Abstract
Common forms of metabolic and cardiovascular diseases involve the interplay of numerous genes as well as important environmental factors. Traditional biochemical and genetic approaches generally attempt to dissect these diseases one gene at a time, for example, by analysis of Mendelian forms or genetically engineered experimental organisms. But, it is also important to understand how the genes interact with each other and the environment, and how these interactions change in disease states. Technological advances, such as the development of expression arrays that allow quantification of all transcript levels in a cell or tissue, have made it feasible to globally monitor molecular phenotypes that underlie disease states. By applying statistical methods, relationships between DNA variation, gene expression patterns, and diseases can be modeled.
Keywords: DNA variation, gene network, complex traits
Common forms of metabolic and cardiovascular diseases are exceptionally complex. Although only a small fraction of the underlying genes has been identified, it seems likely that common or rare variants of hundreds or even thousands of genes will be involved (1, 2). And, of course, environmental factors such as overnutrition, sedentary lifestyle, and smoking play a crucial role. Most biochemical and genetic studies to date, such as those involving transgenic animals, have focused on identifying and characterizing the individual genes that contribute to disease (3). Likewise, genome-wide association and other genetic approaches typically focus on finding the specific genes responsible for the association (4). While these approaches continue to be informative, it is also important to address the interactions between genes and the environment (5). In this brief review, we discuss how systems-based approaches involving the integration of genomic, molecular and physiological/clinical data, can complement traditional approaches to address the complexity of these disorders.
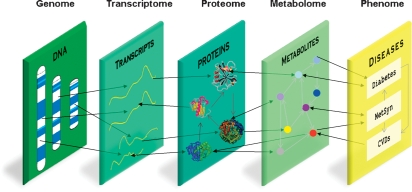
A systems perspective on disease envisions the integration of multiple elements, from genome through phenotype as depicted in Fig. 1. The technological advances that allow large-scale and high-throughput quantification of elements at each level are critical. Thus far, only genome and transcriptome come near the level necessary, and so we focus on these as exemplary of the systems approach.
Fig. 1.
A systems perspective of complex disease encompasses integrated analysis of quantitative measurements at each level depicted.
IDENTIFYING DISEASE-ASSOCIATED GENES BY INTEGRATING GENETIC AND GENE EXPRESSION INFORMATION
Candidate gene studies and recent genome-wide association studies have identified an impressive list of genes contributing to complex disorders such as atherosclerosis (6), hyperlipidemia (7, 8), obesity (9), and diabetes (10), but altogether these account for a minority of the genetic effect on disease development. This suggests that many genes carrying small to modest effect contribute to complex disease, as has long been postulated. The established biology underlying these disorders has identified at least hundreds of genes for each. Genetic studies in humans and animal models have made significant contributions to elucidating the pathogenesis of complex diseases, though the number of specific genes identified has been only a fraction of the total expected to be involved for any given disease. Accordingly, one of the major challenges in studying complex disease is to understand how genes that carry causal variants interact with each other, and with “downstream” genes, to regulate disease expression.
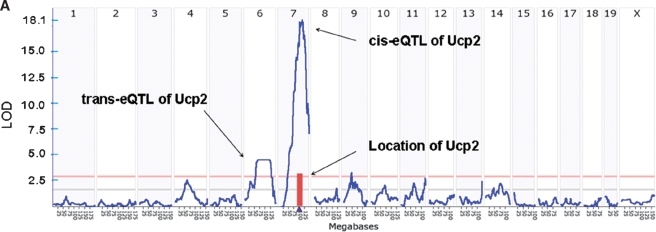
Traditional genetic studies in humans or animal models establish the relationship between genotype and phenotype, but provide little inference as to the intervening biology or, correspondingly, guidance for selecting likely causative genes. Incorporating global gene expression analyses into genetic studies has significantly enhanced both these aspects (1, 11, 12). Gene expression can be measured as a quantitative trait and has been observed to be highly heritable (12–14). Linkage or association analysis can be employed to identify genetic loci or single nucleotide polymorphisms perturbing abundance and activity of gene products. The linkage-based identification of loci for gene expression is termed eQTL (expression quantitative trait locus) mapping, and the same concept can be extended to discover single nucleotide polymorphisms associated with transcript abundance, termed eSNP (expression single nucleotide polymorphism) (15, 16). Observation of eQTL for a gene indicates that genetic factors are partially responsible for its transcript abundance. Based on proximity between genetic factors influencing gene expression and the location of the gene, eQTL can be categorized as cis- or trans-eQTL. When an eQTL localizes closely to the location of the gene coding the transcript, it is likely that the causative genetic variations resides within the gene or its regulator elements and directly influence transcription, or transcript stability, acting in a cis manner, and thus termed cis-eQTL. Conversely, when eQTL does not encompass the physical location of the gene and flanking regions, the gene is defined as being regulated in trans manner, termed trans-eQTL (Fig. 2A). In several genetic crosses between inbred strains of mice, hundreds or thousands of expressed genes in a given tissue have at least one eQTL, including both cis and trans ones. Among all observed eQTLs in crosses of several hundred or more mice, the total number of trans-eQTL is approximately 10 times higher than cis-eQTL.
