Abstract
There is an intrinsic complexity in the pathogenesis of common diseases. The concept of gene–environment interaction is receiving support from emerging evidence coming primarily from studies involving diet and cardiovascular disease (CVD) and its various risk factors. The accumulating evidence shows that common variants at candidate genes for lipid metabolism, inflammation, and obesity are associated with altered plasma levels of classic and new biomarkers of metabolic syndrome and CVD risk. Major contributors to this knowledge have been a series of large population studies containing phenotype-rich databases and dietary information to which genetic data have been added. Although this approach has provided strong evidence supporting the concept of gene–diet interactions modulating CVD risk factors, the strength of the individual effect is very small, and the replication among studies is rather disappointing. Current population studies are starting to incorporate experimental and analytical approaches that could provide more solid and comprehensive results. However, other limitations, such as the size of the populations required to examine higher-level interactions, are still major obstacles to translating this knowledge into practical public health applications. Nevertheless, data from numerous molecular and genetic epidemiological studies provide tantalizing evidence suggesting that gene–environment interactions, i.e., the modulation by a genetic polymorphism of a dietary component effect on a specific phenotype (e.g., cholesterol levels and obesity), can interact in ways that increase the risk for developing chronic disease, including susceptibility to developing the metabolic syndrome. Once further experience is gained from patients and/or individuals at high risk, more personalized genetic-based approaches may be applied toward the primary prevention and treatment of CVDs and other complex inflammatory diseases.
Keywords: Diet, disease susceptibility, gene–environment interactions, genetics, inflammation, metabolic syndrome
The significant contribution of environmental factors to the susceptibility and causation of multifactorial diseases is becoming better appreciated. More than ever, health care professionals (HCPs) have embraced the link between proper diet and good health for the prevention and treatment of numerous disorders. In the most extreme case, malnutrition in the developing world clearly demonstrates the link between nutritional deficiencies and overall health. However, the comparatively prosperous developed countries are also suffering from malnutrition as a population, most clearly in the form of obesity and its associated disorders, such as diabetes, cardiovascular disease (CVD), and metabolic syndrome (MetS). Yet, the resulting dietary and treatment guidelines to address these disease states, typically the promotion of low-cholesterol, low-fat, low-salt food choices, do not consider the dramatic differences in an individual’s responses to such interventions. 1 HCPs only have to examine their typical patient load to realize that such interventions are not working precisely as predicted. Despite the mechanisms responsible for the interindividual differences in treatment response being far from fully understood, they are not unexpected, considering the variation in responses to treatment when examining pharmacologic or behavioral therapies. The presence of a genetic component in explaining the variations to such intervention has only recently been investigated, despite the fact that there are well-known examples of inherited metabolic diseases, such as phenylketonuria and galactosemia.2 As discussed below, the evidence clearly demonstrates that a number of gene–environment interactions, notably via diet, modulate the risk for developing MetS, diabetes, and CVD through their associations with multiple genes responsible for obesity, lipid levels, and markers of inflammation.
GENE–ENVIRONMENT INTERACTIONS: VARIATIONS IN TREATMENT RESPONSE
In a group of hypercholesterolemic subjects, diet restrictions of fat, saturated fat, and cholesterol in an ad libitum diet led to only modest losses in body weight.3 A similar dietary intervention therapy demonstrated a very large range of diet responsiveness, with changes in low-density lipoprotein (LDL) cholesterol ranging from +5% to −40%.4 It is well understood that such dietary-based therapies, even under the strictest states of compliance, result in modest benefits, along with a great variability of response among the patient population.3,4 The wide interindividual variations witnessed in such controlled conditions may indicate a potential role for genetic variability in outcome determination. Gene–environment interaction is defined as the modulation by a genetic polymorphism of a dietary component effect on a specific phenotype (e.g., cholesterol levels and obesity).1 In terms of gene–environment interactions for common multifactorial diseases, one of the clearest evidence of their involvement has been identified with the factors involved in the inflammatory response.5,6
GENE–ENVIRONMENT INTERACTIONS: THE EXAMPLE OF TYPE 2 DIABETES
To better understand how far we have come as HCPs in understanding the genetic components of a multifactorial disease and the role of environmental influences in its pathology, it is necessary to show how we came to understand the pathobiology of type 2 diabetes.
