Abstract
Lipids fulfill multiple and diverse functions in cells. Establishing the molecular basis for these functions has been challenging due to the lack of catalytic activity of lipids and the pleiotropic effects of mutations that affect lipid composition. By combining molecular genetic manipulation of membrane lipid composition with biochemical characterization of the resulting phenotypes, the molecular details of novel lipid functions have been established. This review summarizes the results of such a combined approach to defining lipid function in bacteria.
Keywords: phosphatidylethanolamine, cardiolipin, phosphatidylglycerol, lactose permease, membrane domains, topogenesis, lipid genetics
THE CHALLENGE IN DEFINING LIPID FUNCTION
Individual lipid classes, such as phospholipids, sphingolipids, sterols, and glycolipids, are composed of a broad spectrum of individual species that differ in the nature of their hydrophobic and hydrophilic moieties so that the complexity of the lipidome may exceed that of the proteome (1). Lipids define the permeability barrier of cells and organelles, are integral components of multisubunit protein complexes, provide the solvent within which membrane proteins fold, support and influence membrane associated processes, provide precursors for macromolecular synthesis, and act as regulatory molecular signals. Uncovering potential roles for lipids using solely biochemical approaches is limited by the lack of inherent catalytic activity so that many putative functions have been based on how a particular lipid affects a reconstituted biological process. Although extensive data exist on the physical organization and chemical properties of lipids in solution, it has not always been possible to translate this information into biological function.
Therefore, verification of function by genetic approaches is particularly important in defining a physiological role for a lipid. However, lipid composition is not encoded by genes but is defined by metabolic pathways dependent on sets of enzymes. Thus, mutations must be made in enzymes of lipid metabolism. Elimination of a major lipid may compromise membrane integrity resulting in cell death before other functions are affected. Changes along a pathway results in changes in minor intermediates that play important roles. In spite of these limitations, molecular genetics when combined with biochemical approaches have defined the molecular basis for unexpected functions of lipids. This review focuses primarily on examples of lipid functions in bacteria as a means for uncovering unexpected roles for lipids that can be generalized to more complex organisms.
BIOCHEMISTRY AND GENETICS OF PHOSPHOLIPIDS SYNTHESIS
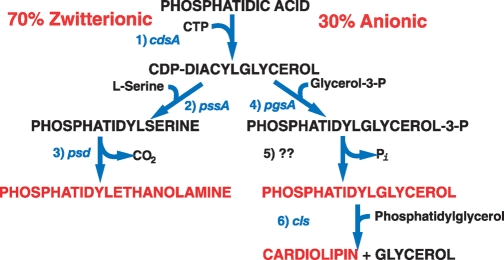
The pathways leading to the major glycerol phosphate-based phospholipids of Escherichia coli (Gram negative) have been established for over 50 years, and the genes encoding all but one of the enzymes have been identified, cloned, and sequenced (Fig. 1) (1). Multiple gene products are identified that dephosphorylate phosphatidylglycerol (PG)-phosphate, but the enzyme primarily responsible for this step is not known (2). The combined phospholipid composition of the inner membrane lipid bilayer and inner leaflet of the outer membrane bilayer varies with growth conditions between 70–80% phosphatidylethanolamine (PE), 20–25% PG, and 5–10% cardiolipin (CL), with the remaining intermediates comprising less than 5%. The outer leaflet of the outer membrane is almost exclusively composed of diglucosamine-based phospholipid (3). The extensive amount of biochemical and genetic information coupled with a simple membrane structure makes E. coli an excellent organism in which to define functions for phospholipids. Bacillus subtilis (Gram positive) has a similar phospholipid composition, but also contains an additional zwitterionic phospholipid lysyl-PG (4) as well as up to 40% neutral glycolipids, such as monoglucosyl diacylglycerol (MGlcDAG) and diglucosyl diacylglycerol, which are synthesized by the transfer of glucose from UDP-glucose to diacylglycerol (5).
Fig. 1.
Pathway for synthesis of the major phospholipids of E. coli. Steps in the pathway are indicated and catalyzed by the following enzymes, which are encoded by the indicated gene: 1) CDP-diacylglycerol synthase; 2) phosphatidylserine (PS) synthase; 3) PS decarboxylase; 4) phosphatidylglycerol (PG)-P synthase; 5) PG-P phosphatase; 6) cardiolipin (CL) synthase.
