov IB, Renard RA, Papadopoulos N, Gale NW, Thurston G, Yancopoulos GD, Wiegand SJ. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci U S A. 2007;104:3219–24. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–89. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- Madge LA, Pober JS. TNF signaling in vascular endothelial cells. Exp Mol Pathol. 2001;70:317–25. doi: 10.1006/exmp.2001.2368. [DOI] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–8. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Min W, Pober JS. TNF initiates E-selectin transcription in human endothelial cells through parallel TRAF-NF-kappa B and TRAF-RAC/CDC42-JNK-c-Jun/ATF2 pathways. J Immunol. 1997;159:3508–18. [PubMed] [Google Scholar]
- Nakatsu MN, Sainson RC, Aoto JN, Taylor KL, Aitkenhead M, Perez-del-Pulgar S, Carpenter PM, Hughes CC. Angiogenic sprouting and capillary lumen formation modeled by human umbilical vein endothelial cells (HUVEC) in fibrin gels: the role of fibroblasts and Angiopoietin-1. Microvasc Res. 2003;66:102–12. doi: 10.1016/s0026-2862(03)00045-1. [DOI] [PubMed] [Google Scholar]
- Plaksin D, Baeuerle PA, Eisenbach L. KBF1 (p50 NF-kappa B homodimer) acts as a repressor of H-2Kb gene expression in metastatic tumor cells. J Exp Med. 1993;177:1651–62. doi: 10.1084/jem.177.6.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainson RC, Aoto J, Nakatsu MN, Holderfield M, Conn E, Koller E, Hughes CC. Cell-autonomous notch signaling regulates endothelial cell branching and proliferation during vascular tubulogenesis. FASEB J. 2005;19:1027–9. doi: 10.1096/fj.04-3172fje. [DOI] [PubMed] [Google Scholar]
- Sainson RC, Johnston DA, Chu HC, Holderfield MT, Nakatsu MN, Crampton SP, Davis J, Conn E, Hughes CC. TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood. 2008;111:4997–5007. doi: 10.1182/blood-2007-08-108597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savan R, Kono T, Igawa D, Sakai M. A novel tumor necrosis factor (TNF) gene present in tandem with theTNF-alpha gene on the same chromosome in teleosts. Immunogenetics. 2005;57:140–50. doi: 10.1007/s00251-005-0768-4. [DOI] [PubMed] [Google Scholar]
- Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–4. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- Suchting S, Freitas C, le Noble F, Benedito R, Breant C, Duarte A, Eichmann A. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci U S A. 2007;104:3225–30. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang H, Lin Y, Li J, Pober JS, Min W. RIP1-mediated AIP1 phosphorylation at a 14-3-3-binding site is critical for tumor necrosis factor-induced ASK1-JNK/p38 activation. J Biol Chem. 2007;282:14788–96. doi: 10.1074/jbc.M701148200. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Connell MC, MacEwan DJ. TNFR1-induced NF-kappaB, but not ERK, p38MAPK or JNK activation, mediates TNF-induced ICAM-1 and VCAM-1 expression on endothelial cells. Cell Signal. 2007;19:1238–48. doi: 10.1016/j.cellsig.2006.12.013. [DOI] [PubMed] [Google Scholar]