Although in the 1960s and 1970s we did not have the technical abilities that modern genetic research now affords us, the observation of disease clustering in families led to the hypothesis of genes in the predisposition of diabetes.7 In the 1980s, investigations focused on individual genes and their polymorphisms and led to a better understanding of its genetic susceptibility, as witnessed with the report of a single mutation in the insulin gene.8 Following a confusing flood of similar single-mutation reports, reports began to surface that offered direct relationships between gene dysfunction and disease pathology, as demonstrated by the relationship between an amino acid substitution in the peroxisome proliferator-activated receptor-gamma (PPAR-γ) and the resulting decreased receptor activity, thus leading to a lower body mass index (BMI) and improved insulin sensitivity.9 More recently, current population studies of thousands of individuals are starting to incorporate far better genetic approaches through increased coverage of the genetic variation at each locus and genome-wide associations. For example, in the last year alone, researchers identified more genes potentially involved in diabetes susceptibility than had been identified in the previous 40 years.10–15 However, what we have learned from these genome-wide studies for diabetes also applies to investigations involving obesity, hypercholesterolemia, or any other multifactorial disease. That is, per-allele effect sizes of all candidate loci identified are modest at best, with odds ratios (ORs) approximately ranging from 1.1 to 1.2, suggesting each gene is probably a minor contributor to the overall pathogenesis, if at all. This is a sobering observation that highlights the need for larger sample sizes if further loci are to be identified at genome-wide levels of significance.16 For example, it has been estimated that >11,500 cases and 11,500 control subjects would be necessary to detect an effect size of 1.15 with 80% power at P = 10−7 for a common single nucleotide polymorphism (SNP) with 20% minor allele frequency.16 Even larger population sizes are required to examine higher-level interactions, i.e., multigene–diet–behavioral factors, and this represents a major obstacle to the translation of this knowledge into practical public health applications. There are imperative quantitative and qualitative needs, including experiment design and data interpretation, which can be fulfilled only through the collaboration of experts in the different fields, from basic science to computational biology and behavioral science.
GENE–ENVIRONMENT INTERACTIONS AND CVD
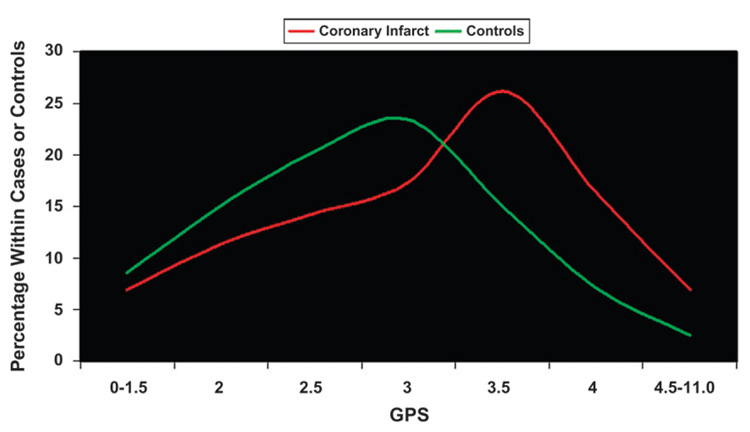
As highlighted by the diabetes genome-wide analyses, individual genes are minor contributors to disease risk when examined individually. Perhaps examining the contribution of many genes together would produce more fruitful results. Such is the case in a recent publication17 that, very simply, looked at the distribution of genetic predisposition score (GPS) values in subjects with coronary infarct compared to control subjects. By examining nine candidate CVD genes and 11 SNPs present at these loci in 200 control subjects and 200 subjects with coronary infarct of the European Prospective Investigation into Cancer and Nutrition (EPIC) study, subjects were assigned a GPS for coronary infarct. If the subject was homozygous for increased risk, heterozygous for increased risk, or did not express the increased risk SNP, the subject was assigned a value of 1, 0.5, or 0, respectively, for each of the 11SNPs. Thus, the risk for coronary infarct increased in proportion to the GPS value. Upon examination of the data, the subjects with coronary infarct had a GPS distribution that skewed toward the higher values, whereas the distribution of the control subjects skewed toward lower values (Fig. 1). Not surprisingly, GPS population distributions differed between ethnic groups, especially compared to those groups with a high degree of obesity, CVD, and diabetes (e.g., high-scoring Hispanic populations versus Mediterranean populations).17 Together, the data suggested that there may be a genetic threshold, which, if crossed, leads to the initiation of disease progression.