Null mutations in genes prior to the synthesis of CDP-diacylglycerol are lethal. Null mutations in genes encoding the remaining reactions are viable and exhibit a variety of phenotypes. Therefore, varying the ratio of zwitterionic and amine containing phospholipids [phosphatidylserine (PS) and PE] to the anionic phospholipids (PG and CL) by genetic manipulation can be used to probe lipid function. However, only reduction in enzyme levels and not overproduction of enzymes affect phospholipid composition and cell properties (6, 7).
Elimination of PG and CL via a ΔpgsA (null) mutant is lethal (8). However, a group of suppressor mutations allows near normal growth of null mutants (9). Null mutants in cls also lack strong phenotypes but display impaired survival in stationary phase, increased sensitivity to some antibiotics (10), and resistance to freeze-thaw cycles (11). Interestingly, Δcls strains still contain trace levels of CL, which may be a result of some promiscuity in the PS synthase reaction (12).
Reduction of PE to 30–40% in temperature sensitive pssA and psd mutants results in filamentous cells and growth arrest (13, 14). However, addition of mmolar amounts of Ca+2 or Mg+2 to the growth supports growth as filamentous cells, which is also the case for ΔpssA mutants completely lacking PS and PE (15). Besides defects in cell division, ΔpssA cells have multiple defects in solute transport across the inner membrane (16) and weakened outer membrane barrier function that allows leakage of periplasmic proteins (17).
Phenotypes of mutants in phospholipid metabolism in B. subtilis differ significantly from E. coli, although they contain similar phospholipids except for low amounts of lysyl-PG. Null psd or pss mutants lacking PE or PE and PS grow normally without divalent metal ion supplementation but show increased levels of MGlcDAG (4), suggesting a compensation for the lack of PE. Mutants of B. subtilis lacking glycolipids show an elongated and coiled morphology, which is greatly accentuated in the absence in PE (4). These morphological defects are attenuated by addition of Mg+2 to the growth medium. Engineering the synthesis of MGlcDAG in PE-lacking E. coli cells significantly lowers the divalent metal ion requirement and prevents filamentous growth supporting a common role for PE and MGlcDAG in cell function (17).
DEFINING FUNCTIONS FOR PE
Properties of cells lacking PE
Divalent cations support the growth of pssA mutants but do not suppress filamentous growth suggesting that PE is required for a late step in cell division. Cell division proteins assemble on the Z-ring scaffolding at potential division sites between multiple genomes along filamentous cells, but the frequency of cell constriction is dramatically reduced and out of sync with cell growth and DNA replication (18). A role for PE in a late step of cell division has also been documented in yeast (19) and somatic cells (20). Although the molecular details of PE involvement in cell division remain to be established, the combination of genetic and biochemical approaches uncovered an unrecognized role for PE.
The divalent metal ion requirement of ΔpssA mutants suggests a change in the physical properties of membranes lacking the nonbilayer lipid PE (1). The order of divalent ion effectiveness in suppressing growth arrest is Ca+2 >Sr+2 >Mg+2, with Ba+2 being ineffective (21). This is the same order of the potency for inducing the nonbilayer phase for CL, which is a major lipid in the mutant. Because Ca+2 and Sr+2 are actively excluded from the cell interior and Mg+2 concentrations in the cytoplasm are high, the divalent metal ion requirement may be for stability of the outer leaflet of the inner or the outer membrane. The leakiness of the outer membrane to proteins may be due to the lack of ethanolamine (derived from PE) decoration of lipopolysaccharide chains (22) resulting in an increase in their negative charge nature.
Membrane phospholipid composition in support of solute transport function
Membranes contain secondary solute transporters that couple the uphill transport of substrates with the downhill movement of protons or other cations in order to concentrate solutes from the growth medium or expel solutes from the cytoplasm. These transporters also move solute down a concentration gradient to equilibrate solutes across the membrane in energy independent transport. The most extensively studied transporter is the lactose permease (LacY) of E. coli, which is a paradigm for secondary transporters throughout nature (23). Reconstitution of uphill transport in liposomes containing LacY was shown to require PE but not PG or CL. The physiological significance of this observation was validated when the same results were shown in wild-type versus ΔpssA cells (16). Therefore, the combination of genetic manipulation and biochemical studies demonstrated a physiological role for PE in supporting the function of LacY.