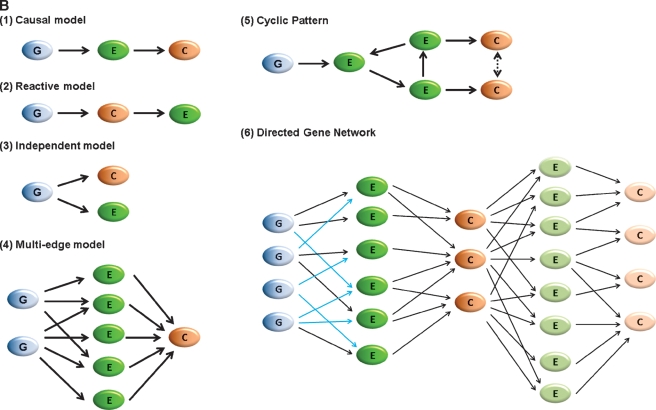
Fig. 2.
Genetic loci controlling transcript levels and their application to causal modeling. A: Example of cis- and trans-acting expression quantitative trait loci (eQTL) of Ucp2 transcript, obtained by integrating genetic and hepatic gene expression data from a genetically randomized population. B: Examples of patterns employed in simplified causality inference model (single edge model, 1–3) and advanced causality network (4–6). Blue dots (G): genetic marker; green dots (E): gene expression patterns; orange dots (C): clinical trait variation. Light green dots (E) and pink dots (C): reactive or secondary gene expression patterns and clinical trait variations respectively. In model 6, black arrows between G and primary E indicate the cis-acting, while the blue ones indicate trans-acting relationships.
Cis-acting eQTL are obvious candidates for genes underlying a phenotypic trait (the quantitative trait gene or QTG) when the eQTL and the trait quantitative trait loci analysis (QTL) coincide. Though causative mutations may not act by altering transcript levels, many do. Applying this concept can significantly reduce the time and effort involved in positional cloning of genes. For example, this was a critical factor in our discovery of Abcc6 as the major causal gene in cardiac calcification in a genetically randomized mouse population (17). Coincidence of trans-eQTL and trait QTL may also be informative for downstream genes involved in trait expression as discussed below.
INTEGRATIVE GENETICS ALLOWS CAUSAL INFERENCE BETWEEN TRAIT-TRANSCRIPT CORRELATIONS
The application of global gene expression analyses to studies of cells and tissues has provided a wealth of data relevant to complex diseases. Finding statistically significant correlations between a trait and particular genes suggests a biologic relationship between them (18, 19). Many databases and analytical tools have been developed to help identify and characterize which functional classes of genes may be involved in a given process. Gene Set Enrichment Analysis (20), Database for Annotation Visualization and Integrated Discovery (21, 22), and Ingenuity Pathway Analysis are analytical tools developed to test the enrichment of particular biological processes and molecular functions of gene sets by examining information collected by databases such as Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway.