Figure 1.
Distribution of cases with coronary infarct and control subjects by GPS. The subjects with coronary infarct (red line) had a GPS distribution that skewed toward the higher values, whereas the distribution of the control subjects (green line) was skewed toward lower values. Data from Trichopoulou et al.17
The reaching of such a threshold may explain why there is disagreement within the scientific literature regarding the replication of findings in gene-association studies, i.e., examining a genetic marker alone does not reveal the whole story. Moreover, there is evidence that environmental factors, in this case, diet, influence the likelihood of disease status in the same coronary infarct population.17 Briefly, the investigators examined the ORs of being a coronary infarct case in relation to the concurrent presence of high (>3.5) or low (<3.5) GPS and adherence to a Mediterranean diet. The healthy Mediterranean diet was characterized by a high intake of plant foods and olive oil, a low intake of meat and dairy products, and a moderate intake of wine. Not surprisingly, those with a high GPS and non-adherence to the Mediterranean diet had the highest ORs for coronary infarct compared to the reference group with good genes (low GPS) and healthy diet (Mediterranean diet compliance). However, by simply adhering to the Mediterranean diet, those with high GPS could significantly lower their OR for coronary infarct by ~60%. In fact, this dietary change was enough to bring the OR in line with that of subjects with a low GPS who were not following a Mediterranean diet.
Some of the genes used to generate the GPS for coronary infarct were genes involved in the inflammatory response, e.g., interleukin (IL)-1β and −6 and tumor necrosis factor-alpha (TNF-α). An ever-growing array of experimental, epidemiological, and clinical studies has revealed an association between variations in the inflammatory responses, dietary factors, and the progression of numerous serious disease states including MetS and CVD.5,6
GENE–ENVIRONMENT INTERACTIONS: INFLAMMATION AND METS
MetS is a constellation of abnormalities, generally considered to include abdominal obesity, high blood glucose/impaired glucose tolerance, dyslipidemia, and high blood pressure, which together increase the risk for overt type 2 diabetes and CVD.5,18 There is increasing evidence from experimental and epidemiologic studies to suggest that at the heart of all these abnormalities is a proinflammatory state.19 The involvement of chronic low-level inflammation underlying the pathobiology of MetS is evident in the finding that as the number of characteristics of MetS increases in a population, the plasma concentrations of proinflammatory markers, high sensitivity C-reactive protein (CRP) and IL-6, increase, whereas the concentration of adiponectin, an adipocyte-derived protein important in glucose regulation and fatty acid (FA) catabolism, decreases.20,21 This article details the paradigm of inflammatory involvement in MetS by highlighting some of the data that implicate gene–environment interactions in defining susceptibility to MetS.