Probing of the molecular basis of the PE requirement led to unexpected new insight into the role of lipids in the assembly and topological organization of membrane proteins. Topogenesis is the process by which integral membrane proteins orient their transmembrane domains (TMs) with respect to the plane of the membrane bilayer. Membrane protein topology can be determined by employing the substituted cysteine accessibility method as applied to TMs, which is reviewed elsewhere (24).
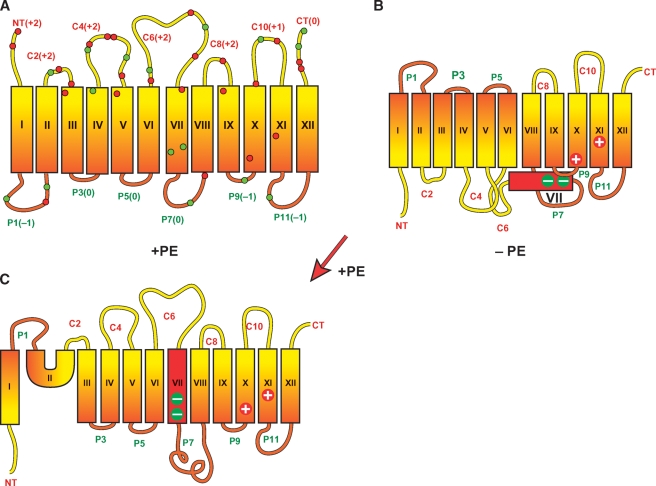
LacY is a 12-TM spanning membrane protein containing independently folding N-terminal and C-terminal 6-TM helical bundles that are connected by a long cytoplasmic domain (C6) (Fig. 2). When assembled in PE-lacking cells, the N-terminal helical bundle is completely inverted with respect to the C-terminal bundle (25). TM VII no longer spans the membrane and is exposed to the periplasmic side of the membrane (26). TM VII contains two negatively charged amino acids that are salt bridged to positive residues in TMs IX and X, which normally stabilize this hydrophilic domain in the membrane. In order for inversion of the N-terminal bundle to take place, a reorganization must occur resulting in an odd number of TMs and disruption of the salt bridges. A D240I mutation in TMVII, which increases the hydrophobicity of TMVII, prevents inversion of the N-terminal bundle (26). Induction of PE synthesis, using a mutant with the pssA gene under control of an inducible promoter (26), after aberrant assembly of LacY in PE-lacking cells, resulted in reinsertion of TMVII and reorientation of most of the N-terminal helical bundle except for TMs I and II. This dramatic reorientation demonstrates that the organization of large membrane domains is dynamic and subject to changes in membrane properties after attaining a compact folded state.
Fig. 2.
Topological organization of lactose permease (LacY) as a function of membrane lipid composition. Topology of LacY initially assembled in phosphatidylethanolamine (PE)-containing (A) or PE-lacking (B) cells is shown. The topology of LacY after induction of PE synthesis postassembly of LacY in PE-lacking cells is also shown (C). LacY is oriented with the cytoplasmic face up and the periplasmic face down. Rectangles (orange and red) indicate helical transmembrane domains (TMs) numbered in Roman numerals from the N-terminus (NT) to the C-terminus (CT). The extramembrane domains connecting the TMs are named based on their topological disposition in PE-containing cells as indicated by the prefix “C” for cytoplasmic (red) or “P” for periplasmic (green). Numbers in parentheses are the net charge of each domain containing positively (red dot) and negatively (green dot) charged residues. The large plus and minus signs indicate the amino acids involved in salt bridges that stabilize TM VII in the membrane.
The phenylalanine (PheP) (27) and γ-aminobutyrate (GabP) (28) permeases also display topological inversions in PE-lacking cells where the N-terminal two-TM hairpin is inverted and TMIII acts as a molecular hinge to the remaining protein. Several other amino acid and sugar permeases lack uphill transport function in PE-lacking cells (Bogdanov, M. and Dowhan, W., unpublished results) indicating that sensitivity of topological to lipid composition is widespread among secondary transporters.