However, as has long been recognized in statistics, correlation does not prove causality. In complex diseases, genes may be correlated with a given trait because they are directly involved in the development of that process (i.e., “causal”) or because the process itself secondarily alters the expression of the genes (“reactive”). A major contribution of integrative genetics has been the development of analytical tools to allow causal inferences to be made between correlated genes and traits when genetic data are incorporated (23, 24). This is elaborated in Fig. 2B, which shows possible relationships between trait, transcript, and genetic location when correlations among these are observed. Because genetic variation is for practical purposes always primary, this can be utilized to order the relationship between transcript and trait.
Several analytical approaches have been proposed to assess potential causality in this setting, and undoubtedly more will be developed given the importance of the problem. Schadt et al. (23) developed the likelihood-based causality model selection procedure and applied it to predicting liver expressed genes causal for abdominal obesity in a mouse intercross setting. Subsequent studies using transgenic or knockout models have validated eight of nine predicted genes. Structural equation modeling has also been applied to assess directionality between correlated traits and transcripts, termed edge orientation. An algorithm based on this approach was developed by Aten et al. (25), termed Network Edge Orientation.
CONSTRUCTING GENE EXPRESSION NETWORKS FOR COMPLEX DISEASES
Rather than uncovering single genes for complex diseases such as atherosclerosis, a systems-based perspective is interested in elucidating the interactions of genes and environment operating on a complex multicellular biological system (26). Such a systems approach involves modeling the relationship among elements of the system, such as transcript levels in the form of a network. Two major modeling approaches have been employed to decipher network patterns underlying complex traits: forward and reverse engineering. Forward approaches apply a set of equations generated a priori from previously defined biologic relationships that are then tested and revised as needed. This approach generally is used with small-scale network formation. The reverse approach does not apply a predefined set of relationships. Rather it utilizes general mathematical tools for network construction and lets the data itself define the relationships among the elements being studied, such as transcript level (27). This approach typically utilizes large data sets and is computationally intensive. In the setting of data obtaining from populations with genetic and or environmental variations, such analyses allow one to infer the relationship and interaction among all such elements. This is valuable for studying complex diseases, because we do not yet adequately understand the relationships between gene expression and trait variability.
Network analysis provides a useful framework to identify and visualize interactions among genes, by creating a graphic model. A network is composed of elements, such as specific gene transcripts (referred to technically as nodes), and connections (relationships) among these (“edges”). Edges can indicate a relationship between genes as at transcript level, protein interaction pattern, and any other measurement that describes a meaningful association between two elements of the system. A gene transcriptional network is composed of individual gene transcripts as nodes, while the edges represent a measure of pair-wise correlation of transcript levels. A given gene can correlate with multiple genes, and a measure of the relative number of such connections is referred to as connectivity (28, 29). An important feature of network constructed with biological data is the “scale-free” nature. In a scale-free network, there is a small number of highly connected genes and many more with far fewer connections. Such highly connected genes are often referred to as hubs. Targeting hub genes have been found to disrupt the structure of gene networks and are more likely to impact biological processes when disrupted in animal models. For example, in our analyses of coexpression networks for activated endothelial cells, Atf4, Xbp1, and Insig1 were identified as hub genes (among others) (30). Targeted knockout of these genes had been shown to be lethal in mouse models (31–34).
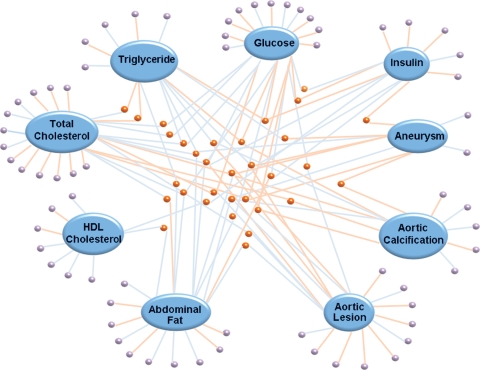
Similar to other biologic networks, genes in coexpression networks are found to organize into “modules,” which are clusters of genes that have higher degree of connectedness with other members of the same module than with genes in different modules (29). Genes composing a module therefore tend to behave more similarly to one another with regard to correlation with phenotypes (Fig. 3) and are often enriched for particular functional categories of genes. Using data-reduction methods such as principle component analysis, the aggregate of genes in a module can be characterized by a single value to use for such analyses (35). One example from our work is the identification of the unfold protein response (UPR) pathway as being important in the response of endothelial cells to oxidized lipids. Coexpression networks were constructed of transcripts induced by oxPAPC in primary endothelial cells isolated from human aorta (30, 36). Two of fifteen modules were strongly correlated with interleukin-8 induction. These modules were enriched for UPR pathway genes, and several of the most highly connected genes were members of the UPR pathway. Knockdown experiment with certain UPR genes revealed the role of the URP pathway in the regulation of interleukin-8 and other cytokines. In other experiments, a hub gene in the UPR module, MGC4504, proved to contribute to an apoptosis response. These findings led to the discovery of a novel gene that is critical for UPR function in this process, based on it being a hub gene in the same module and closely associated with known UPR genes. By traditional analyses, it mostly would not have been recognized as being particularly important.