A number of gene–environmental interactions have been drawn from an ongoing genetic/diet interaction study that examines how gene–environment interactions influence susceptibility to developing MetS.22 Known as the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study, its aim is to characterize the genetic basis of the variable response of triglycerides (TGs) following two dietary challenges, one that raises TGs acutely through diet (via fat load) and one that lowers TGs through drug therapy (via fenofibrate treatment), over several weeks.22 Twelve hundred subjects, aged 18 to 92 years, have been recruited from two genetically homogeneous and predominantly white centers (Salt Lake City and Minneapolis). The subjects in this study have been separated into those with and without MetS and are phenotypically distinct.23 For example, BMI, waist circumference, saturated FA content in erythrocyte membrane, and notably, the levels of CRP, IL-6, and TNF-α, are all higher in subjects with MetS compared to those without MetS (P <0.001). In contrast, levels for total polyunsaturated FA (PUFA), (n-3) PUFA (particularly the marine-derived [n-3] PUFAs, docosahexaenoic acid and eicosapentaenoic acid), and (n-6) PUFA in erythrocyte membranes are lower in MetS than in non-MetS subjects (P <0.05). Most interestingly, a recently published report23 identified an association not just between the levels of IL-1β and the risk for MetS, but rather between the genetic variants, or alleles, of IL-1β and the risk for MetS. The human IL-1β gene is a member of the IL-1 cluster on the long arm of chromosome 2 and is flanked by the IL-1α and IL-1Ra genes.24 Once adjusted for age, gender, alcohol, smoking status, and pharmaceutical treatment of confounding disorders, such as diabetes, hypertension, and hypercholesterolemia, specific polymorphisms along the IL-1β gene among GOLDN participants were significantly associated with an increased risk for developing MetS (SNP 6054G>A, OR = 2.22 for GG and 1.79 for GA versus 1.00 for AA; P = 0.004).23 Knowing the existence of allelic associations with the risk for disease development, could environmental interactions, such as diet, counter this increased risk? To test this, data for these same GOLDN participants were stratified by PUFA erythrocyte membrane level, and, thus presumably PUFA dietary intake level, where a high PUFA intake is associated with decreases in inflammatory markers.25 Briefly, it was found that GG and GA subjects with low erythrocyte membrane PUFA content (<5.6%) and presumably low PUFA dietary intake continued to have a significantly increased MetS risk compared to AA subjects (OR = 3.29 for GG and 1.95 for GA versus 1.00 for AA; P <0.0001). However, for those participants with high PUFA content (>5.6%), the increased risk for MetS was not evident among any of these polymorphisms (OR = 1.46 for GG and 1.58 for GA versus 1.00 for AA; P = 0.549).23 Clearly, the data suggest that the increased genetic predilection toward the development of MetS could be eliminated by a diet rich in (n-3) PUFA, supporting the concept that more customized dietary recommendations may be successfully used to prevent chronic diseases.
Another report20 derived from the GOLDN study investigated the impact of TG-lowering fenofibrate treatment on risk factors for CVD. Fenofibrate is a peroxisome proliferator-activated receptor-alpha (PPAR-α) agonist and is a member of the fibrate class commonly used for the management of dyslipidemia. A wealth of data has suggested that fibrates target the atherogenic “lipid triad” (high TGs and low high-density lipoprotein [HDL] with small and dense LDL particles) and inflammation.26 Because both phenotypes are important components of diabetes and MetS, potentially linking these two metabolic disorders to CVD,6 fibrates are hypothesized to be candidates for treating the dyslipidemia associated with diabetes and MetS, effectively reducing CVD risk in these high-risk populations.27 However, outcomes from clinical studies for this family of lipid-lowering drugs have been mixed; a study28 showed that genetic variants at genes involved in lipid metabolism modulate the lipid response to fenofibrate. A recent study20 examined the role of CRP genetic variations on the anti-inflammatory effect of fenofibrate. Specifically, the effects of common CRP polymorphisms on baseline plasma CRP levels and the effect of these variants on the response of CRP to an anti-inflammatory 3-week fenofibrate treatment in GOLDN participants with MetS were investigated. Also, the role of CRP in atherogenesis has been well described.29 CRP integrates the action of several activated cytokines and is a significant predictor of clinical cardiovascular events in healthy individuals and patients with CVD, independent of traditional lipid-based risk factors. Furthermore, CRP has been associated with multiple risk factors for CVD, including obesity, insulin resistance, and hypertension, and is a predictor for the risk for MetS.29 In GOLDN subjects, although plasma CRP levels displayed extensive interindividual variability, SNP analyses showed that the CRP locus contributes to the levels of circulating CRP as well as to the variation of response to fenofibrate treatment among subjects with MetS.20 Specifically, carriers for the rare alleles represented byAor T of m301G>A>T exhibited higher CRP levels than carriers of the common G allele. Furthermore, the rare allele of i178T>A was associated with high CRP levels, whereas the rare allele of 3u2131C>T was associated with low CRP levels. With regard to 3-week fenofibrate intervention among participants with MetS, different CRP alleles had significantly different plasma CRP responses to treatment. For example, G allele carriers for the m301G>A>T SNP displayed a greater reduction of CRP levels than non-carriers. Similarly, TT individuals with the i178T>A SNP had a greater reduction in CRP levels than TA and AA subjects. Clearly, in this gene–environment interaction, the data suggest that resistance to the anti-inflammatory effect of the drug fenofibrate is dependent on CRP allelic expression among subjects with MetS.