Lipid-protein charge balance hypothesis
A systematic investigation of the charge of the lipid bilayer surface and the charge of the normally cytoplasmic domains of lipid-sensitive membrane proteins reveals that lipid-protein charge interactions are a determinant of membrane protein topology. A completely anionic lipid membrane surface results from lack of PE (no net charge). Introduction of the neutral glycolipids MGlcDAG (29) or diglucosyl diacylglycerol (30) into a PE-lacking cell restores wild-type topological organization to LacY. Phosphatidylcholine also supports wild-type topology of LacY in vivo (30) and in proteoliposomes (31). Therefore, a primary role for PE appears to be dilution of high negative charge density due to PG and CL.
A complementary result was found when the charge nature of the cytoplasmic domains of LacY is systematically changed. All extra-membrane domains of LacY (Fig. 2) follow the positive inside (i.e., cytoplasmic) rule as do the majority of integral membrane proteins (32). The cytoplasmic domains of LacY contain a mixture of positively and negatively charged residues. Neutralizing any one of the multiple negatively charged residues in the cytoplasmic face of the N-terminal bundle in a position independent manner or addition of a single positively charged residue to this face of the bundle prevents inversion in PE-lacking cells (26). Addition of negatively charged residues to the cytoplasmic face of the N-terminal bundle induces inversion in PE-containing cells but requires addition of multiple negative charges. The results are consistent with the dominant effects of positively charged residues over negatively charged residues as topological determinants in normal cells. The results also support a charge interaction between the lipid headgroups and the charges of the cytoplasmic loops in determining topology (i.e., decreasing the negative charge density of the membrane surface or increasing the positive charge of the cytoplasmic domains work in a complementary manner to maintain normal topology). The results also suggest that PE dampens the translocation potential of negatively charged residues thereby strengthening the positive inside rule. Thus an important physiological role for PE is to allow the presence of negatively charged residues in cytoplasmic domains for functional purposes without affecting topology.
DEFINING ANIONIC LIPID FUNCTION
Anionic lipids are in lower amounts than zwitterionic and neutral lipids as membrane components throughout nature. Many of the functions assigned to these anionic lipids are as membrane docking platforms for amphitropic proteins. In bacteria, there is a preference for a particular anionic lipid species, but, in general, any anionic lipid will partially or fully support function (9). Cells nearly devoid of CL appear normal. Complete lack of PG and CL can largely be compensated by an increase in anionic lipid precursors. Therefore, defining specific roles for anionic lipids is a more difficult undertaking and relies heavily on strong in vitro biochemical effects correlated with weak phenotypes evident in mutants with reduced anionic lipid content.
Lipid domains
Biological membranes are not composed of homogeneous and static distributions of proteins and lipids but rather are a dynamic array of lipid and protein subdomains that fluctuate with time in size, composition, and location (33). Primary focus has been on the role of lipid enrichment at the septal and polar regions of bacteria.
The CL-specific fluorescent dye 10-N-nonyl acridine orange (NAO) localizes at the septal and polar regions of living E. coli cells (34) and mini-cells (small membrane vesicles devoid of DNA) that bud from the polar regions of E. coli ΔminCDE (three component system responsible for positioning Z-ring at septum) mutants are highly enriched in CL (35). Similar localization of NAO is observed in B. subtilis (36) and Pseudomonas putida (37). Mutants of E. coli lacking PE display multiple sites of NAO binding in the region of aborted septa and along the surface of the filamentous cells (34). NAO localization is not observed in CL synthase mutants. Biophysical studies strongly support a separation of PE and PG into different domains in E. coli membranes (38).
Therefore, lipid-enriched domains clearly exist in bacteria and provide a means to localize and organize molecular assemblies on the membrane surface. These domains are enriched in anionic phospholipids, which provide a binding site for amphitropic proteins containing positively charged amphipathic helices. Many amphitropic proteins bind anionic lipids in a functional manner in vitro and are also localized to the septal and polar regions of cells, providing evidence for a functional role of anionic lipid domains.
Cell processes stimulated by anionic lipids
Mutants with reduced PG/CL levels show a reduced rate of SecA protein-dependent protein translocation across the cytoplasmic membrane (39). This correlates with strong binding of SecA to and stimulation of its ATPase activity by liposomes containing increasing amounts of anionic phospholipids (40). Reconstitution of translocation of proteins across E. coli inner membrane vesicles shows a strong dependence on anionic phospholipid content, which was varied by regulated expression of the pgsA gene (40).