Fig. 3.
Network construction approach reveals molecular signatures associated with complex diseases. Association network comprising gene modules (small nodes) and edges (connecting lines) was constructed by k-mean clustering method with gene expression profile in whole livers of a genetically randomized mouse population. Blue and orange edges represent positive and negative correlation between modules. Phenotypes related to metabolic and cardiovascular diseases were selected in this figure to indicate that examining molecular signatures at transcript level can reveal trait interconnectedness. Purple and orange modules represent unique and common modules among relevant phenotypes in the network (unpublished work).
One promising approach to reconstruct a directed gene network is the Bayesian Modeling approach. Instead of examining strictly the gene connectivity and module formation, Bayesian modeling is useful in leveraging genetic information to infer causality among genes in the directed network (39). As a probabilistic model approach, Bayesian network reconstruction utilizes posterior probability to map traits with particular markers to exploit the increased information from joint mapping of correlated transcripts. Yang et al. have incorporated the genetic data into Bayesian networks for hepatic gene expression from genetically randomized mouse populations, using the likelihood-based causality model selection approach described above for causal gene detection (Yang et al., in press). This provides a significant improvement over the gene coexpression network constructed without the genetic data by incorporating predicted causal relationships among genes. Such “directed” networks can elucidate the mechanisms underlying phenotypes by which causal regulators give rise to changes in expression activity of various genes. More recently, using expression data from a genetically randomized yeast population, Zhu et al. (40) integrated noisy protein interaction data collected from various sources as well as genetic information into gene expression network by applying Bayesian modeling approach.
Recently, a genome-wide functional network for mouse population has also been established and validated, with which Bayesian integrative modeling brings the protein interaction pattern together with gene expression profiles to illustrate the network including probabilistic functional linkages among over 20,000 genes (38). In addition to Bayesian probabilistic models, there are various ways to integrate biological information, especially transcriptional regulatory mechanisms into gene network underlying complex diseases. Success in identifying critical regulators have been demonstrated in various organisms by employing integrative genetics approaches (41). Furthermore, the involvement of tissue specificity and sex effect in complex disease pathogenesis can also provide further information in personalized medicine in the assist of systems biology approaches.
We are a long way from understanding complex diseases from a systems perspective. However, the use of high-throughput global gene expression assays in the context of genetic analyses has shown how an integrative genetics approach can reveal higher order interactions for traits as complex as diabetes and heart disease (1, 2, 5, 8). As analogous methods for the metabolomic and proteomic elements are developed, progressively richer models of complex disease will be developed (42, 43).
Published, JLR Papers in Press, November 24, 2008.