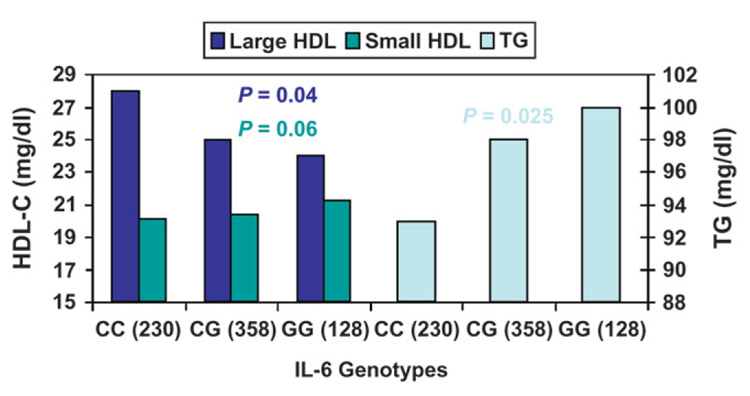
Similar to CRP, it was found that functional polymorphisms associated with the proinflammatory IL-6 gene (e.g., IL6-174C>G) are responsible for modulating serum levels of IL-6.21 However, owing to the fact that these factors are highly interrelated, these same IL-6 polymorphisms also modulate various serum lipids levels associated with MetS, including large HDL particle levels and TG levels (Fig. 2).21 The physiological significance of such modulatory effects is great, because baseline TG levels are affected, as are postprandial TG levels, whose elevated levels represent an emerging CVD risk factor.18 It was shown that the minor G allele at IL6-174C>G was associated with a significantly greater postprandial response of TG-rich lipoproteins, including chylomicrons and total very-low density lipoprotein (VLDL), compared to noncarriers. 21 Together, these combined effects could be responsible for the observed association of this genetic variant with increased CVD risk.
Figure 2.
Baseline lipid measurements are modulated by polymorphisms in IL-6. Polymorphisms associated with the proinflammatory IL-6 gene (e.g., IL6-174C>G) are responsible for modulating serum levels of IL-6. Additionally, these same polymorphisms modulate various serum lipids levels associated with MetS, including large HDL particle levels and TG levels. P values were adjusted for age, gender, BMI, physical activity, smoking, alcohol intake, drugs for diabetes, hypertension and hypercholesterolemia, and hormone use in women. Data from Shen et al.21
It is worth examining TG metabolism further, specifically the role of perilipins, which are a family of proteins that coat intracellular lipid droplets of adipocytes. The expression of perilipin seems primarily to be in adipocytes and steroidogenic cells, where its major role is in the regulation of intracellular lipolysis in adipocytes by hormone-sensitive lipase, and it is a major candidate gene for obesity in humans, in which specific polymorphisms have been associated.30 This is not surprising considering its role in defining the release of free FAs (FFAs), and it was recently demonstrated that postprandial TG metabolism is modified by the presence of genetic variation in the perilipin locus.31 Specifically, data from GOLDN participants showed that minor C and A alleles at PLIN1 and PLIN4, respectively, are associated with a lower postprandial response, which may result in a lower atherogenic risk for carriers. There are dietary consequences to these findings, because similar perilipin gene variations were shown to determine varied susceptibility to insulin resistance in Asian women when consuming a high–saturated fat, low-carbohydrate diet.32 In a separate study,33 the association between perilipin haplotypes and serum concentrations of lipid and inflammation-related markers (FFA, TG, and adiponectin) was demonstrated. The perilipin locus is a significant determinant of body weight, obesity risk, and other MetS-related traits. Because this locus is associated with postprandial plasma lipid metabolism as well as markers of inflammation, it clearly can be considered a significant gene–diet interaction that modulates the risk for MetS.