The ATP-bound form of DnaA protein initiates a single round of DNA replication, which results in the inactive ADP bound form that is rejuvenated to the DnaA-ATP form through interaction with anionic phospholipids, which is common to many bacteria (41). Down-regulation of pgsA gene expression results in cell arrest that is suppressed by mutations that bypass the need for DnaA (42). A mutation that increases the affinity of DnaA protein for anionic lipids (43) suppresses the growth arrest of mutants with low anionic phospholipid content.
Cell division in E. coli is initiated by polymerization of FtsZ protein into the Z-ring at mid-cell, which is dependent on exclusion of FtsZ from the polar anionic lipid domains by MinCDE proteins (44). MinD shows self-association and high affinity cooperative binding to anionic lipids dependent on ATP (45). Binding of MinE to MinD induces ATP hydrolysis and release of MinD from the membrane. MinC also associates with MinD and is an inhibitor of Z-ring formation. This cycle results in oscillation of the MinCD complex from pole to pole where it is of highest concentration thus restricting Z-ring formation at mid-cell. Similar interaction of MinD and anionic lipids has been observed in Gram positive bacteria (33). In E. coli lacking PE, the dwell time of MinD on the membrane is increased and its cycling pattern is disrupted, which results in misplaced oscillating focal points that follow the pattern of the CL-enriched domains detected by NAO (45). This colocalization of MinD and CL domains in the mutant further supports a physiological role for anionic lipids in MinD function. Biophysical studies showed that the affinity of the MinD-ATP for liposomes containing a mixture of zwitterionic and anionic phospholipids is dramatically increased when phase separation of these lipids is induced (46, 47). The above results demonstrate a strong correlation using biochemical and genetic approaches between the existence of anionic lipid domains and the organization and function of several amphitropic ATPases responsible for a broad range of membrane-associated processes.
CONCLUSIONS
Through a combination of genetic and biochemical approaches, the scope of functions for membrane lipids has been broadened beyond simply defining membrane barrier function and providing a solvent for membrane proteins. Initial clues have come from both approaches, but only the combination of both approaches has provided definitive evidence for the unexpected physiological roles of lipids. Genetic manipulation of cells has uncovered many unsuspected roles for lipids that have led to the design of biochemical approaches to further define the molecular detail of lipid involvement. Studies in bacterial provide a rationale for investigations of similar roles in more complex organisms.
Abbreviations
CL, cardiolipin
LacY, lactose permease
MGlcDAG, monoglucosyl diacylglycerol
MinCDE, three component system responsible for positioning Z-ring at septum
NAO, 10-N-nonyl acridine orange
PG, phosphatidylglycerol
PE, phosphatidylethanolamine
PS, phosphatidylserine
TM, transmembrane domain
The work of the author was supported in part by National Istitutes of Health grant GM20478 and the John S. Dunn, Sr. Research Foundation.
Published, JLR Papers in Press, October 30, 2008.
References
- 1.Dowhan, W., M. Bogdanov, and E. Mileykovskaya. 2008. Functional roles of lipids in membranes. In D. E. Vance and J. E. Vance, editors. Biochemistry of Lipids. Lipoproteins and Membranes, 5th edition. Elsevier Press, Amsterdam. 1–37.