References
- 1.Lusis A. J., A. D. Attie, and K. Reue. 2008. Metabolic syndrome: from epidemiology to systems biology. Nat. Rev. Genet. 9 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watkins H., and M. Farrall. 2006. Genetic susceptibility to coronary artery disease: from promise to progress. Nat. Rev. Genet. 7 163–173. [DOI] [PubMed] [Google Scholar]
- 3.Flint J., W. Valdar, S. Shifman, and R. Mott. 2005. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 6 271–286. [DOI] [PubMed] [Google Scholar]
- 4.Kruglyak L. 2008. The road to genome-wide association studies. Nat. Rev. Genet. 9 314–318. [DOI] [PubMed] [Google Scholar]
- 5.Gibson G. 2008. The environmental contribution to gene expression profiles. Nat. Rev. Genet. 9 575–581. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y., J. Rollins, B. Paigen, and X. Wang. 2007. Genetic and genomic insights into the molecular basis of atherosclerosis. Cell Metab. 6 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suviolahti E., H. E. Lilja, and P. Pajukanta. 2006. Unraveling the complex genetics of familial combined hyperlipidemia. Ann. Med. 38 337–351. [DOI] [PubMed] [Google Scholar]
- 8.Wang J., M. R. Ban, G. Y. Zou, H. Cao, T. Lin, B. A. Kennedy, S. Anand, S. Yusuf, M. W. Huff, R. L. Pollex, et al. 2008. Polygenic determinants of severe hypertriglyceridemia. Hum. Mol. Genet. 17 2894–2899. [DOI] [PubMed] [Google Scholar]
- 9.Bell C. G., A. J. Walley, and P. Froguel. 2005. The genetics of human obesity. Nat. Rev. Genet. 6 221–234. [DOI] [PubMed] [Google Scholar]
- 10.Frayling T. M. 2007. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat. Rev. Genet. 8 657–662. [DOI] [PubMed] [Google Scholar]
- 11.Johannes F., V. Colot, and R. C. Jansen. 2008. Epigenome dynamics: a quantitative genetics perspective. Nat. Rev. Genet. 9 883–890. [DOI] [PubMed] [Google Scholar]
- 12.Phillips P. C. 2008. Epistasis–the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9 855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emilsson V., G. Thorleifsson, B. Zhang, A. S. Leonardson, F. Zink, J. Zhu, S. Carlson, A. Helgason, G. B. Walters, S. Gunnarsdottir, et al. 2008. Genetics of gene expression and its effect on disease. Nature. 452 423–428. [DOI] [PubMed] [Google Scholar]
- 14.Dixon A. L., L. Liang, M. F. Moffatt, W. Chen, S. Heath, K. C. Wong, J. Taylor, E. Burnett, I. Gut, M. Farrall, et al. 2007. A genome-wide association study of global gene expression. Nat. Genet. 39 1202–1207. [DOI] [PubMed] [Google Scholar]
- 15.Schadt E. E., C. Molony, E. Chudin, K. Hao, X. Yang, P. Y. Lum, A. Kasarskis, B. Zhang, S. Wang, C. Suver, et al. 2008. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6 e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake T. A., E. E. Schadt, and A. J. Lusis. 2006. Integrating genetic and gene expression data: application to cardiovascular and metabolic traits in mice. Mamm. Genome. 17 466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng H., I. Vera, N. Che, X. Wang, S. S. Wang, L. Ingram-Drake, E. E. Schadt, T. A. Drake, and A. J. Lusis. 2007. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc. Natl. Acad. Sci. USA. 104 4530–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal E., N. Friedman, N. Kaminski, A. Regev, and D. Koller. 2005. From signatures to models: understanding cancer using microarrays. Nat. Genet. 37 (Suppl): S38–S45. [DOI] [PubMed] [Google Scholar]
- 19.Park J., and A. L. Barabasi. 2007. Distribution of node characteristics in complex networks. Proc. Natl. Acad. Sci. USA. 104 17916–17920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W., B. T. Sherman, Q. Tan, J. R. Collins, W. G. Alvord, J. Roayaei, R. Stephens, M. W. Baseler, H. C. Lane, and R. A. Lempicki. 2007. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8 R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W., B. T. Sherman, Q. Tan, J. Kir, D. Liu, D. Bryant, Y. Guo, R. Stephens, M. W. Baseler, H. C. Lane, et al. 2007. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35 W169–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schadt E. E., J. Lamb, X. Yang, J. Zhu, S. Edwards, D. Guhathakurta, S. K. Sieberts, S. Monks, M. Reitman, C. Zhang, et al. 2005. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 37 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Degnan J. H., J. Lasky-Su, B. A. Raby, M. Xu, C. Molony, E. E. Schadt, and C. Lange. 