CONCLUSIONS
The evidence clearly demonstrates that a number of gene–environment interactions modulate the risk for diabetes, MetS, and CVD through their associations with multiple risk factors, such as obesity, lipid levels in the fasting and postprandial state, and markers of inflammation. Data from numerous molecular and genetic epidemiological studies indicate that gene–environment interactions, via dietary changes, can interact in ways that increase the risk for developing chronic disease. This genetic predisposition or vulnerability toward MetS and other disorders can be substantially decreased with targeted dietary advice and constitutes the basis of nutrigenomics,34 a difficult, but not impossible, task of identifying the interplay between human genetic variation and environmental factors, which ultimately may lead to the identification of disease-causative genes and the influential role of nutrients.35
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants NIH/National Heart, Lung, and Blood Institute (NHLBI) HL54776 and NIH/National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) DK075030 and by contract 58-1950-9-001 from the U.S. Department of Agriculture’s Agricultural Research Service. The initial draft of this manuscript was developed by a medical writer (Axon Medical Communications Group, Toronto, Ontario) based on content provided solely by the authors. The final manuscript submitted was under the sole control of the authors.
REFERENCES
- 1.Ordovas JM, Corella D. Nutritional genomics. Annu Rev Genomics Hum Genet. 2004;5:71–118. doi: 10.1146/annurev.genom.5.061903.180008. [DOI] [PubMed] [Google Scholar]
- 2.Corella D, Ordovas JM. Integration of environment and disease into ‘omics’ analysis. Curr Opin Mol Ther. 2005;7:569–576. [PubMed] [Google Scholar]
- 3.Schaefer EJ, Lichtenstein AH, Lamon-Fava S, et al. Body weight and low-density lipoprotein cholesterol changes after consumption of a low-fat ad libitum diet. JAMA. 1995;274:1450–1455. doi: 10.1001/jama.1995.03530180044028. [DOI] [PubMed] [Google Scholar]
- 4.Schaefer EJ, Lichtenstein AH, Lamon-Fava S, et al. Efficacy of a National Cholesterol Education Program Step 2 diet in normolipidemic and hypercholesterolemic middle-aged and elderly men and women. Arterioscler Thromb Vasc Biol. 1995;15:1079–1085. doi: 10.1161/01.atv.15.8.1079. [DOI] [PubMed] [Google Scholar]
- 5.Haffner SM. The metabolic syndrome: Inflammation, diabetes mellitus, and cardiovascular disease. Am J Cardiol. 2006;97(2A):3A–11A. doi: 10.1016/j.amjcard.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105:1135–1143. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- 7.Swift M. Diabetes-predisposing genes. Lancet. 1973;2:497. doi: 10.1016/s0140-6736(73)92089-8. [DOI] [PubMed] [Google Scholar]
- 8.Rotwein P, Chyn R, Chirgwin J, Cordell B, Goodman HM, Permut MA. Polymorphism in the 5′-flanking region of the human insulin gene and its possible relation to type 2 diabetes. Science. 1981;213:1117–1120. doi: 10.1126/science.6267694. [DOI] [PubMed] [Google Scholar]
- 9.Deeb SS, Fajas L, Nemoto M, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 10.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 12.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 14.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 15.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeggini E. A new era for type 2 diabetes genetics. Diabet Med. 2007;24:1181–1186. doi: 10.1111/j.1464-5491.2007.02274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trichopoulou A, Yiannakouris N, Bamia C, Benetou V, Trichopoulos D, Ordovas JM. Genetic predisposition, non-genetic risk factors and coronary infarct. Arch Intern Med. 2008;168:891–896. doi: 10.1001/archinte.168.8.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanefeld M, Fischer S, Julius U, et al. Risk factors for myocardial infarction and death in newly detected NIDDM: the Diabetes Intervention Study, 11-year follow-up. Diabetologia. 1996;39:1577–1583. doi: 10.1007/s001250050617. [DOI] [PubMed] [Google Scholar]