- 2.Funk C. R., L. Zimniak, and W. Dowhan. 1992. The pgpA and pgpB genes of Escherichia coli are not essential: evidence for a third phosphatidylglycerophosphate phosphatase. J. Bacteriol. 174 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raetz C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salzberg L. I., and J. D. Helmann. 2008. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J. Bacteriol. 190 7797–7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorasch P., F. P. Wolter, U. Zahringer, and E. Heinz. 1998. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol. Microbiol. 29 419–430. [DOI] [PubMed] [Google Scholar]
- 6.Ohta A., K. Waggoner, K. Louie, and W. Dowhan. 1981. Cloning of genes involved in membrane lipid synthesis. Effects of amplification of phosphatidylserine synthase in Escherichia coli. J. Biol. Chem. 256 2219–2225. [PubMed] [Google Scholar]
- 7.Ohta A., K. Waggoner, A. Radominska-Pyrek, and W. Dowhan. 1981. Cloning of genes involved in membrane lipid synthesis: effects of amplification of phosphatidylglycerophosphate synthase in Escherichia coli. J. Bacteriol. 147 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heacock P. N., and W. Dowhan. 1989. Alterations of the phospholipid composition of Escherichia coli through genetic manipulation. J. Biol. Chem. 264 14972–14977. [PubMed] [Google Scholar]
- 9.Matsumoto K. 2001. Dispensable nature of phosphatidylglycerol in Escherichia coli: dual roles of anionic phospholipids. Mol. Microbiol. 39 1427–1433. [DOI] [PubMed] [Google Scholar]
- 10.Tropp B. E. 1997. Cardiolipin synthase from Escherichia coli. Biochim. Biophys. Acta. 1348 192–200. [DOI] [PubMed] [Google Scholar]
- 11.Sleight S. C., C. Orlic, D. Schneider, and R. E. Lenski. 2008. Genetic basis of evolutionary adaptation by Escherichia coli to stressful cycles of freezing, thawing and growth. Genetics. 180 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishijima S., Y. Asami, N. Uetake, S. Yamagoe, A. Ohta, and I. Shibuya. 1988. Disruption of the Escherichia coli cls gene responsible for cardiolipin synthesis. J. Bacteriol. 170 775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawrot E., and E. P. Kennedy. 1978. Phospholipid composition and membrane function in phosphatidylserine decarboxylase mutants of Escherichia coli. J. Biol. Chem. 253 8213–8220. [PubMed] [Google Scholar]
- 14.Raetz C. R. H. 1976. Phosphatidylserine synthetase mutants of Escherichia coli: genetic mapping and membrane phospholipid composition. J. Biol. Chem. 251 3242–3249. [PubMed] [Google Scholar]
- 15.DeChavigny A., P. N. Heacock, and W. Dowhan. 1991. Phosphatidylethanolamine may not be essential for the viability of Escherichia coli. J. Biol. Chem. 266 5323–5332. [PubMed] [Google Scholar]
- 16.Bogdanov M., and W. Dowhan. 1995. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J. Biol. Chem. 270 732–739. [DOI] [PubMed] [Google Scholar]
- 17.Wikstrom M., J. Xie, M. Bogdanov, E. Mileykovskaya, P. Heacock, A. Wieslander, and W. Dowhan. 2004. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J. Biol. Chem. 279 10484–10493. [DOI] [PubMed] [Google Scholar]
- 18.Mileykovskaya E., Q. Sun, W. Margolin, and W. Dowhan. 1998. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J. Bacteriol. 180 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto K., S. Kobayashi, R. Fukuda, M. Umeda, T. Kobayashi, and A. Ohta. 2004. Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity. Genes Cells. 9 891–903. [DOI] [PubMed] [Google Scholar]
- 20.Emoto K., H. Inadome, Y. Kanaho, S. Narumiya, and M. Umeda. 2005. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J. Biol. Chem. 280 37901–37907. [DOI] [PubMed] [Google Scholar]
- 21.Rietveld A. G., V. V. Chupin, M. C. Koorengevel, H. L. Wienk, W. Dowhan, and B. de Kruijff. 1994. Regulation of lipid polymorphism is essential for the viability of phosphatidylethanolamine-deficient Escherichia coli cells. J. Biol. Chem. 269 28670–28675. [PubMed] [Google Scholar]
- 22.Reynolds C. M., S. R. Kalb, R. J. Cotter, and C. R. Raetz. 2005. A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J. Biol. Chem. 280 21202–21211. [DOI] [PubMed] [Google Scholar]