2008. Genomics and genome-wide association studies: an integrative approach to expression QTL mapping. Genomics. 92 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aten J. E., T. F. Fuller, A. J. Lusis, and S. Horvath. 2008. Using genetic markers to orient the edges in quantitative trait networks: the NEO software. BMC Syst. Biol. 2 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sieberts S. K., and E. E. Schadt. 2007. Moving toward a system genetics view of disease. Mamm. Genome. 18 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schadt E. E., and P. Y. Lum. 2006. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Reverse engineering gene networks to identify key drivers of complex disease phenotypes. J. Lipid Res. 47 2601–2613. [DOI] [PubMed] [Google Scholar]
- 28.Barabasi A. L., and Z. N. Oltvai. 2004. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 5 101–113. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, B., and S. Horvath. 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4: Article17. [DOI] [PubMed]
- 30.Gargalovic P. S., M. Imura, B. Zhang, N. M. Gharavi, M. J. Clark, J. Pagnon, W. P. Yang, A. He, A. Truong, S. Patel, et al. 2006. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc. Natl. Acad. Sci. USA. 103 12741–12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reimold A. M., A. Etkin, I. Clauss, A. Perkins, D. S. Friend, J. Zhang, H. F. Horton, A. Scott, S. H. Orkin, M. C. Byrne, et al. 2000. An essential role in liver development for transcription factor XBP-1. Genes Dev. 14 152–157. [PMC free article] [PubMed] [Google Scholar]
- 32.Carter S. L., C. M. Brechbuhler, M. Griffin, and A. T. Bond. 2004. Gene co-expression network topology provides a framework for molecular characterization of cellular state. Bioinformatics. 20 2242–2250. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka T., T. Tsujimura, K. Takeda, A. Sugihara, A. Maekawa, N. Terada, N. Yoshida, and S. Akira. 1998. Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells. 3 801–810. [DOI] [PubMed] [Google Scholar]
- 34.Engelking L. J., G. Liang, R. E. Hammer, K. Takaishi, H. Kuriyama, B. M. Evers, W. P. Li, J. D. Horton, J. L. Goldstein, and M. S. Brown. 2005. Schoenheimer effect explained–feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J. Clin. Invest. 115 2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langfelder P., and S. Horvath. 2007. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 1 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gargalovic P. S., N. M. Gharavi, M. J. Clark, J. Pagnon, W. P. Yang, A. He, A. Truong, T. Baruch-Oren, J. A. Berliner, T. G. Kirchgessner, et al. 2006. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 26 2490–2496. [DOI] [PubMed] [Google Scholar]
- 37.Lee D. S., J. Park, K. A. Kay, N. A. Christakis, Z. N. Oltvai, and A. L. Barabasi. 2008. The implications of human metabolic network topology for disease comorbidity. Proc. Natl. Acad. Sci. USA. 105 9880–9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan Y., C. L. Myers, R. Lu, I. R. Lemischka, C. J. Bult, and O. G. Troyanskaya. 2008. A genomewide functional network for the laboratory mouse. PLOS Comput. Biol. 4 e1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J., M. C. Wiener, C. Zhang, A. Fridman, E. Minch, P. Y. Lum, J. R. Sachs, and E. E. Schadt. 2007. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLOS Comput. Biol. 3 e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J., B. Zhang, E. N. Smith, B. Drees, R. B. Brem, L. Kruglyak, R. E. Bumgarner, and E. E. Schadt. 2008. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 40 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S. I., D. Pe'er, A. M. Dudley, G. M. Church, and D. Koller. 2006. Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc. Natl. Acad. Sci. USA. 103 14062–14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrara C. T., P. Wang, E. C. Neto, R. D. Stevens, J. R. Bain, B. R. Wenner, O. R. Ilkayeva, M. P. Keller, D. A. Blasiole, C. Kendziorski, et al. 2008. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS Genet. 4 e1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaibub Neto E., C. T. Ferrara, A. D. Attie, and B. S. Yandell. 2008. Inferring causal phenotype networks from segregating populations. Genetics. 179 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]