- 19.Ordovas J. Diet/genetic interactions and their effects on inflammatory markers. Nutr Rev. 2007;65:S203–S207. doi: 10.1111/j.1753-4887.2007.tb00363.x. [DOI] [PubMed] [Google Scholar]
- 20.Shen J, Arnett DK, Parnell LD, et al. Association of common C-reactive protein (CRP) gene polymorphisms with baseline plasma CRP levels and fenofibrate response: The GOLDN Study. Diabetes Care. 2008;31:910–915. doi: 10.2337/dc07-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen J, Arnett DK, Parnell LD, et al. The effect of IL6-174C/G polymorphism on postprandial triglycerides metabolism in the GOLDN study. J Lipid Res. 2008;49:1839–1845. doi: 10.1194/jlr.P700033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.GOLDN. [Accessed March 12, 2008];Genetics Of Lipid Lowering Drugs and Diet Network. Available at: http://www.biostat.wustl.edu/goldn/
- 23.Shen J, Arnett DK, Peacock JM, et al. Interleukin 1 beta genetic polymorphisms interact with polyunsaturated fatty acids to modulate risk of the metabolic syndrome. J Nutr. 2007;137:1846–1851. doi: 10.1093/jn/137.8.1846. [DOI] [PubMed] [Google Scholar]
- 24.Nicklin MJ, Weith A, Duff GW. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics. 1994;19:382–384. doi: 10.1006/geno.1994.1076. [DOI] [PubMed] [Google Scholar]
- 25.Simopoulos AP. Omega-3 fatty acids in inflammation and autoimmune diseases. J Am Coll Nutr. 2002;21:495–505. doi: 10.1080/07315724.2002.10719248. [DOI] [PubMed] [Google Scholar]
- 26.Yin WH, Jen HL, Chen JW, Lin SJ, Young MS. Differential effects of peroxisome proliferator-activated receptor ligands and sulfonylurea plus statin treatment on plasma concentrations of adipokines in type 2 diabetes with dyslipidemia. Diabetes Metab. 2006;32:229–235. doi: 10.1016/s1262-3636(07)70273-2. [DOI] [PubMed] [Google Scholar]
- 27.Barter PJ, Rye KA. Is there a role for fibrates in the management of dyslipidemia in the metabolic syndrome? Arterioscler Thromb Vasc Biol. 2008;28:39–46. doi: 10.1161/ATVBAHA.107.148817. [DOI] [PubMed] [Google Scholar]
- 28.Brisson D, Ledoux K, Bosse Y, et al. Effect of apolipoprotein E, peroxisome proliferator-activated receptor alpha and lipoprotein lipase gene mutations on the ability of fenofibrate to improve lipid profiles and reach clinical guideline targets among hypertriglyceridemic patients. Pharmacogenetics. 2002;12:313–320. doi: 10.1097/00008571-200206000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Verma S, Szmitko PE, Ridker PM. C-reactive protein comes of age. Nat Clin Pract Cardiovasc Med. 2005;2:29–36. doi: 10.1038/ncpcardio0074. [DOI] [PubMed] [Google Scholar]
- 30.Qi L, Shen H, Larson I, et al. Gender-specific association of a perilipin gene haplotype with obesity risk in a white population. Obes Res. 2004;12:1758–1765. doi: 10.1038/oby.2004.218. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Martinez P, Yiannakouris N, Lopez-Miranda J, et al. Postprandial triacylglycerol metabolism is modified by the presence of genetic variation at the perilipin (PLIN) locus in 2 white populations. Am J Clin Nutr. 2008;87:744–752. doi: 10.1093/ajcn/87.3.744. [DOI] [PubMed] [Google Scholar]
- 32.Corella D, Qi L, Tai ES, et al. Perilipin gene variation determines higher susceptibility to insulin resistance in Asian women when consuming a high-saturated fat, low-carbohydrate diet. Diabetes Care. 2006;29:1313–1319. doi: 10.2337/dc06-0045. [DOI] [PubMed] [Google Scholar]
- 33.Jang Y, Kim OY, Lee JH, et al. Genetic variation at the perilipin locus is associated with changes in serum free fatty acids and abdominal fat following mild weight loss. Int J Obes (Lond) 2006;30:1601–1608. doi: 10.1038/sj.ijo.0803312. [DOI] [PubMed] [Google Scholar]
- 34.DeBusk RM, Fogarty CP, Ordovas JM, Kornman KS. Nutritional genomics in practice: Where do we begin? J Am Diet Assoc. 2005;105:589–598. doi: 10.1016/j.jada.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Kaput J, Ordovas JM, Ferguson L, et al. The case for strategic international alliances to harness nutritional genomics for public and personal health. Br J Nutr. 2005;94:623–632. doi: 10.1079/bjn20051585. [DOI] [PubMed] [Google Scholar]