- 23.Guan L., and H. R. Kaback. 2006. Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35 67–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogdanov M., W. Zhang, J. Xie, and W. Dowhan. 2005. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAMTM): application to lipid-specific membrane protein topogenesis. Methods. 36 148–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogdanov M., P. N. Heacock, and W. Dowhan. 2002. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 21 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogdanov M., J. Xie, P. Heacock, and W. Dowhan. 2008. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J. Cell Biol. 182 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W., M. Bogdanov, J. Pi, A. J. Pittard, and W. Dowhan. 2003. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J. Biol. Chem. 278 50128–50135. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W., H. A. Campbell, S. C. King, and W. Dowhan. 2005. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the γ-aminobutyric acid permease (GabP) of Escherichia coli. J. Biol. Chem. 280 26032–26038. [DOI] [PubMed] [Google Scholar]
- 29.Xie J., M. Bogdanov, P. Heacock, and W. Dowhan. 2006. Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J. Biol. Chem. 281 19172–19178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wikström M., A. A. Kelly, A. Georgiev, H. M. Eriksson, M. R. Klement, M. Bogdanov, W. Dowhan, and A. Wieslander. 2009. Lipid-engineered Escherichia coli membranes reveal critical headgroup size for protein function. J. Biol. Chem. 284 954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X., M. Bogdanov, and W. Dowhan. 2002. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J. 21 5673–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson J., B. Persson, and G. von Heijne. 2005. Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins. 60 606–616. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto K., J. Kusaka, A. Nishibori, and H. Hara. 2006. Lipid domains in bacterial membranes. Mol. Microbiol. 61 1110–1117. [DOI] [PubMed] [Google Scholar]
- 34.Mileykovskaya E., and W. Dowhan. 2000. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182 1172–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koppelman C. M., T. Den Blaauwen, M. C. Duursma, R. M. Heeren, and N. Nanninga. 2001. Escherichia coli minicell membranes are enriched in cardiolipin. J. Bacteriol. 183 6144–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai F., M. Shoda, R. Harashima, Y. Sadaie, H. Hara, and K. Matsumoto. 2004. Cardiolipin domains in Bacillus subtilis Marburg membranes. J. Bacteriol. 186 1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernal P., J. Munoz-Rojas, A. Hurtado, J. L. Ramos, and A. Segura. 2007. A Pseudomonas putida cardiolipin synthesis mutant exhibits increased sensitivity to drugs related to transport functionality. Environ. Microbiol. 9 1135–1145. [DOI] [PubMed] [Google Scholar]
- 38.Vanounou S., A. H. Parola, and I. Fishov. 2003. Phosphatidylethanolamine and phosphatidylglycerol are segregated into different domains in bacterial membrane. A study with pyrene-labelled phospholipids. Mol. Microbiol. 49 1067–1079. [DOI] [PubMed] [Google Scholar]
- 39.Kusters R., W. Dowhan, and B. de Kruijff. 1991. Negatively charged phospholipids restore prePhoE translocation across phosphatidylglycerol-depleted Escherichia coli inner membranes. J. Biol. Chem. 266 8659–8662. [PubMed] [Google Scholar]
- 40.Lill R., W. Dowhan, and W. Wickner. 1990. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 60 271–280. [DOI] [PubMed] [Google Scholar]
- 41.Boeneman K., and E. Crooke. 2005. Chromosomal replication and the cell membrane. Curr. Opin. Microbiol. 8 143–148. [DOI] [PubMed] [Google Scholar]
- 42.Xia W., and W. Dowhan. 1995. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. USA. 92 783–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aranovich A., A. H. Parola, and I. Fishov. 2007. The reactivation of DnaA (L366K) requires less acidic phospholipids supporting their role in the initiation of chromosome replication in Escherichia coli. FEBS Lett. 581 4439–4442. [DOI] [PubMed] [Google Scholar]
- 44.Mileykovskaya E., and W. Dowhan. 2005. Role of membrane lipids in bacterial division-site selection. Curr. Opin. Microbiol. 8 135–142. [DOI] [PubMed] [Google Scholar]
- 45.Mileykovskaya E., I. Fishov, X. Fu, B. D. Corbin, W. Margolin, and W. Dowhan. 2003. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo J. Biol. Chem. 278 22193–22198. [DOI] [PubMed] [Google Scholar]
- 46.Mazor S., T. Regev, E. Mileykovskaya, W. Margolin, W. Dowhan, and I. Fishov. 2008. Mutual effects of MinD-membrane interaction: I. Changes in the membrane properties induced by MinD binding. Biochim. Biophys. Acta. 1778 2496–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazor S., T. Regev, E. Mileykovskaya, W. Margolin, W. Dowhan, and I. Fishov. 2008. Mutual effects of MinD-membrane interaction: II. Domain structure of the membrane enhances MinD binding. Biochim. Biophys. Acta. 1778 2505–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]